Abstract
In this study, we report a familial inversion of chromosome 18, inv(18)(p11.31q21.33), in both members of a consanguineous couple. Their first child had inherited one balanced pericentric inversion along with a recombinant chromosome 18 resulting in dup(18q)/del(18p), and had mild dysmorphic features in the absence of mental and developmental retardation. The second child had received two recombinant chromosomes 18, from the mother a derivative chromosome 18 with dup(18p)/del(18q) and from the father a derivative chromosome 18 with dup(18q)/del(18p). The aberration was prenatally detected; however, as the two opposite aneuploidies were thought to compensate each other, the family decided to carry on with the pregnancy, knowing that uniparental disomy for the segments outside the inversion could have an adverse influence on the development of the child. Uniparental disomy was confirmed by SNP arrays. The child, who has been followed up until the age of 20 months, is healthy and normal. It seems to be the first reported case with two opposite recombinant chromosomes that compensate each other and lead to segmental uniparental disomy for two segments on the chromosome, one maternal and the other paternal.
Keywords: segmental uniparental disomy, chromosome 18, inversion chromosome 18
Introduction
Uniparental disomy (UPD) for only a few chromosomes has been shown to have a definite phenotypic effect due to uniparental inheritance of imprinted regions (chromosomes 7, 14 and 15 maternally inherited and chromosomes 6, 11, 14, 15 and 20 paternally inherited).1, 2, 3, 4, 5 For some chromosomes (eg, UPD16mat), it is not clear whether there are phenotypic effects due to imprinting. The consequence of UPD of chromosome 18 is unclear.
There have been several reports of patients with a recombinant chromosome 18 resulting from a pericentric inversion. Most of these patients had severe mental retardation and multiple congenital anomalies with severe morbidity and often early lethal outcome.6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 There are a few reports of patients with recombinant dup(18q)/del(18p) resulting from inversions, with very mild phenotypic findings and behavioral problems.18, 19 The first offspring of the family described in this study is a further case of dup(18q)/del(18p), with very mild dysmorphic features and normal developmental and borderline mental ability.
Subjects and methods
Family history
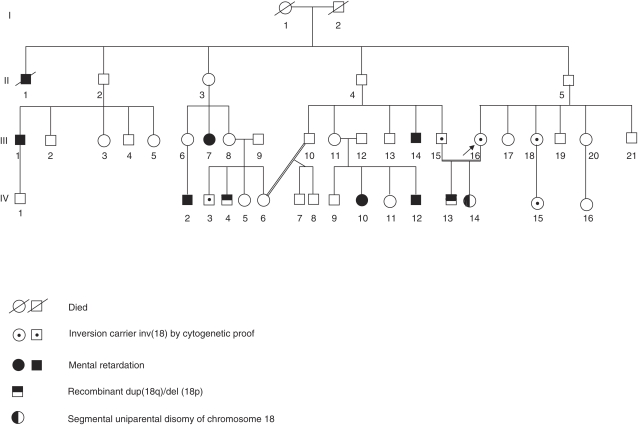
The couple (Figure 1, III-15 and III-16) came to our genetics center for a prenatal chromosome investigation for their second pregnancy because of chromosomal abnormality in the lady's sister (Figure 1, III 18). The sister had come to our center because of a history of mental retardation in many members of her family (Figure 1, II-1, III-1, III-7 III-14, IV-2, IV-4, IV-10, IV-12). Chromosomal study of the sister and her fetus (Figure 1, IV15) revealed an inv(18)(p11.2q21.3).
Figure 1.
Family pedigree.
On the basis of the history of a familial chromosome aberration and the consanguinity of the couple, chromosomal study was requested for the mother (Figure 1, III-16), the father (Figure 1, III-15), their first child, Case 1 (Figure 1, IV-13), and the fetus, Case 2 (Figure 1, IV-14).
Subjects
Parents
Both parents are normal and healthy. The mother's weight and height are 53 kg and 162 cm, respectively, and the father's weight and height are 98 kg and 180 cm, respectively.
