SUMMARY
Mammalian lipid homeostasis requires proteolytic activation of membrane-bound sterol regulatory element binding protein (SREBP) transcription factors through sequential action of the Golgi Site-1 and Site-2 proteases. Here, we report that while SREBP function is conserved in fungi, fission yeast employs a different mechanism for SREBP cleavage. Using genetics and biochemistry, we identified four genes defective for SREBP cleavage, dsc1–4, encoding components of a transmembrane Golgi E3 ligase complex with structural homology to the Hrd1 E3 ligase complex involved in endoplasmic reticulum-associated degradation. The Dsc complex binds SREBP and cleavage requires components of the ubiquitin-proteasome pathway: the E2 conjugating enzyme Ubc4, the Dsc1 RING E3 ligase and the proteasome. dsc mutants display conserved aggravating genetic interactions with components of the multivesicular body pathway in fission yeast and budding yeast, which lacks SREBP. Together, these data suggest that the Golgi Dsc E3 ligase complex functions in a post-ER pathway for protein degradation.
INTRODUCTION
Mammalian cells control lipid homeostasis through the ER membrane bound, basic helix-loop-helix leucine zipper transcription factors called sterol regulatory element binding proteins (SREBPs) (Brown et al., 2000). SREBPs contain two transmembrane segments and have a membrane topology that places the N- and C-termini in the cytosol separated by a short ∼30 amino acid luminal loop. SREBP cleavage activating protein (Scap) binds the C-terminal domain of SREBP and forms a stable complex (Espenshade and Hughes, 2007). When membranes are replete with sterols, SREBP remains inactive through the ER retention of Scap. Upon depletion of cellular cholesterol, Scap escorts SREBP to the Golgi via COPII transport vesicles where the SREBP N-terminal transcription factor domain is liberated by the sequential action of two Golgi resident proteases. Site-1 protease, a subtilisin-like serine protease, cleaves in the SREBP luminal loop yielding an SREBP N-terminal domain tethered to the membrane through a single transmembrane segment (Brown et al., 2000). Subsequent intramembrane cleavage by the Site-2 protease, a zinc metalloprotease, releases the soluble SREBP N-terminal transcription factor that travels to the nucleus and upregulates transcription of genes required for lipid synthesis and uptake from the environment. Increased cholesterol supply feeds back to inhibit SREBP-Scap transport to the Golgi, thus maintaining homeostasis.
Fungi contain SREBPs that function as hypoxic transcription factors (Bien and Espenshade, 2010). The fission yeast Schizosaccharomyces pombe codes for two SREBP homologs, Sre1 and Sre2 (Hughes et al., 2005). Sre1 binds to yeast Scap, called Scp1, and the mechanism of sterol regulation is conserved. However, Sre1 functions beyond the control of lipid homeostasis and is a principal hypoxic transcription factor (Todd et al., 2006). Production of ergosterol, the fission yeast equivalent of cholesterol, is highly oxygen consumptive. Under low oxygen conditions, ergosterol biosynthesis is inhibited and Sre1 precursor is cleaved to produce the active N-terminal transcription factor Sre1N. Thus, ergosterol synthesis serves as an indirect measure of oxygen availability to the cell. In addition, oxygen acts independently to regulate the degradation of Sre1N (Hughes and Espenshade, 2008). Together these two mechanisms cooperate to regulate production of Sre1N, which is essential for low oxygen growth (Hughes et al., 2005; Todd et al., 2006). Sre2 lacks the C-terminal Scp1 binding domain and does not bind to Scp1 (Hughes et al., 2005). Although the Sre2 transcriptional program has not been determined, Sre2 is not regulated by sterols and is cleaved constitutively. How Sre1 and Sre2 are proteolytically activated is unknown.
The basidiomyceteous fungus Cryptococcus neoformans is a human opportunistic pathogen that contains a Site-2 protease required for proteolytic activation of C. neoformans Sre1 (Bien et al., 2009). However, sequence database searches failed to identify Site-2 protease homologs in fungal ascomycetes encoding SREBP, such as S. pombe and the pathogen Aspergillus fumigatus (Bien and Espenshade, 2010). To identify the Sre1 cleavage machinery, we screened the S. pombe non-essential haploid deletion collection for genes required to generate Sre1N. We identified four genes defective for SREBP cleavage, dsc1–4, which define the Golgi Dsc E3 ligase complex. We find that Sre1 and Sre2 cleavage requires the ubiquitin-proteasome system and our data suggest that the Dsc complex may serve a general function in post-ER protein degradation.
RESULTS
Sre1 cleavage occurs in the cytosol and requires dsc genes
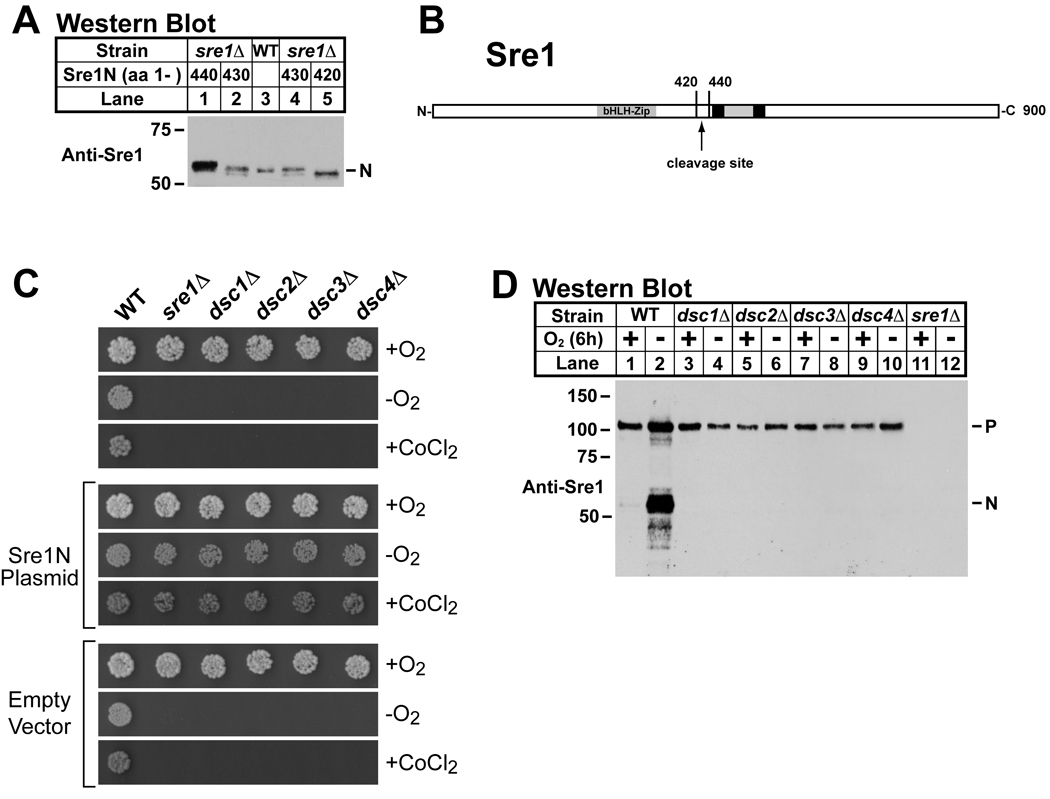
The absence of a Site-2 protease homolog in S. pombe suggested that Sre1 cleavage occurs through an alternative mechanism. We examined the site of Sre1 cleavage by comparing the mature nuclear form of Sre1 (Sre1N) to C-terminal truncations of known size. In mammalian cells, Site-2 protease cleaves SREBP at a site within the first transmembrane segment, which for Sre1 is predicted to begin at amino acid 445. Expression of truncated versions of Sre1N in sre1Δ cells revealed that endogenous Sre1N migrated as a protein of ∼429 amino acids, smaller than expected if cleavage occurred in the membrane (Figure 1A). These data suggest that Sre1N is cleaved at a cytosolic position through a mechanism different than mammalian SREBP.
