Abstract
Pseudomonas aeruginosa is responsible for chronic and acute infections in humans. Chronic infections are associated with production of fimbriae and the formation of a biofilm. The two-component system Roc1 is named after its role in the regulation of cup genes, which encode components of a machinery allowing assembly of fimbriae. A non-characterized gene cluster, roc2, encodes components homologous to the Roc1 system. We show that cross-regulation occurs between the Roc1 and Roc2 signalling pathways. We demonstrate that the sensors RocS2 and RocS1 converge on the response regulator RocA1 to control cupC gene expression. This control is independent of the response regulator RocA2. Instead, we show that these sensors act via the RocA2 response regulator to repress the mexAB-oprM genes. These genes encode a multidrug efflux pump and are upregulated in the rocA2 mutant, which is less susceptible to antibiotics. It has been reported that in cystic fibrosis lungs, in which P. aeruginosa adopts the biofilm lifestyle, most isolates have an inactive MexAB-OprM pump. The concomitant RocS2-dependent upregulation of cupC genes (biofilm formation) and downregulation of mexAB-oprM genes (antibiotic resistance) is in agreement with this observation. It suggests that the Roc systems may sense the environment in the cystic fibrosis lung.
Introduction
Bacteria constantly probe the surrounding environment to adapt their colonization strategy be it in the environment or within a host. An important molecular device to achieve sampling of environmental signals is the so-called two-component regulatory system (TCS) (Stock et al., 2000). This system consists of a sensor, which is a histidine kinase capable of autophosphorylation on a conserved histidine residue, and a response regulator on which the phosphate is transferred. The phosphate is loaded onto a conserved aspartate residue in the conserved receiver domain of the response regulator, and the phosphorylation event results in activation of the output domain of the regulator. Frequently, the output domain is a DNA binding domain, which contributes directly to the control of gene expression. In some cases, the output domain carries an enzymatic activity (Galperin and Nikolskaya, 2007).
The distribution of TCSs in bacterial genomes is quasi-universal but the number of genes encoding TCSs can vary from a few to hundreds. Obviously, the more the bacterial strain copes with a complex environment, or is versatile, the more TCSs are needed to contribute to its adaptation. This is possible because histidine kinases have extremely variable detection or input domains, which are involved in the recognition of a specific stimulus (Galperin and Nikolskaya, 2007). Environmental signals can be related to oxygen availability, nutrient limitation, phosphate limitation or osmolarity, but sensors can also detect antimicrobial peptides (Otto, 2009), signalling molecules such as homoserine lactones (Freeman and Bassler, 1999) and many as yet unknown molecules. However, despite the variability of the input domain, the phosphorylation cascade resulting from the detection of the signal is a conserved process, and the structure and sequence of the catalytic machinery found in histidine kinases displays high similarity. Similarly, the receiver domain of response regulators is highly conserved.
Pseudomonas aeruginosa belongs to a category of versatile bacteria that encounter different environments, infect various hosts and have a broad catabolic potential. The PAO1 genome sequence analysis revealed the presence of about 130 genes encoding TCS components (Rodrigue et al., 2000). In theory, a signalling cascade is very specific, with one sensor talking to its cognate response regulator. This specificity contributes to the avoidance of unwanted cross-talk between TCSs that may result in inappropriate adaption (Laub and Goulian, 2007; Skerker et al., 2008). The molecular basis of this specificity is becoming to be more extensively understood, and involves a limited number of key residues involved in the interaction between sensors and regulators (Skerker et al., 2008; Casino et al., 2009). However, one could imagine that complex environmental signals could be characterized by a series of sensors that feed multiple signals into one single response regulator, and thus provide an integrated response. Among the many P. aeruginosa TCSs, only a few have been characterized in great detail, and the signalling molecules have not always been identified. A large number of signalling pathways involving chemotaxis (Garvis et al., 2009) and TCSs (Gooderham and Hancock, 2009) have been described as having a key role during the infection process in P. aeruginosa. The GacAS/LadS/RetS system is an example of a regulatory pathway for which the signal is unknown (Kay et al., 2006; Ventre et al., 2006; Goodman et al., 2009). It constitutes an intricate network of several sensors, and is a major player in the control of P. aeruginosa virulence. In particular, the RetS and GacS sensor signalling pathways terminate on the GacA response regulator, which in turn modulates expression of small RNAs (Brencic et al., 2009). The input is thus variable but the output is conserved, with a final antagonistic control on genes involved in biofilm formation, such as the pel genes (Vasseur et al., 2005), and genes involved in virulence and cytotoxicity such as the type III secretion genes. Another P. aeruginosa signalling pathway, for which the signal is unknown, is the Roc1 system (Kulasekara et al., 2005). This time, one sensor, RocS1, feeds onto two response regulators, RocA1 and RocR. In this case the detection of a single signal may lead to a diversified response. The Roc system is also a major player in controlling the balance between biofilm formation and cytotoxicity (Kuchma et al., 2005). In particular, it positively controls the expression of cup genes involved in fimbrial assembly (Ruer et al., 2007), and negatively controls genes involved in the type III secretion system (T3SS) (Kuchma et al., 2005). Interestingly, there are additional sensors, which are coded by rocS1 paralogous genes, namely rocS2 and rocS3 (Kulasekara et al., 2005).
In the present study we analysed the potential cross-regulation between these systems and how this impacts P. aeruginosa gene expression. We demonstrated that RocS1 and RocS2 could signal to both the RocA1 and RocA2 response regulators. However, the responses arising from the activation of either response regulator are totally different. Our analysis further highlights an intriguing adaptive combination, which suggests that biofilm formation and antibiotic resistance could be controlled antagonistically.
Results
RocS2 and RocS1 control cupC gene expression in a RocA1-dependent but RocA2-independent manner
We previously reported that transposon insertions within the P. aeruginosa PA3946-PA3948 genetic locus, or roc1 locus, resulted in elevated cupC gene expression (Kulasekara et al., 2005; Ruer et al., 2007). This upregulation resulted from overproduction of the sensor kinase RocS1 (PA3946) or the response regulator RocA1 (PA3948), a TCS (Fig. S1). In the same study, it was also shown that transposon insertions in the intergenic region of the PA3044-PA3045 locus, or roc2 locus, increased cupC gene expression levels. It is noticeable that the sensor kinases RocS1 (PA3946) and RocS2 (PA3044) are paralogues (45% identity), likewise with the pair of response regulators RocA1 (PA3948) and RocA2 (PA3045) (59% identity) (Fig. S1). The roc1 locus also codes for RocR (PA3947), a response regulator with an EAL-containing C-terminal domain identified as a phosphodiesterase output domain (Fig. S1) (Rao et al., 2008). It has previously been shown that RocR antagonizes the activity of RocA1 (Kulasekara et al., 2005).
In this study, we further analysed the role of the Roc2 system. We cloned the rocS2 and rocA2 genes into the broad host range vector pMMB67HE42, yielding pMMB67-rocS2 and pMMB67-rocA2 respectively (Table 1). We then introduced either one of the recombinant plasmids into the P. aeruginosa PAK strain containing a cupC-lacZ transcriptional fusion at the chromosomal attB site (Table 1). Strikingly, overproduction of RocS2 in this strain resulted in a 40-fold increase in β-galactosidase activity, as compared with the strain harbouring the vector control, pMMB67HE (Fig. 1A). In contrast, overproduction of RocA2 from pMMB67-rocA2 had no effect on cupC-lacZ transcription (Fig. 1A). In order to verify that RocA2 has no role in cupC gene expression, we engineered a rocA2 deletion mutant in the PAK strain, PAKΔrocA2 (Table 1), and introduced the cupC-lacZ fusion into the chromosome. Upon RocS2 overproduction, induction of cupC gene expression was still observed in the rocA2 mutant (Fig. 1A). This confirmed that RocA2 is not involved in the induction of cupC genes by RocS2. Because RocA2 and RocA1 are paralogues, we then investigated whether RocS2 could act through RocA1 to activate cupC expression. As for rocA2, we engineered a rocA1 mutant in the PAK strain carrying the cupC-lacZ fusion. Upon introduction of pMMB67-rocS2 into the rocA1 mutant, no activation of the fusion could be observed and β-galactosidase levels were similar to those observed in a strain containing the vector control (Fig. 1A). Thus, we concluded that cupC gene expression occurs through RocA1 not only upon activation by RocS1 (Kulasekara et al., 2005) but also via RocS2 signalling.