Case 1
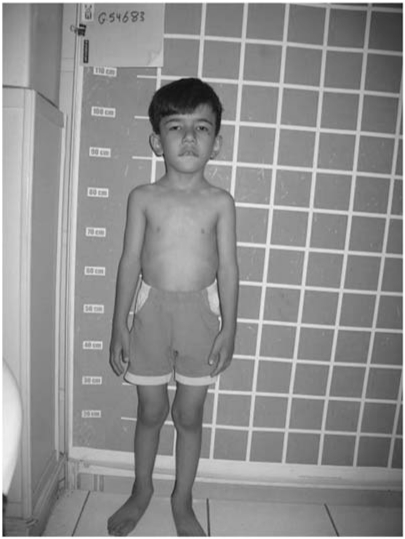
Delivery occurred at 37 weeks through elective cesarian section following an uneventful pregnancy. Birth weight, birth length and head circumference were 2750 g (50th centile), 48 cm (25th centile) and 34.5 cm (80th centile), respectively. Milestones were achieved on time. He held his head at 3 months, rolled over at 5 months, stood at 12 months and started walking at 13 months. He spoke his first words at 10 months. He was examined at our center at the age of 5 years. His weight, height and head circumference were 17 kg (10th–25th centile), 105 cm (10th centile) and 50 cm (3rd–10th centile), respectively. He had deep set eyes, tented upper lip, constantly open mouth, pronounced dental caries and hallux valgus, but no other dysmorphic features (Figure 2). He stammers.
Figure 2.
Case 1, no major dysmorphic features are present.
Psychometric testing using the Wechsler Intelligence Scale for Children-Revised, The Vineland Adaptive Behavior Scales and graphic test was carried out. The proband scored 71 in the practical and 78 for verbal performance. His performance in short-term memory, long-term memory, verbal learning and conceptual thinking was within the normal range, whereas processing speed, understanding of spatial relationship, visual attention and concentration fell below the normal range. The Vineland Adaptive Behavior Scales, Second Edition (Vineland-II), measures the personal and social skills of individuals. He scored 94 in the Vineland test, which was in the normal range; however, the practical and verbal IQ was borderline.
His behavioral and personality testing showed that he has difficulty coping with social interactions, which cause him anxiety and hesitation. He has tendency toward depression. He has low self-esteem, and feels the need for acceptance and has difficulty in acceptance of failure.
Case 2
The second child, a girl, was born by repeat cesarean section at 40 weeks of gestation. Birth weight, length and head circumference were 3200 g, 52 cm (both 50th centile) and 35 cm (75th centile), respectively. At the last examination, at the age of 20 months, her weight (12.2 kg), length (80 cm) and head circumference (47 cm) were at the 75th, 25th and 25th centile, respectively.
She had mild jaundice during neonatal period, which was treated by phototherapy.
Her milestones have been achieved on time up to now. She smiled at 2 months, held her head upright at 2 months, sat without support at 5.5 months, crawled at 6 months and stood without support at 9 months. She expressed her first words at 11 months, two-word sentences at 17 months and began climbing stairs at 17 months. She did not have any dysmorphic features and appeared to be a completely healthy girl. She has been routinely checked by her pediatrician who says that the child is growing and developing well.
The Denver Developmental Screening Test, a test for screening cognitive and behavioral problems in pre-school children, was used. She scored 101, which is within the normal range. She has good personal and social behavior. Cognition skills, visual and auditory memory, fine and gross motor skills, language concentration and tolerance are all within the normal range.