Figure 1. Sre1 cytosolic cleavage requires dsc genes.
(A) Western blot probed with anti-Sre1 IgG of lysates from wild-type (lane 3) and sre1Δ cells expressing truncated versions of Sre1 (aa 1–440, aa 1–430, or aa 1–420, lanes 1, 2, 4 and 5). (B) Diagram of Sre1 where black boxes represent predicted transmembrane domains. The protein is inserted into the membrane in a hairpin fashion, placing both the N- and C-termini in the cytosol. Arrow denotes cleavage site. (C) Wild-type and mutant yeast (200 cells) containing no plasmid, a plasmid expressing Sre1N (aa 1 –440), or empty vector were grown on rich medium or rich medium containing cobalt chloride (CoCl2) in the presence of oxygen or on rich medium in the absence of oxygen. (D) Western blot probed with anti-Sre1 IgG of lysates from wild-type and indicated mutants grown for 6h in the presence or absence of oxygen. P and N denote Sre1 precursor and cleaved nuclear forms, respectively. See also Figure S1.
To identify genes required for Sre1 cleavage, we screened the S. pombe non-essential haploid deletion collection for mutants that phenocopy sre1Δ cells (Kim et al., 2010). We reasoned that cells lacking Sre1 cleavage machinery would not activate Sre1 and thus would also fail to grow under anaerobic conditions or in the presence of the hypoxia mimetic, CoCl2 (Hughes et al., 2005; Lee et al., 2007). From a collection of 2626 mutants, we identified 28 genes required for growth both in the absence of oxygen and in the presence of CoCl2. Secondary screening revealed that four mutants were fully rescued by expression of Sre1N under both growth conditions indicating that the defect lay upstream of Sre1N function (Figure 1C). Rescued dsc mutant strains expressed wild-type levels of Sre1N under low oxygen (Supplemental Figure S1). To test whether the deletion mutants had a defect in Sre1 proteolysis, we assayed Sre1 cleavage after growing cells at low oxygen for 6h (Figure 1D). Sre1 cleavage was robust under these conditions in wild-type cells, but each of the four mutants failed to generate Sre1N (Figure 1D, lanes 3–10). The presence of full-length Sre1 precursor in these mutants and the ability of exogenously expressed Sre1N to rescue the mutant growth phenotypes suggested that these genes function in Sre1 cleavage. Thus, we named these genes dsc1–4, defective for SREBP cleavage 1–4.
Dsc1 is the homolog of the Golgi E3 ligase Tul1
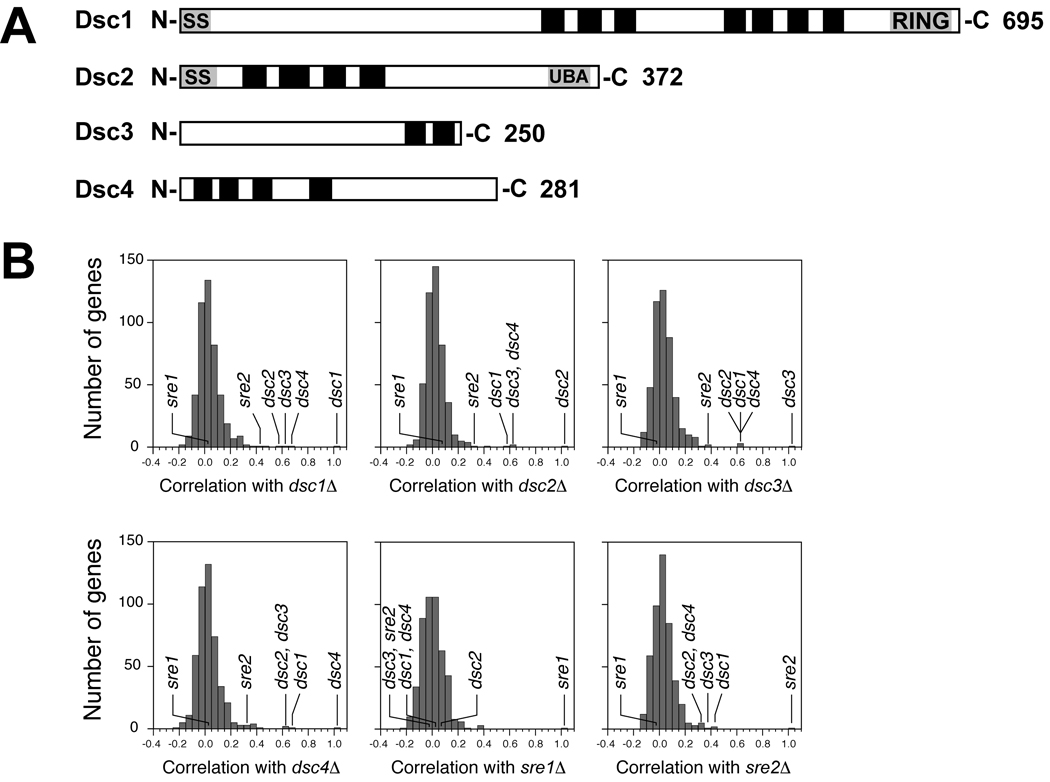
dsc1–4 were uncharacterized in S. pombe and correspond to SPBC947.10, SPAC1486.02c, SPAC20H4.02, and SPAC4D7.11. Bioinformatic analyses predicted that each Dsc protein contains multiple transmembrane segments and that Dsc1 and Dsc2 contain N-terminal signal sequences (Figure 2A). Notably, Dsc1 is the fission yeast homolog of the S. cerevisiae Golgi ubiquitin E3 ligase Tul1 and contains a RING domain at its C-terminus (Supplemental Figure S2)(Reggiori and Pelham, 2002). Tul1 is required to sort membrane proteins with polar transmembrane segments into multivesicular bodies for delivery to the yeast vacuole. Conserved domain database searches with Dsc2 identified a ubiquitin-associated (UBA) domain and sequence similarity to the Der1 family of ER proteins involved in ER associated degradation (Hartmann-Petersen et al., 2003; Hirsch et al., 2009). In addition, Psi-Blast analysis with Dsc3 revealed similarity to ubiquitin-like (UBL) domain containing proteins, such as Rad23 and Herp (Supplemental Figure S2)(Hirsch et al., 2009). Dsc4 is a conserved fungal protein with no recognizable functional domains.