Table 1.
Bacterial strains and plasmids used in this work
| Strains/Plasmid | Relevant characteristicsa | Source/Reference |
|---|---|---|
| Strain | ||
| Escherichia coli | ||
| TG1 | supEΔ(lac-proAB) thi hsdRΔ5 (F' traD36 rpoA+B+lacIqZΔM15) | Lab collection |
| SM10 (λpir) | thi-1 thr-1 leuB6 supE44 tonA21 lacY1 recA-::RP4-2-Tc::Mu Kmr | Lab collection |
| TOP10F' | F'[lacIq Tn10(tetR)]mcrAΔ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 deoR nupG recA1 araD139Δ (ara-leu)7697 galU galK rpsL (Strr) endA1λ | Invitrogen |
| CC118 (λpir) | Host strain for pKNG101 replication; Δ(ara-leu) araDΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1 Rfr (λpir) | Lab collection |
| DHMI | cya-854 recA1 gyrA96 (NaI) thi1 hsdR17 spoT1 rfbD1 glnV44(AS) | Karimova et al. (1998) |
| Pseudomonas aeruginosa | ||
| PAK | Wild type | D. Bradley |
| PAKΔrocA1 | In frame deletion of rocA1 (PA3948) in PAK | Kulasekara et al. (2005) |
| PAKΔrocR | In frame deletion of rocR (PA3947) in PAK | Kulasekara et al. (2005) |
| PAKΔrocA2 | In frame deletion of rocA2 (PA3045) in PAK | This study |
| PAKΔrocA1-rocA2 | In frame deletion of rocA1 (PA3948) and rocA2 (PA3045) in PAK | This study |
| PAK::cupC-lacZ | PAK with a cupC-lacZ fusion integrated on the chromosome at the attB site | This study |
| PAKΔrocA1::cupC-lacZ | rocA1 deletion mutant in PAK::cupC-lacZ | This study |
| PAKΔrocA2::cupC-lacZ | rocA2 deletion mutant in PAK::cupC-lacZ | This study |
| PAKΔrocR::cupC-lacZ | rocR deletion mutant in PAK::cupC-lacZ | This study |
| PAK::cupB-lacZ | PAK with a cupB-lacZ fusion integrated on the chromosome at the ctx att site | This study |
| PAKΔrocA1::cupB-lacZ | rocA1 deletion mutant in PAK::cupB-lacZ | This study |
| PAKΔrocA2::cupB-lacZ | rocA2 deletion mutant in PAK::cupB-lacZ | This study |
| PAKΔrocA1-rocA2::cupB-lacZ | rocA1-rocA2 double deletion mutant in PAK::cupB-lacZ | This study |
| Plasmid | ||
| pRK2013 | ColE1 mob+traRK2ΔrepRK2repE- Kmr | Figurski and Helinski (1979) |
| pCR2.1 | TA cloning vector for PCR products, lacZα ColE1 f1 ori Apr Kmr | Invitrogen |
| pMMB67HE | Broad host range vector, tac promoter Apr | Lab collection |
| pMMB67EH-GW | Broad host range vector, tac promoter Gmr | Lab collection |
| pET-Dest42 | Destination vector for gateway technology, T7 promoter, Apr | Invitrogen |
| pMMB67HE42 | pMMB67HE containing the gateway cassette from pET-Dest42 Apr | E. Termine |
| pMMB67-rocS2 | pMMB67HE harbouring the rocS2 gene from the Gateway library | This study |
| pMMB67-rocA2 | pMMB67HE harbouring the rocA2 gene from the Gateway library | This study |
| pMMB67-rocS1 | pMMB67EH-GW harbouring the rocS1 gene from the Gateway library | Kulasekara et al. (2005) |
| pKNG101 | Suicide vector for P. aeruginosa. sacB, Smr | Kaniga et al. (1991) |
| pKNGΔrocA2 | pKNG101 containing 560 bp upstream and 505 bp downstream from the rocA2 gene DNA fragment | This study |
| miniCTX-lacZ | Vector for unmarked integration into P. aeruginosa att site. oriT, FRT, int, Tcr | Hoang et al. (2000) |
| miniCTX-cupC-lacZ | cupC1 promoter cloned into miniCTX-lacZ (BamHI/XhoI) | This study |
| miniCTX-cupB-lacZ | cupB1 promoter cloned into miniCTX-lacZ (BamHI/XhoI) | This study |
| pKT25 | Cloning and expression vector, encodes the T25 fragment (amino acids 1–224 of CyaA), Kmr | Karimova et al. (1998) |
| pUT18c | Cloning and expression vector, encodes the T18 fragment (amino acids 225–399 of CyaA), Apr | Karimova et al. (1998) |
| pKT25-torR | DNA fragment encoding the D2 domain of TorR cloned into pKT25 | Kulasekara et al. (2005) |
| pUT18c-torS | DNA fragment encoding the Hpt domain of TorS cloned into pUT18c | Kulasekara et al. (2005) |
| pKT25-rocS1 | DNA fragment encoding the Hpt domain of RocS1 cloned into pKT25 | Kulasekara et al. (2005) |
| pUT18c-rocA1 | DNA fragment encoding the D2 domain of RocA1 cloned into pUT18c | Kulasekara et al. (2005) |
| pUT18c-rocR | DNA fragment encoding the D2 domain of RocR cloned into pUT18c | Kulasekara et al. (2005) |
| pKT25-rocS2 | DNA fragment encoding the Hpt domain of RocS2 cloned into pKT25 | This study |
| pUT18c-rocA2 | DNA fragment encoding the D2 domain of RocA2 cloned into pUT18c | This study |
| pUT18c-trpO | DNA fragment encoding the D2 domain of PA0034 cloned into pUT18c | This study |
| pKT25-gacS | DNA fragment encoding the H1-D1-Hpt domain of GacS cloned into pKT25 | This study |
| pUT18c-gacA | DNA fragment encoding the D2 domain of GacA cloned into pUT18c | This study |
Apr, ampicillin resistance; Smr, Streptomycin resistance; Kmr, Kanamycin resistance, Tcr, Tetracycline, Gmr, Gentamicin resistance.
Fig. 1.
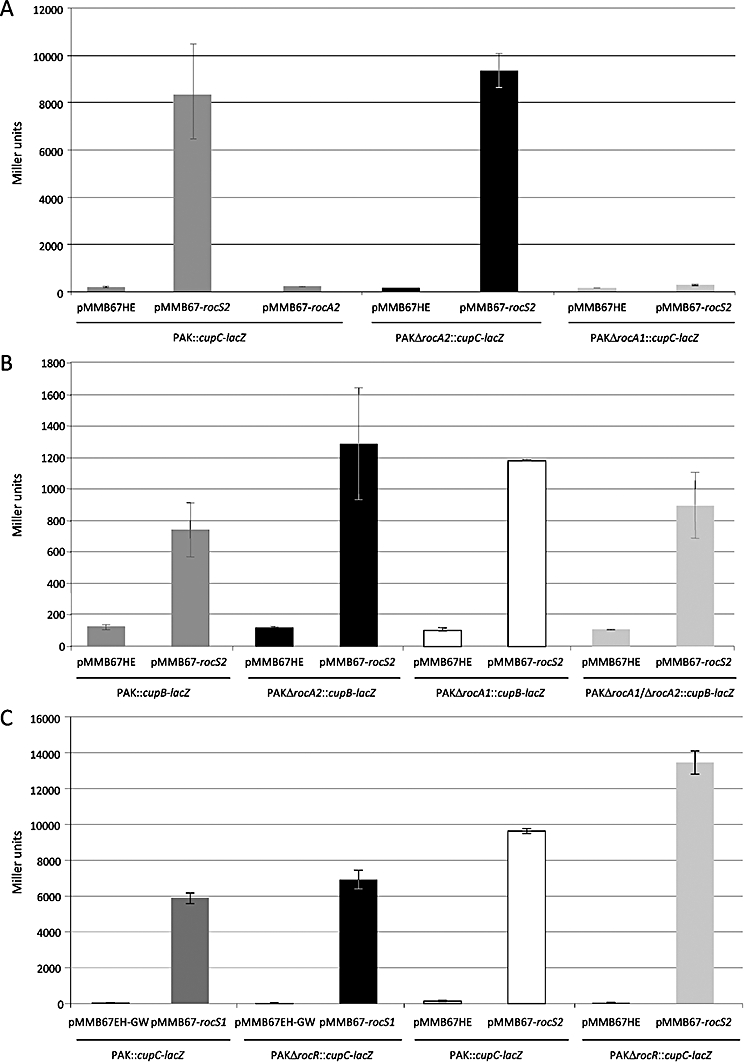
β-galactosidase activity (Miller Units) of PAK wild type or isogenic deletion mutants carrying chromosomal transcriptional fusions at the attB site (cupB-lacZ or cupC-lacZ) as well as a pMMB67HE vector, either empty or overexpressing rocS2, rocA2 or rocS1 as indicated. Data are the average of biological duplicates, and error bars indicate one standard deviation of the mean.
