To the editor:
Adhesion complexes in cells growing on planar substrates have been studied for over three decades. From these studies, several classes of adhesions have been described based on size, location, morphology, dynamics or molecular composition, and their dual role as signalling centres and linkages that connect the extracellular matrix (ECM) with the cytoskeleton has also emerged1. In contrast to this large and growing understanding of adhesion on two-dimensional (2D) substrates, little is known about the adhesions formed during three-dimensional (3D) growth of cells, including whether they even exist. Immunostaining and microscopy of fixed samples2–8 and dynamic imaging at low spatiotemporal resolution9 have shown the presence of large, elongated adhesions on cells in various 3D model systems. However, attempts to visualize adhesions in living cells growing in 3D with resolution similar to that routinely used in 2D have not been successful10. This has led to the recent conclusion that the adhesions defined and characterized in 2D cultures either do not exist in cells in 3D, or are too small or short-lived to be observed11.
One major limitation of imaging live cells in 3D culture is background fluorescence caused by the overexpression of genetically encoded, fluorescently tagged proteins. Having saturated all available association sites, excess fusion proteins accumulate in the cytoplasm resulting in diffuse background fluorescence. In cells on 2D substrates, this cytoplasmic background is less detrimental because of the thinness of the lamellae in migrating cells. Moreover, background fluorescence can be reduced in 2D studies by using TIRF (total internal reflection fluorescence) microscopy, which excludes fluorescence above approximately 100 nm from the substrate. In contrast, protrusions formed on 3D matrices may be thicker12 and must be visualized using wide-field, confocal or multi-photon microscopy, which section a minimum thickness of 500–800 nm (ref. 13). Therefore, overexpression of an adhesion-specific fluorescently tagged protein can be more detrimental to imaging in 3D than in 2D and result in diffuse cytoplasmic fluorescence that masks the signal of molecules localized to adhesions.
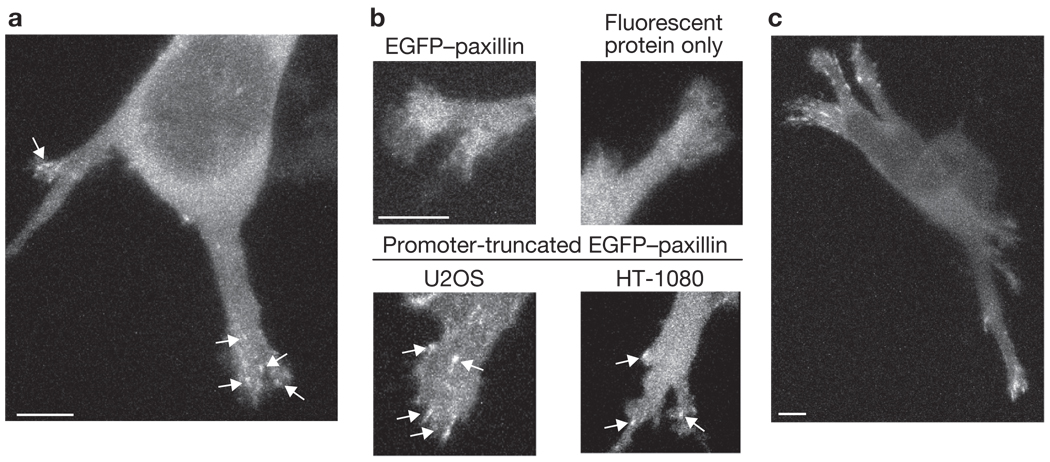
To address this issue, we used an EGFP (enhanced green fluorescent protein)–paxillin construct under the control of a truncated CMV (cytomegalovirus) promoter that was originally developed to express GFP–β-actin at very low levels14. The truncated CMV promoter reduces expression from the plasmid and therefore results in lower protein levels than those achievable by simply minimizing plasmid copy number. U2OS osteosarcoma cells were transfected with EGFP–paxillin in this vector, seeded within fibrillar collagen gels, and imaged with a laser scanning confocal microscope between 3–5 h after seeding. The collagen fibres were simultaneously imaged in reflectance mode. All images were taken several millimetres from the lateral edges of the gels. The cells were mostly multi-polar with three or more protrusions of variable size and shape that extended out from the cell body, spanned multiple focal planes, and exhibited numerous adhesions (Fig. 1a and Supplementary Information, Fig. S1a–f). Adhesions were rarely observed in areas more proximal to the cell body; however, as these areas of the cell were generally much thicker than the distal ends of the protrusion, and thus had higher cytoplasmic background fluorescence, we cannot exclude their existence. We observed adhesions in cells at depths up to the limit of the working distance of our objective (approximately 350 µm). Regardless of depth, at least 50% of the cells with a detectable level of the low-expressing EGFP–paxillin exhibited adhesions (Supplementary Information, Fig. S1g, m). Cells in which adhesions were not visible tended to have fluorescence intensities that were either relatively high (high background) or relatively low (low overall signal; Supplementary Information, Fig. S1h, n).