Result
Cytogenetic study
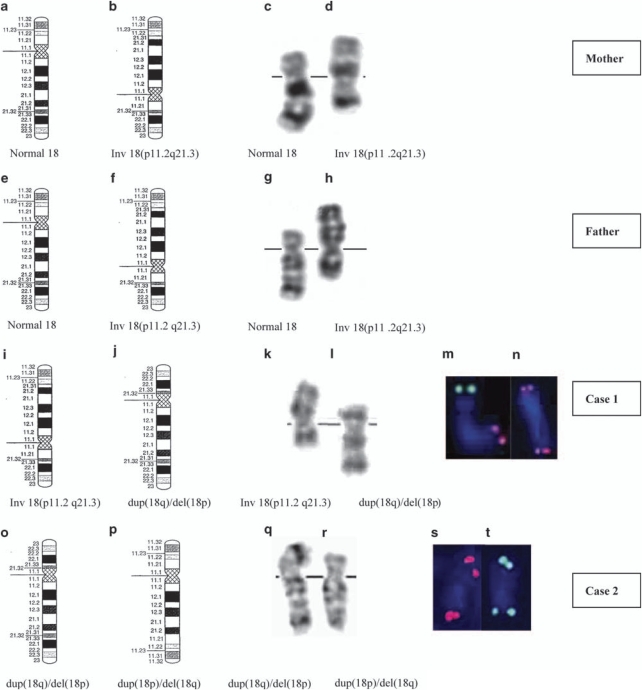
Chromosomal study was carried out on peripheral blood at 450-band resolution using GTG banding. Karyotypes of the mother (Figure 1, III-16) and the father (Figure 1, III-15) revealed an inv(18)(p11.2q21.3) (interpretation from the banding patterns). Chromosomal study of Case 1 (Figure 1, IV-13) revealed an inversion chromosome 18 similar to that of the parents and the other chromosome 18 similar to one of the fetus' chromosomes 18, with a dup(18q)/del(18p). Microsatellite and SNP array analysis of the son clearly indicated that the imbalance is maternal in origin and it is therefore reasonable to conclude that the balanced inversion is paternal. The karyotype of Case 1 can therefore be described as 46,XY,inv(18)(p11.2q21.3)pat,rec(18)dup(q)inv(18)(p11.2q21.3)mat (Figure 3).
Figure 3.
Idiograms of chromosome 18 (a, b, e, f, i, j, o, p), representative G-banded karyotypes (c, d, g, h, k, l, q, r) and FISH results with the use of Cytocell 18pter (green) and 18qter (red) probes (m, n, s, t).
Chromosomal investigation on amniocytes from Case 2 (Figure 1, IV-14) showed a dup(18q)/del(18p) in one chromosome 18 and a dup(18p)/del(18q) in the other. After microsatellite and SNP array analysis (Table 1), the karyotype of the fetus was described as 46,XX,rec(18)dup(p)inv(18)(p11.2q21.3)mat,rec(18)dup(q)inv(18)(p11.2q21.3)pat.
Table 1. Summary of the microsatellite and SNP array data obtained for the siblings and their parents.
| III-16 father | III-21 mother | IV-13 son | IV-14 daughter | |||||
|---|---|---|---|---|---|---|---|---|
| Chromosome 18 markersa /region | MS | SNP | MS | SNP | MS | SNP | MSb | SNP |
| 18pter (0 Mb) | — | No copy number abnormalities | — | No copy number abnormalities | — | Deletion of the maternal allele | — | Maternal (h)UPD, no indication for isodisomy |
| D18S498 (p11.32, 0.934 Mb) | b, c | a, b | c: mat. del. | a, b (mat hUPD) | ||||
| D18S481 (p11.31, 3.05 Mb) | a, c | b, c | a: mat. del. | b, c (mat hUPD) | ||||
| D18S63 (p11.31, 3.43 Mb) | a, d | b, c | a: mat. del. | b, c: mat hUPD | ||||
| D18S54 (p11.31, 3.65 Mb) | b, b | a, c | b: mat. del. | a, c: mat hUPD | ||||
| 18p11.31 (4.700 Mb) | — | — | — | — | ||||
| p11.31-centromere-q21.33 | — | — | — | Biparental inheritance | — | Biparental inheritance | ||
| 18q21.33 (59.275 Mb) | — | — | — | Duplication of the maternal allele | — | Paternal (h)UPD, no indication for isodisomy | ||
| D18S386 (q22.1, 63.94 Mb) | a, b | a, b | a, b, b: dup. | a, b (pat hUPD) | ||||
| D18S390 (q22.3, 67.53 Mb) | a, b | a, a | a, a | a, b (pat hUPD) | ||||
| D18S1009 (q23, 72.19 Mb) | c, d | a, b | b, b, c: mat. dup. | c, d: pat hUPD | ||||
| D18S844 (q23, 72.48 Mb) | a, c | b, c | b, b, c: mat. dup. | a, c (pat hUPD) | ||||
| D18S554 (q23, 73.17 Mb) | a, b | a, a | a, a | a, b (pat hUPD) | ||||
| 18q23 (74.133 Mb) | — | — | — | — | ||||
| Paternal iUPD | ||||||||
| 18q23 (75.493 Mb) | — | — | — | — | ||||
Abbreviations: hUPD, heterodisomy; iUPD, isodisomy; mat. del., maternal origin of the deletion; mat. dup., maternal origin of the duplication; MS, results obtained by microsatellite analysis; SNP, results obtained by SNP arrays.