Figure 2. E-MAP analysis reveals a conserved function for the Dsc proteins.
(A) Diagrams of Dsc proteins where black boxes represent predicted transmembrane domains. Predicted signal sequences and conserved domains are indicated. (B) Histograms of correlation coefficients calculated by comparing the genetic interactions of 471 query mutants mated against a library of 2,277 nonessential deletions. Histograms for dsc1Δ, dsc2Δ, dsc3Δ, dsc4Δ, sre1Δ and sre2Δ are shown and relevant genes are indicated. See also Figures S2 and S3.
To investigate the function of Dsc1–4 in fission yeast we performed quantitative, high throughput genetic screening using the E-MAP (Epistatic Miniarray Profile) approach (Roguev et al., 2007). Each dsc gene deletion was introduced into 2277 single gene deletion strains and the growth of the double mutant was quantified. Each screen yielded 2277 genetic interaction scores that constitute a genetic signature for that gene deletion (Supplemental Table 1). Strong correlation among signatures indicates that the genes function in a related process (Collins et al., 2007; Schuldiner et al., 2005). Cluster analysis was performed using a set of 471 gene signatures, including dsc1–4, sre1, and sre2 (Roguev et al., 2007). Figure 2B shows histograms plotting the correlation coefficients for the genes of interest and each of the 471 genes. The dsc genes were most strongly correlated with each other, indicating that these genes likely function in a related process (Supplemental Tables 2 and 3). In addition, the dsc genes were highly correlated with sre2. Finally, the dsc genes showed no correlation with sre1, possibly because the screens were performed in the presence of oxygen where sre1 is not required for growth.
E-MAP analysis also yields information regarding genetic interactions between individual gene deletions (Schuldiner et al., 2005). Gene ontology analysis revealed that genes showing aggravating genetic interactions with the dsc genes were highly enriched for components of the endosomal sorting complexes required for transport (ESCRT) pathway, which function to target proteins to the vacuole via the multivesicular body sorting pathway (Table 1) (Iwaki et al., 2007; Raiborg and Stenmark, 2009). Notably, the Dsc1 homolog in S. cerevisiae Tul1 has been shown to function in this pathway. To test whether the dsc genes are required for protein targeting to the vacuole via the multivesicular body pathway, we assayed vacuolar sorting of the model substrate Ub-GFP-Cps1 (Iwaki et al., 2007). Dsc deletion mutants properly sorted Ub-GFP-Cps1 to the vacuolar lumen, while cells lacking the ESCRT 0 component Sst4 were completely defective (Supplemental Figure S3). Similar aggravating genetic interactions have been observed between ESCRT pathway mutants and the S. cerevisiae dsc1–3 homologs: TUL1, YOL073C, and YOR223W (Supplemental Table 4)(Costanzo et al., 2010). Dsc4 has no apparent homolog in S. cerevisiae. Given the absence of a SREBP homolog in S. cerevisiae, these data collectively indicate that dsc1–4 function in a conserved process that extends beyond Sre1 cleavage.
Table 1.
GO Term Enrichment for E-MAPs
| Query Gene | GO Term | P Value | Frequency | Array Genes With Enriched GO Term |
|---|---|---|---|---|
| dsc1+ | endosome transport (GO:0016197) | 9.1E-06 | 8/56 |
did4+, SPAPB21F2.02, vps29+,SPCC162.06c, vps20+, sst4+, SPAC17G6.05c, sft2+ |
| ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway (GO:0043162) |
1.0E-04 | 5/56 |
did4+, SPCC162.06c, vps20+, sst4+ SPAC17G6.05c |
|
| dsc2+ | protein targeting to vacuole during ubiquitin-dependent protein catabolic process via the MVB pathway (GO:0043328) |
5.8E-05 | 4/30 | vps20+, did4+, sst4+, vps25+ |
| ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway (GO:0043162) |
4.0E-04 | 4/30 | vps20+, did4+, sst4+, vps25+ | |
| endosome transport via multivesicular body sorting pathway (GO:0032509) | 7.9E-04 | 4/30 | vps20+, did4+, sst4+, vps25+ | |
| late endosome to vacuole transport via multivesicular body sorting pathway (GO:0032511) |
7.9E-04 | 4/30 | vps20+, did4+, sst4+, vps25+ | |
| dsc3+ | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway (GO:0043162) |
2.3E-04 | 5/62 |
did4+, SPCC162.06c, vps20+, SPAC17G6.05c vps25+ |
| dsc4+ | protein targeting to vacuole during ubiquitin-dependent protein catabolic process via the MVB pathway (GO:0043328) |
5.4E-06 | 5/56 | did4+, vps20+, sst4+, SPAC17G6.05c, vps25+ |
| ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway (GO:0043162) |
1.1E-04 | 5/56 | did4+, vps20+, sst4+, SPAC17G6.05c, vps25+ | |
| endosome transport via multivesicular body sorting pathway (GO:0032509) | 2.9E-04 | 5/56 | did4+, vps20+, sst4+, SPAC17G6.05c, vps25+ | |
| late endosome to vacuole transport via multivesicular body sorting pathway (GO:0032511) |
2.9E-04 | 5/56 | did4+, vps20+, sst4+, SPAC17G6.05c, vps25+ | |
Array genes with interaction scores ≤ −5.0 were analyzed using AmiGO GO term enrichment software with default values (ver. 1.7; GO database release 10-04-2009). E-MAP array genes present in the GO database (2237 genes) served as the background set for analysis. GO process terms with p value < 0.001 are shown.