Figure 1.
Cell-matrix adhesions are detected in 3D collagen gels. (a) Z-projection of a multi-polar U2OS cell expressing EGFP–paxillin under the control of a truncated promoter, with adhesions on the distal sections of the protrusions (arrows). See Supplementary Information, Fig. S1a–f for more images. (b) Z-projection images of protrusions from cells expressing the indicated construct. The cell expressing a low level of normal EGFP–paxillin (upper left) has a diffuse cytoplasmic fluorescence, similar in appearance to the cell expressing the fluorescent protein TagRFP-T alone (upper-right). Adhesions (arrows) are detectable in U2OS cells (bottom left) and HT-1080 cells (bottom right) transfected with a plasmid encoding promoter-truncated EGFP–paxillin, which expresses at a lower level than EGFP–paxillin and decreases background cytoplasmic fluorescence. (c) Z-projection of a U2OS cell expressing promoter-truncated EGFP–vinculin. Scale bars, 5 µm.
High-spatial-resolution imaging of the protrusions revealed distinct adhesions (Fig. 1b, lower panels) with a median length of 1 µm (0.8–1.45 µm, 25th–75th percentiles; n = 72 adhesions). Adhesions were also observed in similar short-term experiments with HT-1080 cells (Fig. 1b; median 1.2 µm; n = 36), with both cell types expressing EGFP–vinculin from the same vector (Fig. 1c), and in overnight cultures of U2OS cells (data not shown). In contrast, U2OS cells transfected with low levels of EGFP–paxillin expressed under the full-length CMV promoter, or even with high levels of the truncated promoter plasmid, had a diffuse cytoplasmic fluorescence similar to that of a fluorescent protein expressed alone (Fig. 1b, upper panels). Finally, we observed adhesions in HT-1080 cells cultured overnight in rat-tail collagen gels (Supplementary Information, Fig. S1i–n).
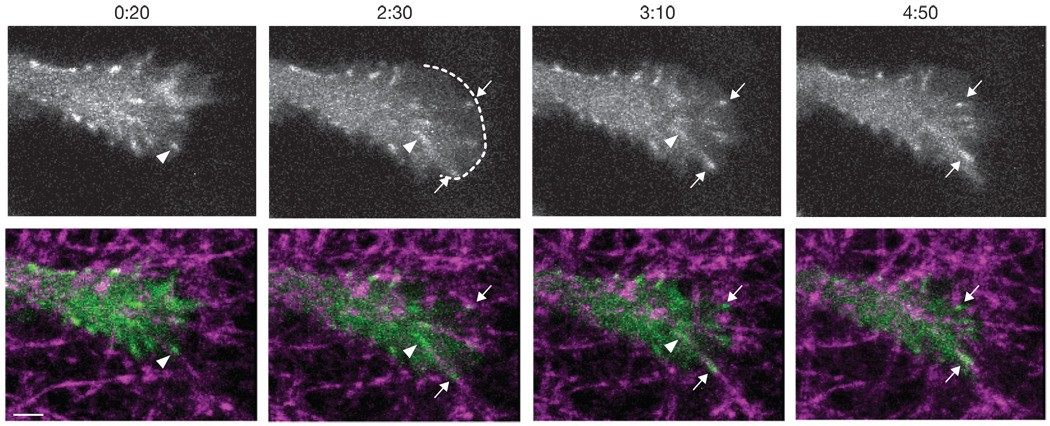
To image adhesion formation, growth, and disassembly, we acquired time-lapse Z-stacks of protrusions from U2OS cells cultured 3–5 h as in Figure 1. As highly dynamic nascent adhesions form and disassemble rapidly with an average lifetime of about 1 min15, we imaged with a 10 s time-interval to be sure of capturing short-lived adhesions. Figure 2 shows frames from a representative time-lapse movie of the protrusion of a U2OS cell transfected with a plasmid encoding the promoter-truncated EGFP–paxillin (Supplementary Information, Video S1). The protrusion undergoes cycles of extension and adhesion formation followed by rearward adhesion movement and matrix fibre deformation. Similar observations were made in six independent experiments and with HT-1080 cells (Supplementary Information, Video S2).