The following microsatellite markers were not informative: D18S59, D18S459, D18S1154, D18S391, D18S453, D18S819, D18S877, D18S57, D18S1162, D18S535, D18S978, D18S858, D18S61, D18S461, D18S70.
In brackets: inferred interpretation.
FISH studies
FISH analysis on the parents showed that both showed one chromosome 18 with normal signal distribution and another chromosome 18 with the signal for 18q subtelomeric region on the short arm of chromosome 18 and the signal for 18p subtelomeric region on the long arm.
FISH analysis on Case 1 showed two signals for the 18q subtelomeric region on one chromosome 18. The other chromosome 18 showed one signal for the 18p subtelomeric region and one signal for the 18q subtelomeric region (Figure 3).
FISH analysis on fetal amniocytes (Case 2) using 18q and 18p subtelomeric Cytocell probes (Cytocell Ltd, Cambridge, UK) showed two signals for the 18p subtelomeric region on one chromosome 18 and two signals for the 18q subtelomeric region on the other chromosome 18 (Figure 3).
Molecular investigation
DNA was extracted from peripheral blood samples of the siblings and their parents (ie, III-15, III-16, IV-13 and IV-14) using conventional techniques.
MLPA analysis of the telomeres (MLPA kit P036, MRC-Holland, Amsterdam, The Netherlands) confirmed the cytogenetic and FISH analysis, showing a deletion at 18pter and a duplication at 18qter for Case 1 (IV-13) (data not shown).
Microsatellite investigations were carried out using standard procedures. Affymetrix SNP_6 arrays (Affymetrix, Santa Clara, CA, USA) were investigated as follows: 500-ng genomic DNA was used for library preparation and PCR amplification according to the manufacturer's instructions. Hybridization of the biotinylated samples was carried out at 50 °C for 16 h. The chips were scanned after signal amplification using streptavidin R-phycoerythrin (SAPE, Invitrogen AG, Basel, Switzerland) and a biotinylated anti-streptavidin antibody (Vector Laboratories Ltd, Peterborough, UK), followed by a second SAPE staining. Quality control and the analysis of raw data were carried out with the Affymetrix Genotyping Console 3.01. Uniparental inheritance was analyzed using the online tool SNPtrio (http://pevsnerlab.kennedykrieger.org/SNPtrio.htm).20
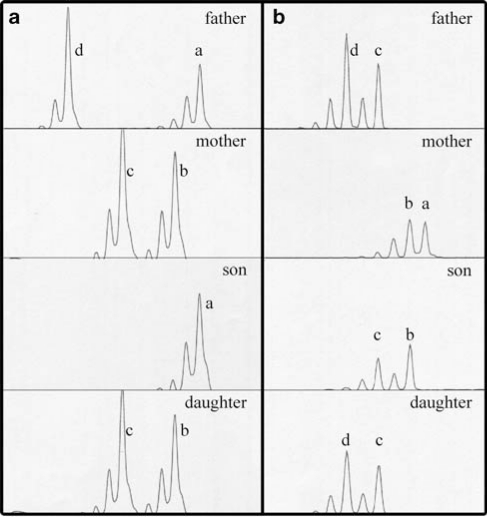
The results obtained by these investigations are summarized in Table 1 and representative examples of the microsatellite analysis are shown in Figure 4. The investigation led to a slight reassignment of the short arm breakpoint (from p11.2 to p11.31) and a more precise determination of the long arm breakpoint (18q21.33). Briefly, Case 1 (IV-13) showed a 4.7-Mb deletion at 18pter-p11.31 of maternal origin; the same region showed a maternal heterodisomy in Case 2 (IV-14). The duplication of ∼18.8 Mb at 18q21.33-18qter (59.275 Mb–18qter) was shown to be of maternal origin in Case 1. The same region showed paternal UPD in Case 2, and was mostly heterodisomic with a 1.36-Mb stretch of isodisomy at 18q23.
Figure 4.