A. rocS2 overexpression induces cupC expression, but this induction is not seen by overexpressing rocA2. The difference between the strain containing the pMMB67HE vector or containing pMMB67-rocS2 is significant with a P-value < 0.001. Furthermore, rocS2 still induces cupC expression in the PAKΔrocA2 mutant (difference between the strain containing the pMMB67HE vector or containing pMMB67-rocS2 is significant with a P-value < 0.001), but the induction is lost in the PAKΔrocA1 mutant indicating that RocA1, but not RocA2, is required for this signalling.
B. rocS2 overexpression also induces cupB expression. This induction is independent of both RocA2 and RocA1 as seen in the single and double deletion mutants. All differences between the strain containing the pMMB67HE vector or containing pMMB67-rocS2 are significant with a P-value < 0.05.
C. cupC expression can be induced by both rocS1 and rocS2, but in both cases the promoter activity is higher in a PAKΔrocR mutant showing that RocR is a repressor of cupC gene expression. The difference between the strains PAK::cupC-lacZ and PAKΔrocR::cupC-lacZ containing pMMB67-rocS2 is significant with a P-value < 0.02.
RocS2 and RocS1 control cupB gene expression in a RocA1- and RocA2-independent manner
It was previously shown that RocS1 not only activates cupC gene transcription but also influences cupB gene expression. We engineered a series of P. aeruginosa PAK strains containing a cupB-lacZ transcriptional fusion at the chromosomal attB site, PAKΔrocA1, PAKΔrocA2 or a double mutant PAKΔrocA1−rocA2 (Table 1). As previously shown with RocS1 (Kulasekara et al., 2005), overproduction of RocS2 resulted in increased levels of cupB gene expression by about sixfold (Fig. 1B). However, and in contrast to what was observed with the cupC genes, upregulation of cupB gene expression persisted in the rocA1, rocA2 and even rocA1-rocA2 double mutant (Fig. 1B). Similar results were obtained when the plasmid carrying the rocS1 gene (pMMB67-rocS1) was introduced in these different strains (Fig. S2). This was unexpected and suggested that induction of cupB gene expression via the RocS1 and RocS2 signalling pathways is independent of both RocA1 and RocA2. It is thus obvious that additional components are likely to be part of the Roc regulatory pathway, and the regulatory component responsible for cupB gene control is yet to be identified.
RocR is a negative regulator of the Roc1 and Roc2 systems
The RocR response regulator has a phosphodiesterase activity (Rao et al., 2008) and was shown previously to have an antagonist effect as compared with RocA1, as RocR represses cupC gene expression (Kulasekara et al., 2005). We tested whether the negative effect could be exerted whether the input came from either RocS1 or RocS2. We engineered a rocR mutant strain (PAKΔrocR) in which the transcriptional cupC-lacZ fusion was inserted at the attB site on the chromosome (Table 1). We then introduced a plasmid carrying either rocS1 or rocS2. In both cases we observe that the induction of cupC gene expression was significantly higher in the rocR mutant background as compared with the parental strain (Fig. 1C). We thus concluded that RocR is a negative regulator not only of the Roc1 system but also of the Roc2 system. This observation further expands the cross-regulation between the Roc1 and Roc2 systems.
RocS1/A1 and RocS2/A2 form a complex and interactive network
We previously reported that activation of RocA1 by RocS1 resulted from a direct interaction between these two components (Kulasekara et al., 2005). Because we identified cross-regulation between Roc1 and Roc2 in the control of cupC gene expression, we performed a systematic analysis to probe the interaction between Roc1 and Roc2 components using the Escherichia coli bacterial two-hybrid method, as previously described (Karimova et al., 1998; Kulasekara et al., 2005). Because RocS1 and RocS2 are unorthodox sensors, we cloned the DNA fragments corresponding to the Hpt domains (Fig. S1) into pKT25 (Karimova et al., 1998). We also cloned the fragments corresponding to the D2 receiver domain of RocA1 and RocA2 response regulators into pUT18c. We then tested tandem interactions between Hpt and D2 domains, and found that RocS1 and RocS2 could interact with both response regulators (Fig. 2). In order to validate this observation, which reveals an extensive cross-regulation between the Roc1 and Roc2 systems, we also used a control showing that neither RocS1 nor RocS2 could interact with the D2 domain of the unrelated response regulator TrpO/PA0034 (Ventre et al., 2004) (Fig. 2). We also tested a P. aeruginosa sensor kinase other than RocS1/RocS2, namely GacS. We showed that GacS strongly interacts with the cognate response regulator GacA, but no interaction is seen with either RocA1 or RocA2 (Fig. 2). We concluded that both RocS2 and RocS1 directly activate RocA1, which results in induction of cupC gene expression. Moreover, we also observed that both RocS1 and RocS2 interact with RocA2, which may result in the induction of as yet unknown target genes.
Fig. 2.
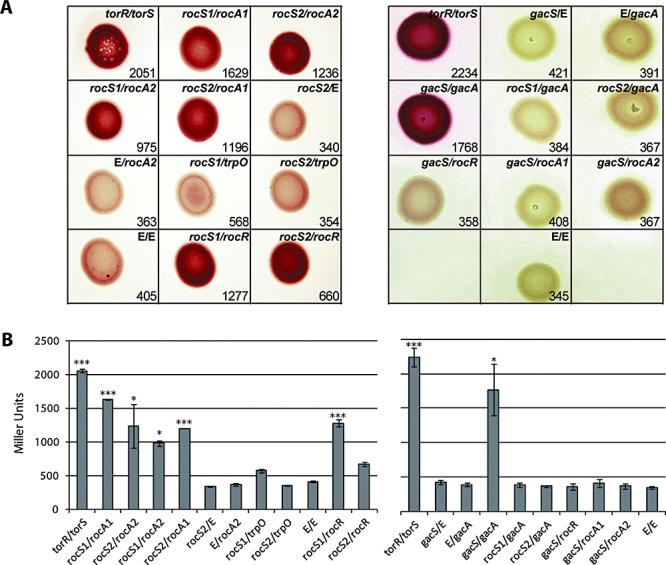
Protein–protein interactions between components of the Roc1 and Roc2 systems as determined by bacterial two-hybrid analysis. Various combinations of recombinant pKT25 and pUT18c plasmids harbouring protein domains of interest (Hpt domains of sensors and D2 domains of response regulators) were co-transformed into E. coli DHM1, and transformants were spotted onto MacConkey agar (A). Dark red colonies indicate a positive interaction. The E. coli two component sensor TorS, together with its cognate response regulator TorR, was used as a positive control. The specificity of interaction was assessed using the unrelated P. aeruginosa TCS GacS/GacA, as well as the unrelated P. aeruginosa response regulator TrpO. The strength of interaction was investigated by measuring the β-galactosidase activity of cells in the respective colonies, and the average activity in Miller Units is indicated next to each colony. (B) Graphic representation of the β-galactosidase activity in panel A. All experiments were carried out in at least duplicates, and error bars represent one standard deviation of the mean. Plasmid combinations of pKT25/pUT18c, respectively, are indicated in panels A and B. It should be noted that E means empty vector. For specific constructs, see Table 1. In panel B, stars indicate a significant difference between a given combination and the control (E/E). Three stars or one star corresponds to P-value < 0.001 or < 0.05 respectively.