Figure 2.
Dynamics of cell-matrix adhesions in 3D culture. Frames are taken from Supplementary Information, Video S1. The top row shows Z-projections of a protrusion end from a U2OS cell that was transfected with a plasmid encoding promoter-truncated EGFP–paxillin. The bottom row shows the image from the top row (green) overlayed with a reflectance image of the collagen fibres (magenta). Time indicates the min:s since the beginning of the movie. At 0:20, a small adhesion (arrowhead) moves rearward, pulling a collagen fibre. At 2:30, a new protrusion (boundary shown by dotted line) pauses, two new adhesions form at the leading edge (arrows), and the earlier adhesion (arrowhead) has elongated while travelling rearward. At 3:10, the two new adhesions (arrows) continue to grow and translocate with the attached collagen fibres. By 4:50, the new adhesions have continued to grow and move rearward while the early adhesion (arrowhead in previous panels) is no longer visible. Scale bar,2 µm
In general, new adhesions formed along collagen fibres at the leading edge of protrusions and travelled rearward, causing fibre deformation. The adhesions had an initial diameter of 0.4 ± 0.07 µm (mean ± s.d.), placing them in the range of focal complexes, but larger than diffraction-limited nascent adhesions1,15. Over the course of the movies, most translocating adhesions grew in intensity and length; adhesions that did not translocate were stable, apparently maintaining isometric force on the matrix. Treatment with a combination of the Rho kinase (ROCK) inhibitor Y-27632 and the myosin light chain kinase (MLCK) inhibitor ML-7 decreased adhesion size similar to their effect in 2D (ref. 1).
Our observations demonstrate that cell-matrix adhesions can form in 3D collagen matrices, show that their dynamics can be studied in living cells, and support previous observations by immunostaining and microscopy in collagen2,4,5,8, fibrin7,8, and cell-derived3,8 3D ECMs. In addition to reducing background fluorescence, factors that affect adhesion size (and therefore signal intensity) may affect adhesion detectability. For example, microenvironment stiffness will affect cell contractility and thus adhesion size and composition2,3,16–18. Therefore, more-pliable regions of collagen gels may have smaller and consequently less readily detected adhesions; if so, it would point to pliability, in addition to dimensionality, as the determining factor. In addition, adhesions are difficult to visualize in certain cell types19. For example, leukocytes do not show prominent adhesions on ICAM-1 coated substrates20 and can migrate in an integrin-independent manner21. The effects of cell contractility on adhesion phenotype have been well-studied in 2D systems; future work will need to determine how these interactions occur in 3D. Reducing background fluorescence overcomes an initial barrier to adhesion visualization in 3D and allows studies of the influence of 3D topography, pliability, matrix protein composition, and cell contractility on adhesion phenotype.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturecellbiology/
Methods
Fluorescent protein constructs
The EGFP–paxillin plasmid has been described previously22. The EGFP–vinculin plasmid was a gift from S. Craig (Johns Hopkins, USA) and the CMV-promoter-truncated EGFP–β-actin was a gift from T. Mitchison (Harvard Medical School, USA)14. All constructs were in Clontech pEGFP vectors. The promoters of EGFP–actin and EGFP–paxillin were excised by digesting with AseI and NheI. The truncated CMV promoter from EGFP–actin was then ligated into the digested EGFP–paxillin plasmid. The same procedure was used to make promoter-truncated EGFP–vinculin, except ApaLI was used instead of AseI due to the presence of a duplicate restriction site within the vinculin gene. The TagRFP-T plasmid was a gift from R. Tsien (UCSD, USA)23.