Dsc proteins form a stable complex
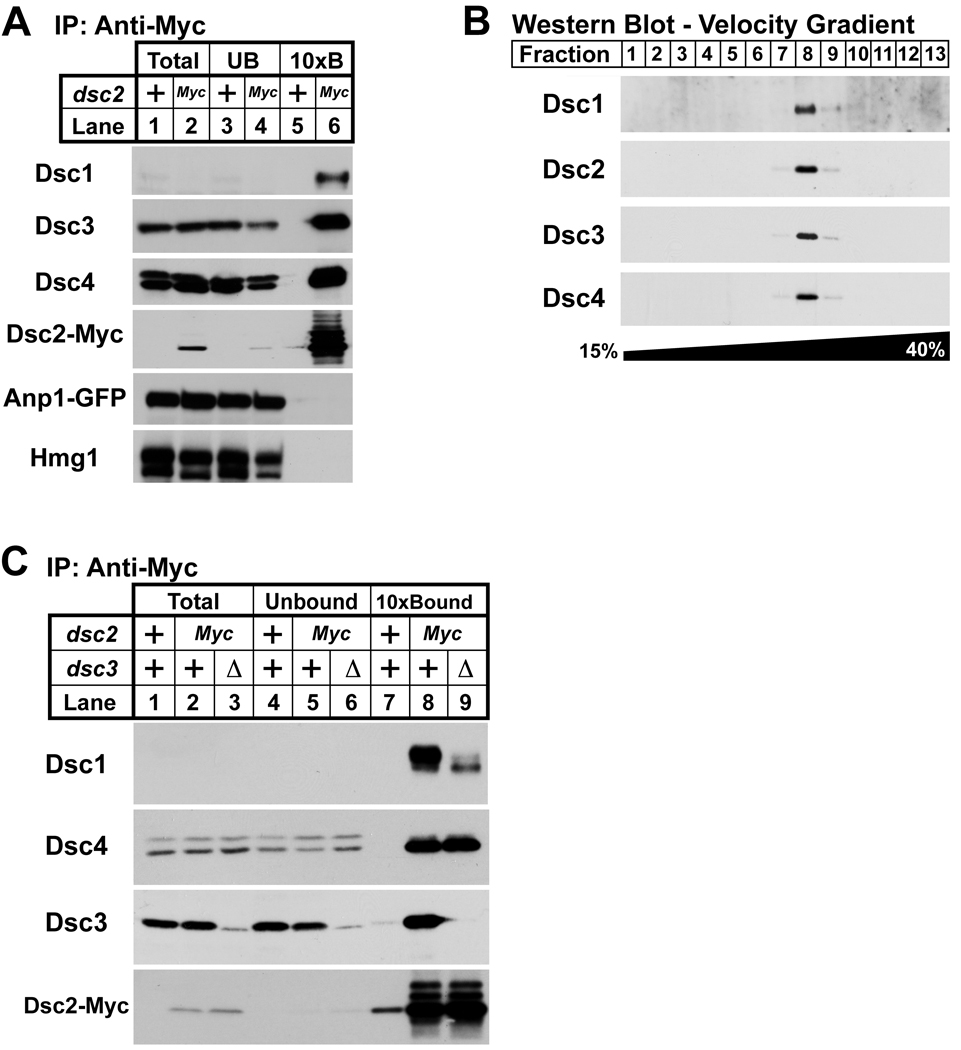
Genes showing highly correlated E-MAP profiles often code for proteins that are physically associated (Beltrao et al., 2010; Collins et al., 2007). To identify Dsc1-interacting proteins, we generated a functional gene fusion of dsc1 to a C-terminal tandem affinity purification (TAP) tag at the endogenous locus (Rigaut et al., 1999). Dsc1-TAP was purified from digitonin-solubilized cell extracts and copurifying proteins were identified by mass spectrometry analysis of the complex eluate. Both Dsc2 and Dsc3 copurified with Dsc1-TAP (Supplemental Table 5). To confirm these results, we performed co-immunoprecipitation using cells expressing a functional dsc2 tagged with 13 Myc epitopes at the endogenous locus. Dsc2-Myc was immunopurified from digitonin solubilized cell extracts, and we assayed for copurifying proteins using antibodies to Dsc1, Dsc3 and Dsc4, as well as the ER resident Hmg1 and the cis-Golgi resident Anp1 (Burg et al., 2008; Vjestica et al., 2008). Dsc2-Myc bound to Dsc1, Dsc3 and Dsc4, but not Hmg1 or Anp1 (Figure 3A, lane 6). To test whether Dsc1–4 form a discrete protein complex, we subjected the Dsc1-TAP eluate to velocity gradient sedimentation and analyzed the fractions for each Dsc protein. Dsc1–4 cofractionated on the gradient, indicating that Dsc1 forms a stable complex with each of the other Dsc proteins (Figure 3B). We refer to Dsc1–4 as the Dsc complex.
Figure 3. Dsc proteins form a complex.
(A) Digitonin solubilized extracts from anp1-GFP and dsc2-Myc anp1-GFP cells were prepared, and proteins associated with Dsc2-Myc were immunopurified with anti-myc IgG-9E10 monoclonal antibody. Equal amounts of total (lanes 1 and 2) and unbound fractions (lanes 3 and 4) along with 10× bound fractions (lanes 5 and 6) were immunoblotted using anti-Dsc1 IgG, anti-Dsc3 serum, anti-Dsc4 serum, rabbit anti-Myc IgG, anti-GFP and anti-Hmg1 IgG. (B) Eluate from tandem affinity purification using dsc1-TAP cells was subjected to velocity centrifugation on a 15%–40% sucrose gradient. Fractions were analyzed by immunoblot using anti-Dsc1 IgG, anti-Dsc2 serum, anti-Dsc3 serum, and anti-Dsc4 serum. (C) Digitonin solubilized extracts from anp1-GFP, dsc2-Myc anp1-GFP, and anp1-GFP dsc2-Myc dsc3Δ cells were prepared, and proteins associated with Dsc2-Myc were immunopurified using anti-myc IgG-9E10 monoclonal antibody. Equal amounts of total (lanes 1, 2 and 3) and unbound fractions (lanes 4, 5 and 6) along with 10× bound fractions (lanes 7, 8 and 9) were immunoblotted using anti-Dsc1 IgG, anti-Dsc4 serum, anti-Dsc3 serum, and rabbit anti-Myc IgG. For (A) and (C), + denotes wild-type allele.