Finally, we previously observed that RocR acts as a negative regulator of the Roc1 system (Kulasekara et al., 2005) and we showed in this study that it also negatively impacts the Roc2 response. We cloned the fragment corresponding to the D2 receiver domain of RocR into pUT18c and tested the interaction with Hpt domains of RocS1 or RocS2. We confirmed the interaction between RocR and RocS1 and found that RocR is also able to directly interact with RocS2 (Fig. 2). This observation further explains the mechanism by which RocR negatively impacts RocS2 signalling.
RocA2 controls expression of the mexAB-oprM genes
Because RocA2 is not involved in cupC gene expression, but interacts with RocS2 and RocS1, we further searched for specific RocA2 target genes. We compared the transcriptomic profile of a PAK or PAKΔrocA2 strain, both overproducing RocS2 from pMMB67-rocS2. This was based on the assumption that transcripts that were altered in PAK but not in PAKΔrocA2 are putative RocA2 target genes. The strains were grown at 37°C in minimal medium, RNA was extracted and cDNA synthesized and labelled using Cy5-dCTP. The cDNA was then hybridized to P. aeruginosa microarrays (see Experimental procedures). Gene expression levels in PAK overproducing RocS2 were directly compared with expression levels observed in PAKΔrocA2 overproducing RocS2. The genes for which expression varies by more than 1.5-fold are described in the Supporting information (Table S1). Using this cut-off, 42 genes were upregulated and 44 genes downregulated in the PAKΔrocA2 strain as compared with PAK. A number of genes involved in iron acquisition, such as fepB, pchG or pvdH are downregulated in PAKΔrocA2, whereas at least five genes involved in T3SS are upregulated in PAKΔrocA2, namely pcr1, orf1, exoT, popB and pcrV (Table S1). In all these cases, the fold change observed was between 1.5 and 1.9. Only eight genes were identified to be upregulated more than threefold, in PAKΔrocA2 (Table S1). Among these eight genes, four are located at a single locus. The mexA, mexB and oprM genes (around eightfold up in PAKΔrocA2) encode components of an efflux pump whereas mexR encodes a regulator of mexAB-oprM gene expression (23-fold up in PAKΔrocA2).
In order to validate the microarray data, we performed quantitative RT-PCR (see Experimental procedures) on the mexA and mexR genes. Upon overexpression of rocS2 we observed a threefold increase in mexA expression, and an almost 12-fold increase for mexR in PAKΔrocA2 as compared with PAK (Fig. 3A). These results confirmed that the mexAB-oprM and mexR genes are part of the RocA2 regulon, and that their expression is negatively controlled by RocA2. Interestingly, when we performed the experiment using rocS1 overexpressing strains, we observed similar results. There is a sixfold and 17-fold upregulation in the PAKΔrocA2 strain for mexA and mexR respectively (Fig. 3B). We thus concluded that in addition to RocS2, RocS1 could also act on RocA2 to repress mex gene expression, which confirms the positive interaction observed between RocS1 and RocA2 using the two-hybrid approach.
Fig. 3.
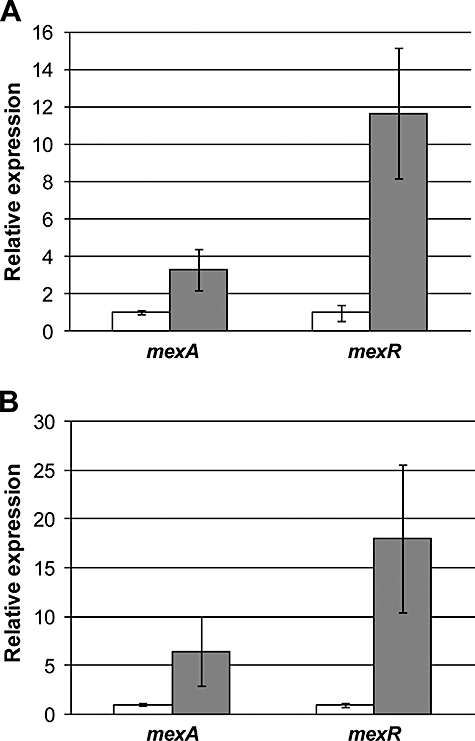
Quantitative RT-PCR analysis of mexA and mexR gene expression. The analysis was done upon overexpression of the sensor-encoding genes rocS2 (A) and rocS1 (B) in a PAK wild type (white bars) and PAKΔrocA2 deletion mutant (grey bars). Expression levels were normalized to the 16SrRNA gene and wild-type levels have been set to 1. The data represent the average of biological triplicates and error bars indicate one standard deviation of the mean. In (A) and (B) the relative expression of mexA is indicated on the left and mexR on the right, as indicated below the figure. In all cases the difference is significant with a P-value < 0.05.
Roc2 is a signalling pathway involved in antibiotic resistance
MexAB-OprM is a multidrug efflux system known to contribute to the natural resistance of P. aeruginosa to several antibiotics. As we showed that RocA2 could repress mexAB-oprM expression, we analysed whether this regulatory mechanism could impact the level of antibiotic resistance in P. aeruginosa strains. We used a series of antibiotic discs to assess the susceptibility of various P. aeruginosa strains (see Experimental procedures). We thus compared the susceptibility of PAK and PAKΔrocA2, both overexpressing rocS2, for compound sulphonamides, cinoxacin and cefotaxime. Interestingly, the level of susceptibility to these antibiotics was decreased in the rocA2 mutant as compared with the PAK strain, as seen by the diameter of bacterial growth inhibition around each disc (Fig. 4 and Table 2). We also determined the minimal inhibitory concentration (MIC) values for some of these antibiotics (see Experimental procedures) and showed that for the rocA2 mutant, the MIC was twofold higher for cinoxacin and eightfold higher for sulfamethoxazole, which is a sulphonamide antibiotic, as compared with the PAK parental strain (Table 3). Finally, we used a specific substrate of the MexAB-OprM pump, namely the β-lactam aztreonam. Again, the rocA2 mutant showed a significant increase in resistance to this antibiotic (Fig. 4 and Table 2). We also compared the antibiotic susceptibility of PAK or PAKΔrocA2, both overexpressing rocS1 from a plasmid that does not carry a carbenicillin resistance gene but has a gentamicin resistance cassette (pMMB67-rocS1). In this context we could show that the rocA2 mutant has a decreased susceptibility towards the β-lactam carbenicillin, when compared with the parental PAK strain (Fig. 4 and Table 2). These observations confirm that the activity of RocA2 prevents MexAB-OprM efflux pump production, and therefore increases susceptibility to antibiotics, including β-lactams such as aztreonam and carbenicillin, which are specific substrates of the MexAB-OprM efflux pump. Consequently, in the absence of RocA2 the P. aeruginosa strains become hyper-resistant to antibiotics.
Fig. 4.