Cell culture and transfection
U2OS cells were cultured in McCoy’s 5A medium supplemented with 10% fetal bovine serum (FBS) and were transfected with Lipofectamine 2000 (Invitrogen) or TransIT-2020 (Mirus Bio). HT-1080 cells were cultured in minimum essential medium (MEM) with Earl’s Salts, non-essential amino acids and 10% FBS, and were transfected with Lipofectamine (Invitrogen) or TransIT-2020. During experiments, the cells were cultured in CCM1 (a CO2-independent medium; Hyclone, Thermo Fisher Scientific) supplemented with 10% FBS. All cell-culture reagents were from Invitrogen unless otherwise indicated.
3D collagen gel preparation and cell seeding
Nutragen (Advanced BioMatrix) pepsin-extracted bovine collagen I solution was mixed with 0.1 M NaOH, 5 × CCM1 and FBS to attain a pH of 7.4 and an overall concentration of 1 × CCM1 and 10% FBS. Approximately 120 × 103 trypsinized cells were added to this solution and the volume was adjusted with CCM1 + 10% FBS to attain a final volume of 300 µl and collagen concentration of 2 mg ml−1. The solution was then pipetted into glass-bottomed dishes and incubated for 3 h at 37 °C. The collagen gels prepared with CCM1 buffer gelled at a slower rate than those prepared with PBS, resulting in many cells sinking to the glass surface before sufficient gelation occurred to encapsulate them. These cells were not imaged or analysed. Where indicated, HT-1080 cells were encapsulated in rat-tail collagen I (Invitrogen) matrices following the same protocol except that the cells were seeded at a density of 30 × 103 per gel and incubated overnight at 37 °C.
Imaging
Samples were imaged on an Olympus Fluoview 1000 laser scanning confocal microscope with a UPlanSApo ×60 (1.20 NA) water-immersion objective and a stage heater that maintained the sample at 37 °C, while the CCM1 buffer maintained the pH at 7.4. EGFP constructs were excited with the 488 nm laser line of an Argon ion laser; the TagRFP-T probe was excited with the 543 nm laser line of a Helium-Neon laser. Collagen fibres were imaged simultaneously in reflectance mode: exciting with the 488 nm laser and collecting the scattered (reflected) light at the same wavelength. Zoom and field-of-view dimensions were adjusted to give a pixel size of 80–90 nm. Z-slices were acquired every 0.33 µm (the axial resolution was determined empirically to be approximately 0.75 µm). To image the leading edge of a typical protrusion (and pad the top and bottom with additional slices to account for Z-drift over the course of the experiment) 10–20 Z-slices were necessary. The shortest possible pixel dwell time (2 µs per pixel) was used. A smaller sub-region of the field-of-view was selected so that one entire Z-stack could be completed within 10 s. Z-stacks were acquired every 10 s for a total of 5 minutes. Laser power was adjusted to minimize photobleaching. Cells selected for imaging were not near or at the glass surface (except when acquiring data for Supplementary Information, Fig. S1g, m) and were not in close proximity to other cells.
For the data shown in Supplementary Information Fig. S1g–n, we performed experiments in which all fluorescent cells in a given area were imaged and the depth of their lowest (closest to the glass) point was recorded. The imaging area was several millimetres away from the lateral edges of the gel and covered a depth ranging from the glass surface (0 µm) to the limit of our ×60 water-immersion objective (approximately 350 µm). Images were given generic names and the depth data were kept in a separate file so that the subsequent analyses could be performed blind. Two separate experimental conditions were used: 1) U2OS cells in 2 mg ml−1 bovine collagen gels cultured for 3–5 h, and 2) HT-1080 cells in 2 mg ml‒1 rat-tail collagen gels cultured for 18–24 h. Three independent experiments were performed for each condition. Excitation and acquisition parameters were kept constant to enable the comparison of protrusion background intensities between experiments.