Hrd1 is a multi-transmembrane domain containing RING E3 ligase that forms a complex with additional proteins to recognize and target misfolded ER proteins for proteasomal degradation through a process known as ER-associated degradation (ERAD)(Hampton and Garza, 2009; Vembar and Brodsky, 2008). As noted above, bioinformatic analyses revealed sequence similarities between Dsc1, Dsc2, and Dsc3 and the Hrd1 ligase complex components Hrd1, Der1 and Usa1 (Herp in mammalian cells). In S. cerevisiae, Hrd1 binding to Der1 requires Usa1 (Carvalho et al., 2006). To examine whether structural parallels exist between these two complexes, we assayed binding of Dsc2-Myc to Dsc1 in dsc3Δ cells. When Dsc2-Myc is purified from wild-type cells, Dsc1, Dsc3, and Dsc4 copurify (Figure 3C, lane 8). Deleting dsc3 decreased Dsc2-Myc binding to Dsc1, but had no effect on Dsc2-Myc and Dsc4 binding (Figure 3C, lanes 8–9). These results begin to define the structural organization of the Dsc complex, which shares properties of the Hrd1 ligase complex.
Sre2 cleavage requires the Dsc complex
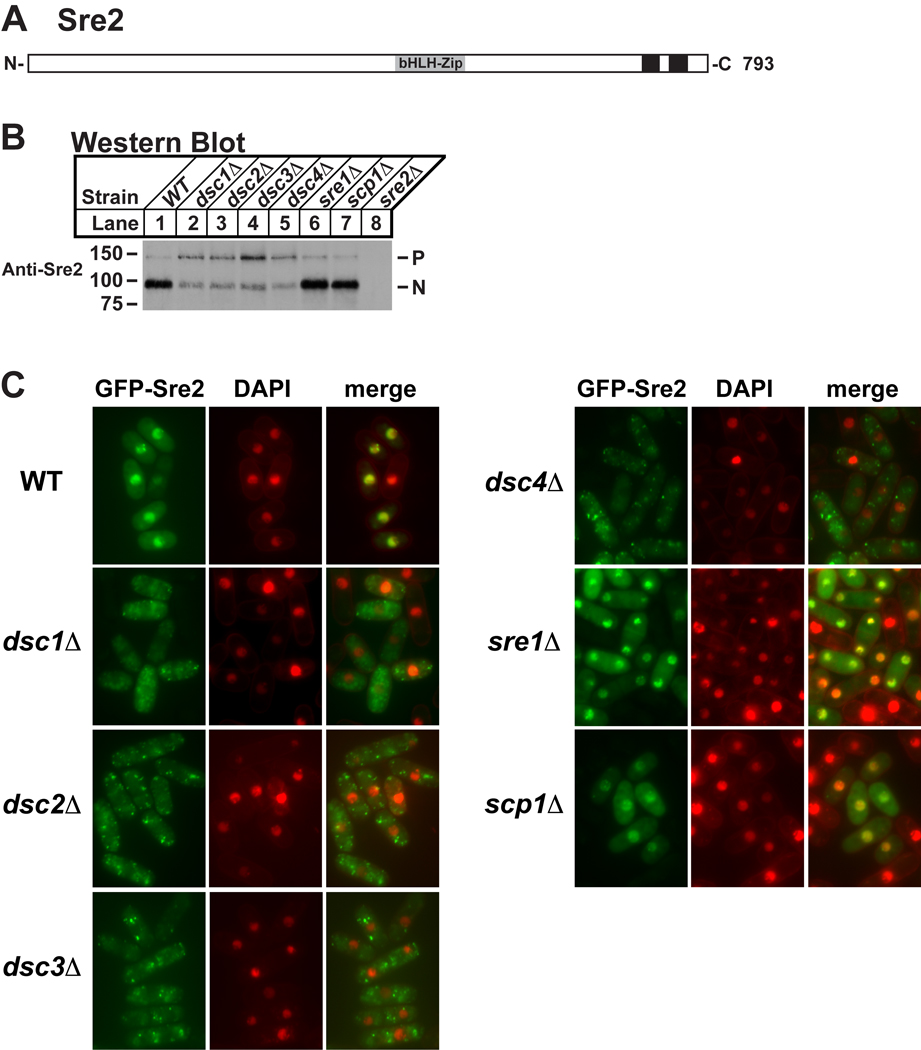
Sre2 is a second S. pombe homolog of mammalian SREBP that has a topology similar to Sre1, but lacks the Scp1 interacting C-terminus (Figure 4A) (Hughes et al., 2005). Consequently unlike Sre1, Sre2 is constitutively cleaved and is unresponsive to perturbations of the sterol biosynthetic pathway that regulate Sre1 cleavage. Based on the role of Dsc1–4 in Sre1 cleavage and the strong functional correlation between the dsc genes and sre2 (Figure 2B), we hypothesized that Sre2 may also require Dsc1–4 for cleavage activation. To facilitate discrimination of the Sre2 membrane-bound precursor from the cleaved N-terminal transcription factor domain, we fused GFP to the C-terminus of Sre2 expressed from the chromosome and assayed Sre2-GFP cleavage using antiserum directed to the Sre2 N-terminus. In wild-type cells in the presence of oxygen, Sre2-GFP is cleaved and the majority of Sre2 exists as the N-terminal form Sre2N with only a small amount present as the precursor form (Figure 4B, lane 1). Deletion of dsc1, dsc2, dsc3, or dsc4 inhibited Sre2-GFP cleavage (Figure 4B, lanes 2–5). Deletion of sre1 or scp1 had no impact on Sre2-GFP cleavage (Figure 4B, lanes 6 and 7).
Figure 4. Sre2 cleavage requires dsc genes.
(A) Diagram of Sre2 where black boxes represent predicted transmembrane domains. (B) Western blot probed with anti-Sre2 serum of lysates from wild-type and indicated deletion mutant cells. P and N denote Sre2-GFP precursor and cleaved nuclear forms, respectively. (C) Wild-type and indicated deletion mutant cells containing a plasmid expressing GFP-sre2 (pAH230) were grown in minimal medium lacking leucine and thiamine for 20h to induce expression after which cells were labeled with DAPI to stain DNA and imaged by fluorescence microscopy.
Although Sre2 cleavage decreased in dsc deletion strains, residual Sre2N was present. To address whether Sre2N was functional in dsc mutants, we expressed an N-terminal GFP fusion protein GFP-Sre2 from a plasmid and visualized its localization in the cell. In wild-type cells, GFP-Sre2N colocalized with nuclear DNA indicating that the Sre2 N-terminus translocates to the nucleus under normal conditions (Figure 4C). In dsc1Δ, dsc2Δ, dsc3Δ and dsc4Δ cells, GFP-Sre2 did not reach the nucleus and instead localized to puncta that are distinct from the nucleus (Figure 4C). Consistent with the cleavage studies on Sre2-GFP (Figure 4B), deletion of sre1 or scp1 had no impact on the localization of GFP-Sre2N. Together these data show that the Dsc complex is required for productive cleavage of Sre2, functions in the presence of oxygen, and that Dsc function is not limited to Sre1.