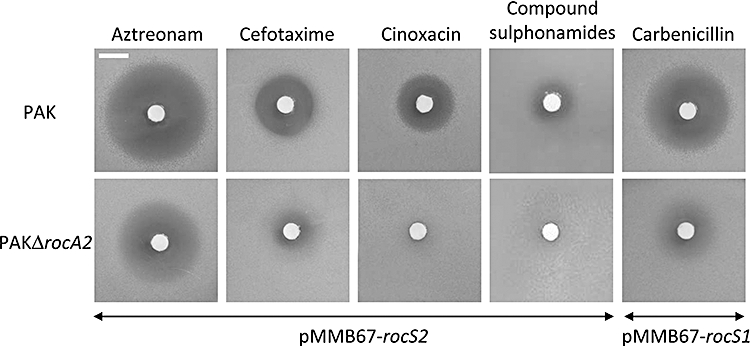
Antibiotic Disc Diffusion Assay. Discs containing Aztreonam (30 µg), Cefotaxime (300 µg) Cinoxacin (100 µg) or compound sulphonamides (300 µg) were applied to M63 media plates overlaid with PAK wild-type or PAKΔrocA2 harbouring pMMB67-rocS2. Discs containing Carbenicillin (100 µg) were applied to M63 media plates inoculated with PAK wild-type harbouring pMMB67-rocS1 or PAKΔrocA2 harbouring pMMB67-rocS1. The bar in the top left panel is a 1 cm scale.
Table 2.
Antibiotic susceptibility assays
| Zone of inhibition (mm)a | |||||
|---|---|---|---|---|---|
| Strain | Aztreonam (30 µg) | Cefotaxime (300 µg) | Cinoxacin (100 µg) | Sulphonamides (300 µg) | Carbenicillin (100 µg) |
| PAK + pMMB67-rocS2 | 15.5 ± 0.7 | 8 ± 0.2 | 8.5 ± 0.8 | 4 ± 0.8 | ND |
| PAKΔrocA2 + pMMB67-rocS2 | 7.5 ± 0.4 | 3 ± 0.2 | Rb | R | ND |
| PAK + pMMB67-rocS1 | 15 ± 0.6 | 8 ± 0.1 | 7 ± 0.5 | 4 ± 0.7 | 13.5 ± 0.6 |
| PAKΔrocA2 + pMMB67-rocS1 | 7.5 ± 0.3 | 2.5 ± 0.1 | R | R | 5.5 ± 0.7 |
Zones of growth inhibition around discs were determined by the disc diffusion assay on M63 minimal media agar plates. The mean and standard deviation of a representative experiment with biological triplicates are given and the experiment has been repeated twice.
R, resistant, no growth inhibition observed.
All differences observed between PAK/pMMB67-rocS2 and PAKΔrocA2/pMMB67-rocS2 or PAK/pMMB67-rocS1 and PAKΔrocA2/pMMB67-rocS1 are significant with a P-value < 0.01.
ND, Not determined.
Table 3.
MIC values
| MIC (µg ml−1)a | ||
|---|---|---|
| Strain | Cinoxacin | Sulfamethoxazole |
| PAK + pMMB67-rocS2 | 62.5 | 31.3 |
| PAKΔrocA2 + pMMB67-rocS2 | 125 | 250 |
MICs were determined for cinoxacin and sulfamethoxazole by a twofold broth dilution method and the lowest concentration where no visible growth was observed is indicated. The assay was done in triplicates.
Discussion
Pseudomonas aeruginosa uses more than 60 different two-component signal transduction systems to adapt to various and changing environments (Gooderham and Hancock, 2009). A significant number of these systems are important for adaptation during the infection process. Among these the GacS/RetS/LadS/HptB pathway has been shown to be important in the switch between acute and chronic infections (Goodman et al., 2004; O'Toole, 2004; Ventre et al., 2006; Bordi et al., 2010). In the chronic infection process, it has been proposed that the development of a biofilm is the most appropriate bacterial lifestyle (Costerton, 2001). Biofilm formation requires several steps, including initial attachment to a surface, cell–cell interaction within microcolonies, formation of mushroom-like structures and finally bacterial dispersal, allowing the process to start all over again (O'Toole et al., 2000; Stoodley et al., 2002). Initial attachment onto a surface has been shown to involve Cup fimbriae (Vallet et al., 2001). However, the production and assembly of these cell surface appendages is tightly controlled and cup genes are typically not expressed in standard laboratory conditions (Kulasekara et al., 2005). It has further been shown that the Roc1 system is an important regulatory pathway in the production and assembly of CupB and CupC fimbriae (Ruer et al., 2007; 2008;). This system involves an unorthodox sensor, RocS1, and two response regulators. The output domain of RocA1 is a helix–turn–helix motif that should provide direct DNA binding properties, whereas the output domain of RocR contains an EAL domain, which is a signature of phosphodiesterase activity (Rao et al., 2008). It was shown that RocA1 positively controls cup gene expression, whereas RocR acts indirectly by modulating the intracellular pool of c-di-GMP. In P. aeruginosa, Roc1 paralogues could be identified, namely Roc2 and Roc3. Interestingly, all three histidine kinases, RocS1, RocS2 and RocS3, were identified as BvgS homologues, and BvgS is a global virulence regulator in Bordetella pertussis (Beier and Gross, 2008).
It has previously been reported that the Roc1 system controls the expression of cupC genes. This came from the identification of transposon insertions in the roc1 region, which resulted in overexpression of rocS1 or rocA1 genes (Kulasekara et al., 2005). To a lesser extent, it was also shown that transposon insertions within the roc2 cluster increased expression of cupC genes. Here we showed that it is indeed overexpression of the rocS2 gene that results in cupC expression. However, it is intriguing that RocS2-dependent activation of cupC is totally independent of the RocA2 response regulator, but exclusively dependent on the RocA1 response regulator. This observation thus suggests the existence of cross-regulation between the Roc1 and Roc2 signalling pathways. Cross-regulation between TCSs is not frequent, at least in vivo, and bacteria have developed means to insulate pathways from unwanted cross-talk (Laub and Goulian, 2007). Importantly, and in contrast to rocS1 and rocS2, the overexpression of rocS3 did not result in cupC gene overexpression, suggesting a degree of specificity in the RocS1 or RocS2 interaction with RocA1.
It was intriguing to observe that RocA2 is not required for RocS2-dependent activation of cupC genes, because we showed using two-hybrid experiments that RocS2 is able to interact with RocA2, in addition to interaction with RocA1. The identification of the RocA2 regulon was thus completed using microarray analysis and we compared gene expression in a wild-type versus rocA2 mutant upon overexpression of rocS2. Among the targets identified, it is noticeable that five T3SS genes were upregulated in the rocA2 mutant. It is a possibility that T3SS genes are common targets both for RocA1 and RocA2, as previous observations by Kuchma and collaborators indicated that lack of both RocA1S1 (named SadAS in their study) resulted in slight derepression of the T3SS genes (Kuchma et al., 2005). Whereas the variation in T3SS gene expression was about 1.5-fold, we identified a series of target transcripts that varied much more, ranging from 3- to 12-fold. These genes, mexAB-oprM and mexR are involved in the regulation and assembly of an efflux pump that contributes to antibiotic resistance in P. aeruginosa. MexAB-OprM is one of the many pumps found in P. aeruginosa (Srikumar et al., 1999). It is likely the pump with the widest substrate specificity, because it is able to provide resistance towards β-lactams, fluoroquinolones and other families of antibiotics (Li et al., 1995; Masuda et al., 2000). The regulation of mexAB-oprM genes has already been extensively documented and underlines the existence of a number of pathways. MexR is the repressor that directly binds the mexA promoter region and prevents transcription of the mexAB-oprM operon (Evans et al., 2001). Interestingly, the armR gene (PA3719), encodes a 53 amino acids-long polypeptide that prevents proper binding of MexR within the mexA promoter region (Wilke et al., 2008). Another repressor, NalD, was also described, which acts by binding within another region of the mexA promoter (Morita et al., 2006). The reason why we observed an upregulation of the gene encoding the MexR repressor, and yet observed an upregulation of the mexAB-oprM operon is unclear. However, it should be noted that most nalB and nalD multidrug resistant mutants display an increased expression of both mexR and mexAB-oprM (Poole et al., 1996; Llanes et al., 2004). One possibility could be that the anti-repressor ArmR is also upregulated but we did not detect such variation in our microarray analysis. Alternatively, a gene encoding a yet unknown anti-MexR protein may be upregulated. MexR has also been reported to be sensitive to oxidative stress and oxidized MexR dissociates from the mexA promoter (Chen et al., 2008). It is a possibility that genes encoding components that contribute to oxidative stress conditions might also be upregulated.