Representative examples of microsatellite analysis. (a) Results for marker D18S63 mapping to 18p11.31. The son, IV-13, lacks a maternal allele, the daughter, IV-14, inherited both maternal alleles but no paternal allele (ie, matUPD18p). (b) Results for marker D18S1009 mapping to 18q23. A clear intensity difference can be seen in the son's alleles (duplication of the maternal allele ‘b', normalized peak area: 1.9 paternal allele: 1 maternal allele), the daughter inherited two paternal alleles but no maternal allele (ie, patUPD18q).
Discussion
This report describes a consanguineous family in which both parents have the same pericentric inversion of chromosome 18. Both their offspring are of interest. Child 1 (IV-13 in the pedigree, Figure 1) has a dup(18q)/del(18p) with very mild features, and borderline cognition, which is rare in patients with chromosomal deletions and duplications.
There have been reports of mild phenotypic findings in patients with dup(18q)/del(18p) with probably similar breakpoints.18, 19 Vermeulen et al19 described a family with pericentric inversion 18, inv(18)(p11.22q23), and three individuals heterozygous for the dup(18p)/del(18q) and dup(18q)/del(18p) recombinant. These individuals exhibited mild learning difficulty and personality disorders. The patient with dup(18q)/del(18p) had an even milder phenotype than the dup(18p)/del(18q) cases, which is in agreement with the observation that 18q deletions go along with more severe mental deficiency than 18p deletions.21 Similarly, mild mental retardation has also been observed occasionally in patients with ‘pure' duplication of 18q21.3-qter, although the majority of these cases show severe mental deficiency.22, 23
Mejia-Baltodano et al18 also reported a patient with minor dysmorphic features, small stature and obesity. Learning difficulty was noticed only after the first year of primary school and a cognitive delay of 2 years was recorded by psychometric testing at 8 years. On the basis of their own observation and that of Mejia-Baltodano, Vermeulen suggested that the paucity of clinical findings in dup(18q)/del(18p) patients could be explained by the fact that the 18pter region contains a lower than average density of genes24 or at least a low number of candidate genes affecting development, maintenance and/or function of the central nervous system. Other hypotheses proposed to explain that the lack of clinical consequences in case of large 18pter deletions is because of the presence of copies of the gene or other genes with similar function located elsewhere in the genome, haplosufficiency of the majority of genes and the possibility of inactivation of the chromosome region as a result of genomic imprinting, so that the deletion of that segment is harmless.25, 26
Case 2 (IV-14 in the pedigree, Figure 1) is of considerable interest in that it is, to the best of our knowledge, the first reported case of UPD 18, although ‘only' segmental. The lack of reports of UPD for chromosome 18 is in contrast to maternal and paternal UPD cases for the chromosomes involved in the two other frequent viable autosomal trisomies, namely 13 and 21.21 Thus, it could be assumed that at least maternal UPD 18 (arising though trisomy rescue events) could be a lethal condition. Interestingly, Case 2 shows that paternal UPD for the distal half of the long arm of chromosome 18 (18q21.33-qter) has no apparent phenotypic effect; this of course does not imply that a maternal UPD of the same segment is also harmless. The segment of 18p showing maternal UPD in Case 2 is very small and seems to be of minor importance for the phenotype, as seen from cases with simple deletions.21 Thus, there seems to be no imbalanced expression of possible imprinted genes having an impact on the phenotype within the above-mentioned segments, at least if manifested up to the age of our proposita (Case 2). Nevertheless, further follow-up including psychometric and personality testing and confirmation from other patients is necessary to confirm the lack of adverse consequences of paternal UPD of the segment 18q21.33-qter.
Another important matter for this family would be genetic counseling issues. Case 2 (IV-14) will not have chromosomally balanced offspring, with three extremely unlikely exceptions: first, that maternal UPD18 would occur (such an offspring would have maternal, and not paternal, UPD for the distal long arm segment of chromosome 18); second, and even more unlikely, if the partner would have the same aberration or that of her brother (Case 1, IV-13), and the offspring would receive der(18)(dup)(q) from the father and der(18)(dup)(p) from her, hence would again have the same chromosome aberration, but this time with the origin of the UPD similar to case 2: maternal for 18pter-p11.31 and paternal for 18q21.33-qter; and finally the possibility of recombination between the two recombinant chromosomes (within the inversion breakpoints) giving rise to normal chromosome 18.