Dsc complex localizes to the Golgi
The Site-1 and Site-2 proteases that cleave SREBP reside in the Golgi apparatus and sterol-regulated cleavage is achieved by controlling access of SREBP to its proteases (Espenshade and Hughes, 2007). In sterol-replete cells, the SREBP-Scap complex is retained in the ER, unable to interact with the proteases, and SREBP is inactive. In sterol-depleted cells, Scap transports SREBP to the Golgi where SREBP is cleaved. This sterol-dependent and Scap-dependent regulation is conserved in fission yeast (Hughes et al., 2005). This conservation along with the knowledge that Dsc1 is the homolog of Golgi-localized Tul1 led us to confirm that the Dsc complex also localizes to the Golgi (Reggiori and Pelham, 2002).
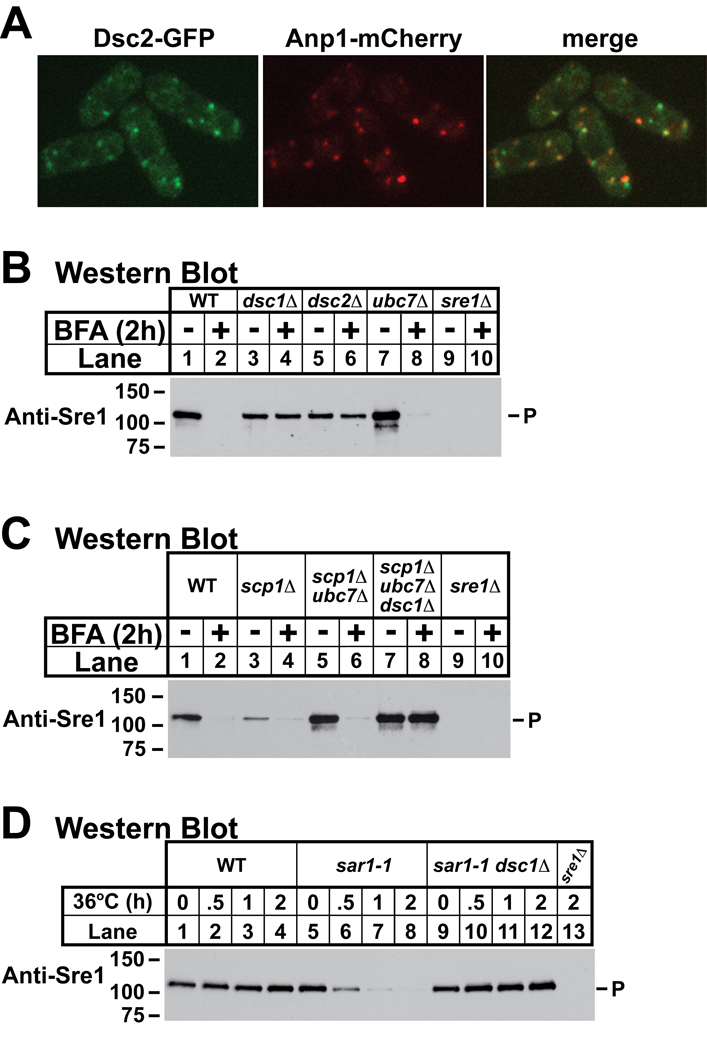
To determine the localization of the Dsc complex, we imaged cells expressing Dsc2 fused to six tandem copies of monomeric GFP using fluorescence microscopy. Dsc2–6xGFP was functional insomuch as this fusion rescued Sre1 cleavage, and Dsc2–6xGFP bound to the other Dsc proteins (data not shown). Dsc2–6xGFP localized to puncta in cells that largely colocalized with the cis-Golgi marker Anp1-mCherry (Figure 5A)(Vjestica et al., 2008). Not all Dsc2–6xGFP puncta overlapped with Anp1 puncta, suggesting that the Dsc complex may not be limited to cis-Golgi cisternae.
Figure 5. Dsc complex resides in the Golgi.
(A) dsc2–6xmGFP anp1-mCherry cells were cultured in the presence of oxygen and imaged by confocal fluorescence microscopy. (B) Western blot probed with anti-Sre1 IgG of lysates from wild-type and indicated mutant cells grown in the presence of oxygen and either brefeldin A (100 µg/ml) or vehicle (2% EtOH) for 2h at 30°C. (C) Western blot probed with anti-Sre1 IgG of lysates from wild-type and indicated mutant cells grown in the presence of oxygen and either brefeldin A (100 µg/ml) or vehicle (2% EtOH) for 2h at 30°C. (D) Western blot probed with anti-Sre1 IgG of lysates from wild-type and indicated mutant cells grown for a time course at 36°C in the presence of oxygen. In each figure, P denotes Sre1 precursor. See also Figures S4 and S5.
As an independent test of whether the Dsc complex resides in the Golgi, we treated cells with the drug brefeldin A (BFA), which induces fusion of the ER and Golgi compartments and results in content mixing between these two organelles (Lippincott-Schwartz et al., 1989). If the Dsc complex resides in the Golgi, treatment with BFA will bypass the requirement for ER-to-Golgi transport of Sre1 and result in unregulated cleavage of Sre1 in the presence of oxygen. We used an identical approach in mammalian cells to demonstrate that SREBP required Scap for transport to the Golgi (DeBose-Boyd et al., 1999). Treatment of cells with BFA induced SREBP cleavage both in the absence of Scap and in the presence of sterol.
Treatment of wild-type cells with BFA for two hours in the presence of oxygen caused Sre1 precursor to disappear, and this disappearance required each of the four dsc genes (Figure 5B, lanes 1–6 and data not shown). Interestingly, Sre1N did not accumulate in BFA-treated cells, perhaps due to the short half-life of Sre1N (∼7 min) in the presence of oxygen (Hughes and Espenshade, 2008). When not bound to Scp1, the fission yeast Scap homolog, Sre1 precursor is degraded by ERAD through the action of the E2 ubiquitin conjugating enzyme Ubc7 (Hughes et al., 2009). The BFA-dependent disappearance of Sre1 did not require this ERAD pathway as Sre1 precursor was absent from BFA-treated ubc7Δ cells (Figure 5B, lanes 7–8). As expected, treatment of wild-type and dsc1Δ cells with BFA resulted in ER localization of the cis-Golgi protein Anp1 (Supplemental Figure S4).