One obvious strategy to analyse whether the observed upregulation of mexAB-oprM results in a relevant phenotype was to test the antibiotic susceptibility of the P. aeruginosa rocA2 mutant overexpressing rocS2 or rocS1 as compared with the parental strain PAK. We have tested a series of antibiotics including sulphonamides and cinoxacin, but also β-lactams such as aztreonam and carbenicillin, which are specific substrates of the MexAB-OprM pump. In all cases the rocA2 mutant showed a striking decrease in sensitivity as compared with the parental strain, which confirms that RocA2 acts as a repressor of mexAB-oprM gene expression. Regulation of antibiotic resistance by TCSs is not frequently observed in P. aeruginosa. However, it has been reported for CzcR/CzcS and CopR/CopS, which control metal and imipenem resistance (Hassan et al., 1999; Perron et al., 2004; Teitzel et al., 2006; Caille et al., 2007), PprB/PprA and GacA/GacS, which control aminoglycoside resistance (Brinkman et al., 2001; Wang et al., 2003), PmrA/PmrB involved in polymyxin B and antimicrobial peptide resistance (McPhee et al., 2003) or ParR/ParS involved in polymyxin B and colistin resistance (Fernández et al., 2010).
A striking feature here is the observation that the RocS2 and RocS1 sensors on one hand induce cupC gene expression and hence biofilm formation via RocA1, while on the other hand they repress antibiotic resistance mechanisms via RocA2. Although the opposition between cytotoxicity and biofilm formation observed through the RetS/LadS/GacS/HptB pathway appears plausible in the context of a chronic infection, the antagonistic regulation of biofilm formation and antibiotic resistance is less obvious. However, previous reports by De Kievit and collaborators have already suggested that efflux pumps may not play a role in the antibiotic resistance observed in P. aeruginosa biofilms (De Kievit et al., 2001). Using transcriptional fusions to gfp they demonstrated that genes encoding multidrug resistance pumps such as mexAB-oprM, were downregulated in the developing biofilm. Even more striking, a recent study by Vettoretti and collaborators suggested that a large proportion of the P. aeruginosa strains isolated from cystic fibrosis (CF) patients (28%) are hyper-susceptible to the β-lactam antibiotic ticarcillin (Vettoretti et al., 2009). In this study it was shown that the lack of resistance is essentially associated with a non-functional MexAB-OprM pump. In other words, in CF patients P. aeruginosa strains grow predominantly as biofilms in the respiratory tract (Bjarnsholt et al., 2009) and at the same time large numbers of these strains appear to become more susceptible to antibiotics. It is largely unclear what the benefit could be for this subpopulation, but likely the lack of this particular pump may provide a not yet understood advantage fitness in the context of CF lungs. Furthermore, other pumps may still provide reasonable levels of antibiotic resistance to cope with the selection pressure imposed by the antimicrobial treatments given to CF patients. Finally, other mechanisms have been shown to contribute biofilm resistance to antibiotics, such as the NdvB-dependent production of cyclic glucans (Mah et al., 2003; Sadovskaya et al., 2010). Nevertheless, our study fully supports the idea that an antagonistic balance exists between biofilm formation and antibiotic resistance in P. aeruginosa, and we show evidence that the TCSs Roc2 and Roc1 are likely to be master regulators in this process (Fig. 5). It also suggests that the Roc systems might be key regulatory systems sensing the particular environment of the CF lungs. In this respect, further studies aimed at the characterization of the stimuli detected by the Roc systems could be of great importance in the development of novel antimicrobials more effective in the treatment of CF patients.
Fig. 5.
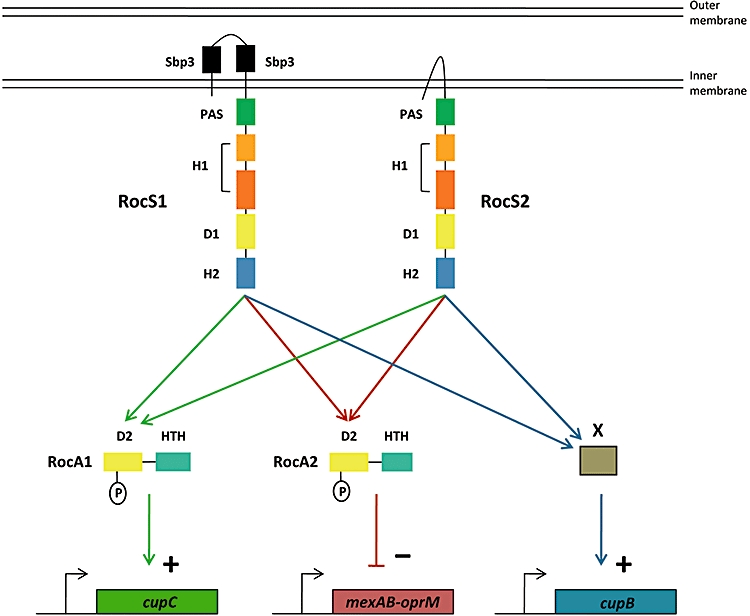
Schematic model showing the cross-regulation between Roc1 and Roc2 signalling pathways. The Roc1 and Roc2 components are represented as in Fig. S1. The RocS1 and RocS2 sensors are shown as integral inner membrane proteins. The positive regulation on the cupC gene expression is shown with green arrows. The negative regulation on the mexAB-oprM genes is shown with brown arrows. The positive regulation on cupB genes is shown with blue arrows and involves a yet unknown regulator indicated as X.
Experimental procedures
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used are described in Table 1. Plasmids were transferred into P. aeruginosa using the conjugative properties of the helper plasmid pRK2013 (Figurski and Helinski, 1979) in triparental matings. P. aeruginosa transconjugants were isolated on Pseudomonas Isolation Agar (Difco) supplemented with the appropriate antibiotics. The following antibiotic concentrations were used for E. coli: ampicillin 50 µg ml−1, kanamycin 50 µg ml−1, streptomycin 50 µg ml−1, tetracycline 25 µg ml−1, gentamicin 25 µg ml−1 and for P. aeruginosa: carbenicillin 300 µg ml−1, streptomycin 2000 µg ml−1, gentamicin 120 µg ml−1, tetracycline 200 µg ml−1. Bacteria were grown in Luria–Bertani (LB) broth or M63 minimal medium supplemented with 1 mM MgSO4, 0.2% glucose and 0.5% casamino acids. Where appropriate, IPTG was added to a final concentration of 1 mM for gene expression in P. aeruginosa.
Strain and plasmid construction
Transcriptional fusions were generated by PCR amplification of the putative cupB and cupC promoter regions, each containing a DNA segment encompassing 472 bp upstream and 137 bp downstream from the translational initiation codon of the cupB1 gene, and 445 bp upstream and 185 bp downstream from the translational initiation codon of the cupC1 gene (primers are listed in Table S2). The promoter fragments were cloned into pCR2.1 and after excision by digestion with BamHI and XhoI, subcloned into mini-CTX-lacZ vector followed by integration of the appropriate fragment into the PAK chromosome at the attB site using established protocols (Hoang et al., 2000).
To construct the rocA2 deletion mutant, PCR was used to generate a 560 bp DNA fragment upstream (Up) from the rocA2 gene bearing a 3′ BamHI restriction site and a 505 bp DNA fragment downstream (Dw) from the rocA2 gene bearing a 5′ BamHI site using the UA3045-LA3045 and UB3045-LB3045 oligonucleotide pairs respectively (Table S2). The Up and Dw fragments were used in a three partner ligation into pCR2.1. The resulting 1065 bp Up-Dw DNA fragment was digested from pCR2.1 using SpeI and ApaI and subcloned into the suicide vector pKNG101 (Kaniga et al., 1991) digested with SpeI and ApaI. The resulting plasmid pKNGΔrocA2 was mobilized into P. aeruginosa and the deletion mutants were selected on LB plates containing 5% sucrose and appropriate antibiotics as previously described (Kaniga et al., 1991). The double rocA1-rocA2 deletion mutant was constructed by conjugating pKNGΔrocA2 into a P. aeruginosa rocA1 deletion (Kulasekara et al., 2005) and proceeding further as described above. The rocS2 and rocA2 genes were obtained from the Gateway library of PAO1 orfs (Labaer et al., 2004) and cloned into the IPTG-inducible expression vector pMMB67HE42, yielding pMMB67-rocS2 and pMMB67-rocA2.