Therefore, Case 2 (IV-14) will have a very high risk of transmitting either dup(18p)/del(18q) or dup(18q)/del(18p) to her offspring if no recombination takes place. There is not enough evidence to predict that if the offspring has dup(18q)/del(18p) the clinical findings will be as mild as those of Case 1 (IV-13); however, as mentioned above, an offspring with del(18q)/dup(18p) would most likely be much more severely affected.
Finally, in case of strong inbreeding in the population from where the family stems, it would theoretically be possible that a ‘new' species could emerge with homozygosity for the balanced inv(18), especially if this chromosome constitution would provide an evolutionary or recreational advantage. From studies of karyotype evolution from the great apes to homo, it is well known that several pericentric inversions distinguish gorilla and chimpanzee from man.27, 28, 29
Acknowledgments
The financial support by the Swiss National Foundation, grant no 320030-113635 to AS is greatly acknowledged.
The authors declare no conflict of interest.
References
- Ledbetter DH, Engel E. Uniparental disomy in humans: development of an imprinting map and its implications for prenatal diagnosis. Hum Mol Genet. 1995;4:1757–1764. doi: 10.1093/hmg/4.suppl_1.1757. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, McCaskill C, Adkins K, Hassold TJ. Systematic search for uniparental disomy in early fetal losses: the results and a review of the literature. Am J Med Genet. 1998;79:366–372. [PubMed] [Google Scholar]
- Kotzot D. Abnormal phenotypes in uniparental disomy (UPD): fundamental aspects and a critical review with bibliography of UPD other than 15. Am J Med Genet. 1999;82:266–274. [PubMed] [Google Scholar]
- Kotzot D. Complex and segmental uniparental disomy updated. J Med Genet. 2008;45:545–556. doi: 10.1136/jmg.2008.058016. [DOI] [PubMed] [Google Scholar]
- Kotzot D, Utermann G. Uniparental disomy (UPD) other than 15: phenotypes and bibliography updated. Am J Med Genet A. 2005;136:287–305. doi: 10.1002/ajmg.a.30483. [DOI] [PubMed] [Google Scholar]
- Andrews T, Gardiner AC, Boon AR. Recombinant chromosome 18 in two offspring of a chromosome 18 inversion heterozygote. Ann Genet. 1982;25:185–188. [PubMed] [Google Scholar]
- Asano T, Ikeuchi T, Shinohara T, Enokido H, Hashimoto K. Partial 18q trisomy and 18p monosomy resulting from a maternal pericentric inversion, inv(18)(p11.2q21.3) Jinrui Idengaku Zasshi. 1991;36:257–265. doi: 10.1007/BF01910544. [DOI] [PubMed] [Google Scholar]
- Ayukawa H, Tsukahara M, Fukuda M, Kondoh O. Recombinant chromosome 18 resulting from a maternal pericentric inversion. Am J Med Genet. 1994;50:323–325. doi: 10.1002/ajmg.1320500405. [DOI] [PubMed] [Google Scholar]
- Jacobs PA, Melville M, Ratcliffe S, Keay AJ, Syme J. A cytogenetic survey of 11,680 newborn infants. Ann Hum Genet. 1974;37:359–376. doi: 10.1111/j.1469-1809.1974.tb01843.x. [DOI] [PubMed] [Google Scholar]
- Kukolich MK, Althaus BW, Sears JW, Mankinen CB, Lewandowski RC. Abnormalities resulting from a familial pericentric inversion of chromosome 18. Clin Genet. 1978;14:98–104. doi: 10.1111/j.1399-0004.1978.tb02113.x. [DOI] [PubMed] [Google Scholar]
- Teyssier JR, Bajolle F. Duplication-deficiency of chromosome 18, resulting from recombination of a paternal pericentric inversion, with a note for genetic counselling. Hum Genet. 1980;53:195–200. doi: 10.1007/BF00273495. [DOI] [PubMed] [Google Scholar]
- Schinzel A. Autosomale chromosomenaberrationen. Arch Genet (Zur) 1979;52:1–4. [PubMed] [Google Scholar]
- Turleau C, de Grouchy J. Trisomy 18qter and trisomy mapping of chromosome 18. Clin Genet. 1977;12:361–371. doi: 10.1111/j.1399-0004.1977.tb00955.x. [DOI] [PubMed] [Google Scholar]