In mammalian cells, Scap is dispensable for SREBP cleavage when cells are treated with BFA, indicating that Scap is required for transport to the Golgi-localized proteases, but not for substrate recognition (DeBose-Boyd et al., 1999). Likewise, Scp1 is required for Sre1 cleavage (Hughes et al., 2005). To determine whether Scp1 is required for the Dsc-dependent cleavage of Sre1, we assayed Sre1 precursor in scp1Δ cells treated with BFA. However, Sre1 is unstable and degraded by ERAD in scp1Δ cells, so we also tested Sre1 cleavage in scp1Δ ubc7Δ cells (Hughes et al., 2009). BFA induced Sre1 cleavage in both scp1Δ and scp1Δ ubc7Δ cells, which had wild-type levels of Sre1 (Figure 5C, lanes 1–6). Importantly, this BFA-dependent cleavage required the Dsc complex (Figure 5C, lanes 7–8). Thus, BFA treatment induces Dsc-dependent Sre1 cleavage in the presence of oxygen and Dsc-dependent Sre1 cleavage does not require Scp1.
As a complementary genetic approach to the BFA studies, we utilized a strain carrying a temperature sensitive allele of sar1 to relocate Golgi proteins to the ER. Sar1 is a small GTPase that controls COPII vesicle formation at the ER (Antonny and Schekman, 2001). Inhibition of Sar1 blocks ER exit of cargo proteins and leads to mixing of the ER and Golgi compartments (Zaal et al., 1999). Consistent with this, growth of sar1-1 cells at the non-permissive temperature caused ER localization of the Golgi protein Anp1 within 30 minutes (Supplemental Figure S5). When grown at the non-permissive temperature Sre1 precursor is stable in wild-type cells, but disappears in a sar1-1 strain (Figure 5D, lanes 1–8). Sre1 cleavage in sar1-1 cells at the non-permissive temperature required the Dsc complex since Sre1 was stable in sar1-1 dsc1Δ cells (Figure 5D, lanes 9–12). These chemical and genetic experiments demonstrate that mixing of the ER and Golgi compartments results in Dsc-dependent cleavage of Sre1. Combined with the Dsc2–6xGFP localization studies, these data indicate that the Dsc complex resides in the Golgi.
Sre1 cleavage requires components of the ubiquitin-proteasome system
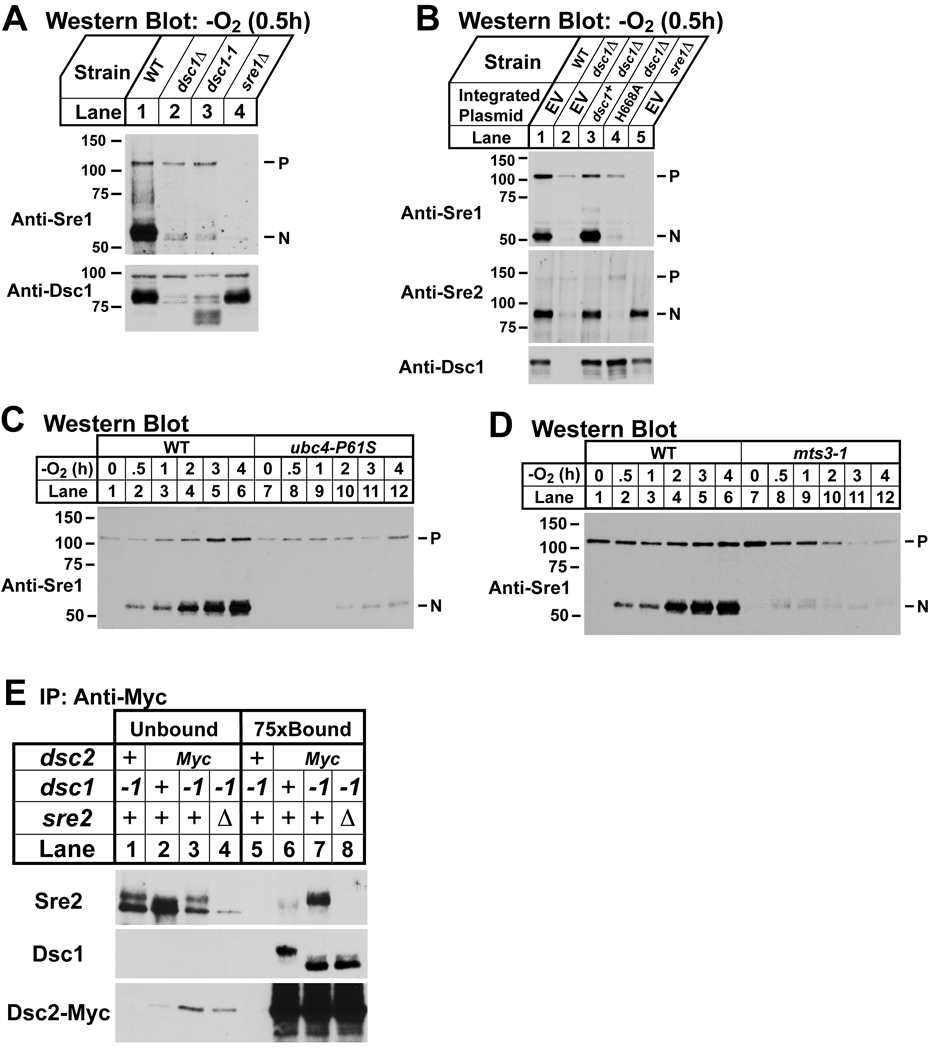
E3 ubiquitin ligases mediate transfer of ubiquitin from an E2 ubiquitin conjugating enzyme to a target substrate (Vembar and Brodsky, 2008). E3 ligases are subdivided into two classes, RING and HECT, based on sequence motifs in their functional domains and the method of ubiquitin transfer. Both Dsc1 and its S. cerevisiae homolog Tul1 contain a canonical RING domain that defines the RING E3 ligases. To test the requirement of the Dsc1 RING domain for Sre1 proteolysis, we assayed cleavage in dsc1–1 cells in which a premature stop codon truncates the last 23 amino acids of Dsc1, removing residues critical for RING function. While wild-type cells showed normal Sre1 cleavage under low oxygen, dsc1–1 and dsc1Δ cells failed to cleave Sre1, demonstrating that the RING domain is required for Sre1 cleavage (Figure 6A). We further tested the RING domain requirement by assaying function of Dsc1 mutated for an essential residue of the RING domain. sre2-GFP or sre2-GFP dsc1Δ cells carrying integrated plasmids expressing wild-type dsc1, dsc1-H668A, or empty vector were assayed for Sre1 and Sre2-GFP cleavage under low oxygen. Expression of wild-type dsc1, but not dsc1-H668A, restored Sre1 and Sre2-GFP cleavage in dsc1Δ cells (Figure 6B). Collectively, these results demonstrate that the RING domain of Dsc1 is required for Sre1 cleavage.