Measurements of β-galactosidase activity
M63 medium supplemented with appropriate antibiotics and IPTG (1 mM) was inoculated to OD600 0.1 with overnight cultures of strains carrying lacZ transcriptional fusions. Cultures were incubated at 37°C with shaking, and samples were harvested at 6 h post inoculation. β-galactosidase assays were carried out as previously described, and activity was expressed in Miller Units (Miller, 1992).
Bacterial two-hybrid assay
DNA fragments encoding the protein domains of interest were cloned into plasmids pKT25 and pUT18c, which each encode for complementary fragments of the adenylate cyclase enzyme, as previously described by Karimova and collaborators (Karimova et al., 1998). DNA fragments encoding the Hpt domain of RocS2 (RocS2-Hpt), the cytoplasmic domain of GacS (GacS-H1-D1-Hpt) and the D2 domain of RocA2 (RocA2-D2) or GacA (GacA-D2) were amplified by PCR using P. aeruginosa PAK genomic DNA. PCR products were cloned into pKT25 (Hpt) and pUT18c (D2) respectively. pKT25 and pUT18c recombinant plasmids were transformed simultaneously into the E. coli DHM1 strain, which lacks adenylate cyclase, and screened for positive interactions. Transformants were spotted on MacConkey agar plates (Difco) supplemented with 1% maltose, in presence of 100 µg ml−1 ampicillin, 50 µg ml−1 kanamycin and 1 mM IPTG. Positive interactions were identified as dark red colonies on MacConkey after 48 h incubation at 30°C followed by 96 h at room temperature. We also used the previously engineered pUT18c derivative encoding RocR-D2 (Kulasekara et al., 2005). The positive controls used in the study were pUT18c or pKT25 derivatives encoding the Hpt domain of TorS (TorS-Hpt), the D2 domain of TorR (TorR-D2), the Hpt domain of RocS1 (RocS1-Hpt) and the D2 domain of RocA1 (RocA1-D2) as previously described by Kulasekara and collaborators (Kulasekara et al., 2005). Finally, a pKT25 derivative encoding TrpO-D2 was engineered and used as a negative control. For quantitative assays, cells were scraped from the plates and resuspended thoroughly in water. β-galactosidase assays were then carried out in the same manner as for liquid cultures.
Microarray analysis
For each strain, microarray experiments were performed in triplicate. Independent overnight cultures of P. aeruginosa PAK and PAKΔrocA2, both harbouring pMMB67-rocS2 were resuspended in M63 medium as described above, to OD600 0.1 in the presence of appropriate antibiotics and 1 mM IPTG. Cells were then grown at 37°C with shaking and samples were harvested after 5 h for RNA extraction. Under these conditions, the final OD600 of both strains was nearly identical. RNAlater® (Ambion) was added immediately to the harvested cells to stabilize RNA and prevent degradation. Cell suspensions were then centrifuged at 4°C and RNA extraction was carried out using Promega SV Total RNA Isolation Kit according to manufacturer's protocol. The protocol was modified such that the DNase I digestion step was carried out twice to reduce amount of contaminating DNA. The integrity of RNA preparations was checked using the Agilent 2100 Bioanalyzer and the expression profiling experiment was carried out at the Microarray Facility, Institute of Infection, Immunity and Inflammation, University of Nottingham, as previously published (Rampioni et al., 2010). The microarrays were designed to contain oligonucleotide probes for all the PAO1 genes including the small RNA genes and were purchased from Oxford Gene Technology (Oxford, UK). Briefly, for each array, 10 µg of RNA was reverse transcribed and labelled with Cy5-dCTP and 2 µg of genomic DNA was labelled with Cy3-dCTP. Samples were hybridized onto the arrays for 16 h. Scanning of the arrays was performed using the Axon 4000B GenePix Scanner (Molecular Devices, Sunnyvale, USA) and data analysis performed using GeneSpring GX10 (Agilent Technologies, Santa Clara, USA). The array data underwent Lowess normalization and genes of altered expression were determined by passing through cut-offs of both a fold change of 1.5 and a paired t-test of P = 0.05.
Quantitative reverse transcription PCR
The same growth conditions as in the microarray analysis described above were used. Quantitative RT-PCR was performed as previously described (Burr et al., 2006). Briefly, for first-stranded cDNA synthesis, 200 ng of total RNA was used in a reverse transcription reaction using Invitrogen SuperScript® II Reverse Transcriptase and random hexamers (Applied Biosystems) and reactions were performed according to the manufacturer's protocol. The expression levels of the mexA and mexR genes were assessed using SYBR Green PCR Master Mix and the 7300 Real Time PCR System apparatus (Applied Biosystems). The primers used for the amplification of mexA are mexAup and mexAdown; those for mexR are mexRup and mexRdown (Table S2). The gene transcription levels were normalized in each strain to the 16S ribosomal RNA gene (16SrRNA) and expressed as ratios to the values of the PAK strain (set to 1). Samples were assayed in triplicate for each condition.
Antibiotic susceptibility assays
For antibiotic disc diffusion assays, 1.5% M63 agar (same composition as liquid M63 medium) supplemented with appropriate antibiotics for selection was overlaid with 0.7% M63 soft agar seeded with bacterial strains (PAK or PAKΔrocA2 carrying pMMB67-rocS2 or pMMB67-rocS1) as indicated in the individual experiments. Filter discs containing antibiotics as indicated were then placed onto the agar overlay, and plates were incubated at 37°C overnight. Antimicrobial sensitivity was determined by measuring the zone of inhibition around the discs. MICs were determined by serial twofold broth dilution method using M63 minimal media with an inoculum size of 105 cells. Growth was assessed visually after 27 h of static incubation at 37°C. The MIC was defined as the lowest concentration of antimicrobial agent that inhibited visible growth. Cinoxacin and sulfamethoxazole (sulphonamide antibiotic) were from Sigma-Aldrich.
Acknowledgments
We thank Victoria Wright and the University of Nottingham for access to microarray facilities and Elise Termine for construction of pMMB67HE42. A. F. is supported by the Royal Society, BBSRC Grant ref N°: BB/F019645/1 and European EST Marie Curie Grant n° MEST-CT-2005-020278. M. S. is supported by a Marie Curie fellowship and a grant from the ‘Fondation pour la Recherche Médicale’ (FRM: FDT20091217603). H. M. is supported by the BBSRC Grant N°: BB/F019645/1. C. B. is supported by ANR grant 05-MIIM-040-01 and institutional grants from the CNRS.