Image possessing and analysis
Images and videos were produced with FV10-ASW Fluoview software (Olympus), MATLAB (MathWorks), and ImageJ (http://rsb.info.nih.gov/ij/) and were not modified beyond the standard adjustment of intensity levels. Adhesion sizes were measured in MATLAB from videos of cells transfected with plasmid encoding promoter-truncated EGFP–paxillin. First, an image of an adhesion (often a maximum Z-projection of two or more Z-slices) was median-filtered using a sliding-block filter with a radius of 10 pixels. This image was then subtracted from the original image to reduce diffuse background signal24. Small, punctate adhesions were fit to a 2D Gaussian function and the full-width-at-half-maximum value was taken as the diameter. Elongated adhesions were measured by thresholding the image and using an automated ellipse-fitting function.
For the data shown in Supplementary Information, Fig. S1g, h, m, n, the number of adhesions in each image was counted and the mean fluorescence intensity of a region of each protrusion (drawn so as to exclude any visible adhesions) was measured. Intensity measurements were performed with the FV10-ASW Fluoview software (Olympus) and were corrected for additional background noise by subtracting the mean intensity of a region drawn outside of the cell. All measurements were performed blind with respect to the depth data (collected during acquisition; see above) and intensity measurements were performed after counting the adhesions in order to prevent biased analyses. The bin ranges for the histograms in Supplementary Information, Fig. S1 were selected based on percentiles (that is, to have close to the same number of data points within each bin) to avoid biased grouping.
Supplementary Material
ACKNOWLEDGEMENTS
We thank C. Choi, M. Vicente-Manzanares, R. Tilghman, and E. Gratton for helpful discussions and technical assistance. The research was supported by the Cell Migration Consortium (U54 GM064346) and GM23244. K.E.K. is supported by a Cancer Training Grant at the University of Virginia Cancer Center (T32 CA009109-34).
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website
AUTHOR CONTRIBUTIONS
K.E.K. performed the experiments and analysed the data. K.E.K. and A.R.H. designed the experiments and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Geiger B, Spatz JP, Bershadsky AD. Nat. Rev. Mol. Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 2.Tamariz E, Grinnell F. Mol. Biol. Cell. 2002;13:3915–3929. doi: 10.1091/mbc.E02-05-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cukierman E, Pankov R, Stevens DR, Yamada KM. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 4.Wolf K, et al. J. Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S, et al. FASEB J. 2002;17:97–99. [Google Scholar]
- 6.Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. J. Cell Biol. 2003;163:583–595. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X, et al. Genes Dev. 2008;22:1231–1243. doi: 10.1101/gad.1643308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hakkinen KM, Harunaga JS, Doyle AD, Yamada KM. Tissue Eng. Part A. doi: 10.1089/ten.tea.2010.0273. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petroll MW, Ma L. Cell Motil. Cytoskeleton. 2003;55:254–264. doi: 10.1002/cm.10126. [DOI] [PubMed] [Google Scholar]
- 10.Knight B, et al. Curr. Biol. 2000;10:576–585. doi: 10.1016/s0960-9822(00)00486-3. [DOI] [PubMed] [Google Scholar]
- 11.Fraley SI, et al. Nat. Cell Biol. 2010;12:598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedl P, Brocker EB. Cell. Mol. Life Sci. 2000;57:41–64. doi: 10.1007/s000180050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hell SW. Nat. Biotechnol. 2003;21:1347–1355. doi: 10.1038/nbt895. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe N, Mitchison TJ. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- 15.Choi CK, et al. Nat. Cell. Biol. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choquet D, Felsenfeld DP, Sheetz MP. Cell. 1997;88:39. doi: 10.1016/s0092-8674(00)81856-5. [DOI] [PubMed] [Google Scholar]
- 17.Pelham RJ, Wang Y-L. Proc. Natl. Acad. Sci. USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyle AD, Wang FW, Matsumoto K, Yamada KM. J. Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedl P, Wolf K. J. Cell Biol. 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith A, et al. J. Cell Biol. 2005;170:141–151. doi: 10.1083/jcb.200412032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lammermann T, et al. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 22.Laukaitis CM, Webb DJ, Donais K, Horwitz AF. J. Cell Biol. 2001;153:1427–1440. doi: 10.1083/jcb.153.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaner NC, et al. Nat. Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zamir E, et al. J. Cell Sci. 1999;112:1655–1669. doi: 10.1242/jcs.112.11.1655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.