- Vianna-Morgante AM, Nozaki MJ, Ortega CC, Coates U, Yamamura Y. Partial monosomy and partial trisomy 18 in two offspring of carrier pericentric inversion 18. J Med Genet. 1976;13:366–370. doi: 10.1136/jmg.13.5.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigi V, Maraschio P, Bosi G, Guerini P, Fraccaro M. Risk for recombinants in pericentric inversion of the (p11 to q21) region of chromosome 18. Hum Genet. 1977;37:1–5. doi: 10.1007/BF00293765. [DOI] [PubMed] [Google Scholar]
- Martin AO, Simpson JL, Deddish RB, Elias S. Clinical implications of chromosomal inversions. A pericentric inversion in no.18 segregating in a family ascertained through an abnormal proband. Am J Perinatol. 1983;1:81–88. doi: 10.1055/s-2007-1000058. [DOI] [PubMed] [Google Scholar]
- Israels T, Hoovers J, Turpin HM, Wijburg FA, Hennekam RC. Partial deletion of 18p and partial duplication of 18q caused by a paternal pericentric inversion. Clin Genet. 1996;50:520–524. doi: 10.1111/j.1399-0004.1996.tb02726.x. [DOI] [PubMed] [Google Scholar]
- Mejia-Baltodano G, Babadilla L, Gonzalez RM, Barros-Nunez P. High recurrence of recombinants in a family with pericentric inversion of chromosome 18. Ann Genet. 1997;40:164–168. [PubMed] [Google Scholar]
- Vermeulen SJ, Speleman F, Vanransbeeck L, et al. Familial pericentric inversion of chromosome 18: behavioral abnormalities in patients heterozygous for either the dup(18p)/del(18q) or dup(18q)/del(18p) recombinant chromosome. Eur J Hum Genet. 2005;13:52–58. doi: 10.1038/sj.ejhg.5201281. [DOI] [PubMed] [Google Scholar]
- Ting JC, Roberson EDO, Miller ND, et al. Visualization of uniparental Inheritance, mendelian Inconsistencies, deletions, parent of origin effects in single nucleotide polymorphism trio data with SNPtrio. Hum Mutat. 2007;28:1225–1235. doi: 10.1002/humu.20583. [DOI] [PubMed] [Google Scholar]
- Schinzel A.A Catalogue of Unbalanced Chromosome Aberrations in ManSecond editionWalter de Gruyter & Co, Berlin and New York; 2001. vol 1, pp717–764. [Google Scholar]
- Brenk CH, Prott EC, Trost D, et al. Towards mapping phenotypical traits in 18p-syndrome by array-based comparative genomic hybridization and fluorescent in situ hybridization. Eur J Hum Genet. 2007;15:35–44. doi: 10.1038/sj.ejhg.5201718. [DOI] [PubMed] [Google Scholar]
- Ceccarini C, Sinibaldi L, Bernardini L, et al. Duplication of 18q21.31-q22.2. Am J Med Genet. 2007;143:343–348. doi: 10.1002/ajmg.a.31588. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Li L, Moore P, Ngo C, et al. Identification of a haplosufficient 3.6-Mb region in human chromosome 11q14.3 → q21. Cytogenet Genome Res. 2002;97:158–162. doi: 10.1159/000066612. [DOI] [PubMed] [Google Scholar]
- Barber JCK. Directly transmitted unbalanced chromosome abnormalities and euchromatic variants. J Med Genet. 2005;42:609–629. doi: 10.1136/jmg.2004.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goidts V, Szamalek JM, de Jong PJ, et al. Independent intrachromosomal recombination events underlie the pericentric inversions of chimpanzee and gorilla chromosomes homologous to human chromosome 16. Genome Res. 2005;15:1232–1242. doi: 10.1101/gr.3732505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Sandig C, Chuzhanova N, et al. Breakpoint analysis of the pericentric inversion distinguishing human chromosome 4 from the homologous chromosome in the chimpanzee (Pan troglodytes) Hum Mutat. 2005;25:45–55. doi: 10.1002/humu.20116. [DOI] [PubMed] [Google Scholar]
- Engel E. A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am J Med Genet. 1980;6:137–143. doi: 10.1002/ajmg.1320060207. [DOI] [PubMed] [Google Scholar]