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Beier D, Gross R. The BvgS/BvgA phosphorelay system of pathogenic Bordetellae: structure, function and evolution. Adv Exp Med Biol. 2008;631:149–160. doi: 10.1007/978-0-387-78885-2_10. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T, Jensen PØ, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, et al. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol. 2009;44:547–558. doi: 10.1002/ppul.21011. [DOI] [PubMed] [Google Scholar]
- Bordi C, Lamy MC, Ventre I, Termine E, Hachani A, Fillet S, et al. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol Microbiol. 2010;76:1427–1443. doi: 10.1111/j.1365-2958.2010.07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol. 2009;73:434–445. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman FS, Macfarlane EL, Warrener P, Hancock RE. Evolutionary relationships among virulence-associated histidine kinases. Infect Immun. 2001;69:5207–5211. doi: 10.1128/IAI.69.8.5207-5211.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr T, Barnard AM, Corbett MJ, Pemberton CL, Simpson NJ, Salmond G. P. Identification of the central quorum sensing regulator of virulence in the enteric phytopathogen, Erwinia carotovora: the VirR repressor. Mol Microbiol. 2006;59:113–125. doi: 10.1111/j.1365-2958.2005.04939.x. [DOI] [PubMed] [Google Scholar]
- Caille O, Rossier C, Perron K. A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J Bacteriol. 2007;189:4561–4568. doi: 10.1128/JB.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- Chen H, Hu J, Chen PR, Lan L, Li Z, Hicks LM, et al. The Pseudomonas aeruginosa multidrug efflux regulator MexR uses an oxidation-sensing mechanism. Proc Natl Acad Sci USA. 2008;105:13586–13591. doi: 10.1073/pnas.0803391105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol. 2001;9:50–52. doi: 10.1016/s0966-842x(00)01918-1. [DOI] [PubMed] [Google Scholar]
- De Kievit TR, Parkins MD, Gillis RJ, Srikumar R, Ceri H, Poole K, et al. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2001;45:1761–1770. doi: 10.1128/AAC.45.6.1761-1770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K, Adewoye L, Poole K. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J Bacteriol. 2001;183:807–812. doi: 10.1128/JB.183.3.807-812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock RE. Adaptive resistance to the ‘last hope’ antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother. 2010;54:3372–3382. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JA, Bassler BL. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol. 1999;31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- Galperin MY, Nikolskaya AN. Identification of sensory and signal-transducing domains in two-component signaling systems. Methods Enzymol. 2007;422:47–74. doi: 10.1016/S0076-6879(06)22003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvis S, Munder A, Ball G, de Bentzmann S, Wiehlmann L, Ewbank JJ, et al. Caenorhabditis elegans semi-automated liquid screen reveals a specialized role for the chemotaxis gene cheB2 in Pseudomonas aeruginosa virulence. PLoS Pathog. 2009;5:e1000540. doi: 10.1371/journal.ppat.1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooderham WJ, Hancock RE. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol Rev. 2009;33:279–294. doi: 10.1111/j.1574-6976.2008.00135.x. [DOI] [PubMed] [Google Scholar]
- Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009;23:249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MT, van der Lelie D, Springael D, Römling U, Ahmed N, Mergeay M. Identification of a gene cluster, czr, involved in cadmium and zinc resistance in Pseudomonas aeruginosa. Gene. 1999;238:417–425. doi: 10.1016/s0378-1119(99)00349-2. [DOI] [PubMed] [Google Scholar]
- Hoang TT, Kutchma AJ, Becher A, Schweizer HP. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid. 2000;43:59–72. doi: 10.1006/plas.1999.1441. [DOI] [PubMed] [Google Scholar]
- Kaniga K, Delor I, Cornelis GR. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay E, Humair B, Dénervaud V, Riedel K, Spahr S, Eberl L, et al. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J Bacteriol. 2006;188:6026–6033. doi: 10.1128/JB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchma SL, Connolly JP, O'Toole GA. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J Bacteriol. 2005;187:1441–1454. doi: 10.1128/JB.187.4.1441-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol. 2005;55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- Labaer J, Qiu Q, Anumanthan A, Mar W, Zuo D, Murthy TV, et al. The Pseudomonas aeruginosa PAO1 gene collection. Genome Res. 2004;14:2190–2200. doi: 10.1101/gr.2482804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- Li XZ, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llanes C, Hocquet D, Vogne C, Benali-Baitich D, Neuwirth C, Plésiat P. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob Agents Chemother. 2004;48:1797–1802. doi: 10.1128/AAC.48.5.1797-1802.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee JB, Lewenza S, Hancock RE. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol. 2003;50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2000;44:3322–3327. doi: 10.1128/aac.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Morita Y, Cao L, Gould VC, Avison MB, Poole K. nalD encodes a second repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J Bacteriol. 2006;188:8649–8654. doi: 10.1128/JB.01342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- O'Toole GA. Microbiology: Jekyll or hide? Nature. 2004;432:680–681. doi: 10.1038/432680a. [DOI] [PubMed] [Google Scholar]
- Otto M. Bacterial sensing of antimicrobial peptides. Contrib Microbiol. 2009;16:136–149. doi: 10.1159/000219377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron K, Caille O, Rossier C, Van Delden C, Dumas JL, Köhler T. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J Biol Chem. 2004;279:8761–8768. doi: 10.1074/jbc.M312080200. [DOI] [PubMed] [Google Scholar]
- Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs DE, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampioni G, Pustelny C, Fletcher MP, Wright VJ, Bruce M, Rumbaugh KP, et al. Transcriptomic analysis reveals a global alkyl-quinolone-independent regulatory role for PqsE in facilitating the environmental adaptation of Pseudomonas aeruginosa to plant and animal hosts. Environ Microbiol. 2010;12:1659–1673. doi: 10.1111/j.1462-2920.2010.02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F, Yang Y, Qi Y, Liang ZX. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J Bacteriol. 2008;190:3622–3631. doi: 10.1128/JB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue A, Quentin Y, Lazdunski A, Mejean V, Foglino M. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol. 2000;8:498–504. doi: 10.1016/s0966-842x(00)01833-3. [DOI] [PubMed] [Google Scholar]
- Ruer S, Stender S, Filloux A, de Bentzmann S. Assembly of fimbrial structures in Pseudomonas aeruginosa: functionality and specificity of chaperone-usher machineries. J Bacteriol. 2007;189:3547–3555. doi: 10.1128/JB.00093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruer S, Ball G, Filloux A, de Bentzmann S. The ‘P-usher’, a novel protein transporter involved in fimbrial assembly and TpsA secretion. EMBO J. 2008;27:2669–2680. doi: 10.1038/emboj.2008.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovskaya I, Vinogradov E, Li J, Hachani A, Kowalska K, Filloux A. High-level antibiotic resistance in Pseudomonas aeruginosa biofilm: the ndvB gene is involved in the production of highly glycerol-phosphorylated beta-(1->3)-glucans, which bind aminoglycosides. Glycobiology. 2010;20:895–904. doi: 10.1093/glycob/cwq047. [DOI] [PubMed] [Google Scholar]
- Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikumar R, Tsang E, Poole K. Contribution of the MexAB-OprM multidrug efflux system to the beta-lactam resistance of penicillin-binding protein and beta-lactamase-derepressed mutants of Pseudomonas aeruginosa. J Antimicrob Chemother. 1999;44:537–540. doi: 10.1093/jac/44.4.537. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- Teitzel GM, Geddie A, De Long SK, Kirisits MJ, Whiteley M, Parsek MR. Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J Bacteriol. 2006;188:7242–7256. doi: 10.1128/JB.00837-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc Natl Acad Sci USA. 2001;98:6911–6916. doi: 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur P, Vallet-Gely I, Soscia C, Genin S, Filloux A. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology. 2005;151:985–997. doi: 10.1099/mic.0.27410-0. [DOI] [PubMed] [Google Scholar]
- Ventre I, Filloux A, Lazdunski A. Two-component signal transduction systems: a key to the adaptive potential of Pseudomonas aeruginosa. In: Ramos J-L, editor. Pseudomonas: Virulence and Gene Regulation. Vol. 2. New York: Kluwer Academic; 2004. pp. 257–288. [Google Scholar]
- Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, et al. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci USA. 2006;103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettoretti L, Plésiat P, Muller C, El Garch F, Phan G, Attrée I, et al. Efflux unbalance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother. 2009;53:1987–1997. doi: 10.1128/AAC.01024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ha U, Zeng L, Jin S. Regulation of membrane permeability by a two-component regulatory system in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2003;47:95–101. doi: 10.1128/AAC.47.1.95-101.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke MS, Heller M, Creagh AL, Haynes CA, McIntosh LP, Poole K, Strynadka NC. The crystal structure of MexR from Pseudomonas aeruginosa in complex with its antirepressor ArmR. Proc Natl Acad Sci USA. 2008;105:14832–14837. doi: 10.1073/pnas.0805489105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