Figure 6. Sre1 cleavage requires components of the ubiquitin-proteasome pathway.
(A) Western blot probed with anti-Sre1 IgG of lysates (top panel) from wild-type and indicated mutants grown for 0.5h at 30°C in anaerobic conditions. Microsomes (bottom panel) were isolated from the cell cultures and immunoblotted using anti-Dsc1 IgG. (B) Western blot probed with anti-Sre1 or anti-Sre2 IgG of lysates (top panels) from sre2-GFP (WT), sre2-GFP dsc1Δ, and sre2-GFP sre1Δ cells carrying an integrated plasmid expressing either empty vector, dsc1+ or dsc1-H668A grown for 0.5h at 30°C in anaerobic conditions. Microsomes (bottom panel) were isolated from the cell cultures, treated with peptide N-glycosidase F, and immunoblotted using anti-Dsc1 IgG. (C) Wild-type and ubc4-P61S cells were grown at the non-permissive temperature (36°C) for 1h and then grown in anaerobic conditions at 36°C for the indicated times. Cell lysates were subjected to immunoblot analysis using anti-Sre1 IgG. (D) Wild-type and mts3-1 cells were grown for 3h at the non-permissive temperature (36°C) to inactivate Mts3 and then shifted to anaerobic conditions at 36°C for the indicated times prior to cell harvest. Cell lysates were immunoblotted using anti-Sre1 IgG. P and N denote Sre1 precursor and cleaved nuclear forms, respectively. (E) dsc1–1, dsc2-Myc, dsc2-Myc dsc1–1 and dsc2-Myc dsc1–1 sre2Δ cells were treated with the reversible, cell-permeable crosslinker DSP (2 mM) for 30 min and detergent solubilized extracts were prepared as described in Experimental Procedures. Proteins associated with Dsc2-Myc were immunopurified using anti-myc IgG-9E10 monoclonal antibody. Unbound (lanes 1– 4) and 75× bound fractions (lanes 5–8) were immunoblotted using anti-Sre2 serum, anti-Dsc1 IgG, and rabbit anti-Myc IgG. + denotes wild-type allele.
The E3 ligase function of S. cerevisiae Tul1 requires the E2 ubiquitin conjugating enzyme Ubc4 and the Tul1 RING domain binds Ubc4 (Reggiori and Pelham, 2002). ubc4 is an essential gene in S. pombe, so we used a previously characterized temperature-sensitive mutant, ubc4-P61S, to test whether Sre1 cleavage requires Ubc4 (Seino et al., 2003). Wild-type and ubc4-P61S cells were grown at the non-permissive temperature under low oxygen for increasing times and Sre1 cleavage was assayed. In wild-type cells, Sre1N appeared at 30 minutes and accumulated during the course of the experiment (Figure 6C, lanes 1–6). Inactivation of Ubc4 blocked Sre1 cleavage and only low levels of Sre1N were present even after 4h at low oxygen (Figure 6C, lanes 7–12). Taken together with the RING domain results, these data demonstrate that the E3 ligase activity of Dsc1 is required for Sre1 cleavage.
Dsc1–4 do not contain domains to suggest that the Dsc complex possesses proteolytic activity (Figure 2A). Based on the requirement of the E2 conjugating enzyme Ubc4 and E3 ligase Dsc1 for Sre1 cleavage and similarities between the Dsc complex and the Hrd1 ligase complex that facilitates ERAD, we next tested whether Sre1 cleavage required the proteasome. To impair proteasome function, we used a temperature-sensitive allele of mts3, which codes for a subunit of the 19S proteasome regulatory cap (Gordon et al., 1996). Wild-type and mts3-1 cells were grown at the non-permissive temperature under low oxygen for increasing times and Sre1 cleavage was assayed. In wild-type cells, Sre1N appeared at 30 minutes and accumulated throughout the course of the experiment (Figure 6D, lanes 1–6). Inactivation of Mts3 blocked Sre1 cleavage and only low levels of Sre1N were present after 1–2h at low oxygen (Figure 6D, lanes 7–12). Sre1 precursor decreased at longer incubation times in mts3-1 cells, possibly due to effects of proteasome inactivation on other pathways.
Sre1 and Sre2 cleavage requires the Dsc complex and specifically the Dsc1 E3 ligase activity. To test whether the Dsc complex directly recognizes the cleavage substrate, we assayed binding between Sre2 and the Dsc complex. We chose Sre2 because its cleavage is not regulated and occurs in the presence of oxygen. We examined binding of Dsc2-Myc and Sre2 in wild-type and dsc1-1 cells with the idea that inactivation of the Dsc1 E3 ligase would increase enzyme-substrate interaction. Sre2 bound to Dsc2 in wild-type cells and binding dramatically increased in dsc1–1 cells. Dsc2-Myc bound equally well to Dsc1 and the truncated Dsc1-1 indicating that loss of RING domain function did not disrupt Dsc1-Dsc2 binding (Figure 6E, lanes 5–8). Finally, the Dsc2-Myc immunoprecipitation was specific as no copurifying Dsc1 or Sre2 was detected in untagged cells (Figure 6E, compare lane 5 and 7). Thus, the Dsc complex binds Sre2. Consistent with Sre2 being a Dsc1 substrate, Sre2 binding increased when the E3 ligase activity of Dsc1 was inhibited.
DISCUSSION
Sre1 is cleaved by a different mechanism than SREBP
Fission yeast lacks a homolog of the Site-2 protease required to release the SREBP transcription factor domain from the membrane, suggesting that Sre1 and Sre2 are proteolytically activated by a different mechanism. Multiple lines of evidence demonstrate that cleavage of these membrane bound transcription factors requires the action of a Golgi E3 ligase, the Dsc complex. First, the size of Sre1N is consistent with cleavage occurring at a cytosolic position and not within the membrane as with Site-2 protease (Figure 1). Second, Dsc1–4 form a discrete membrane complex and each subunit is required for Sre1 and Sre2 cleavage (Figures 1 and 4). Third, Dsc1 is the homolog of the S. cerevisiae Golgi protein Tul1, and independent experimental approaches indicate that the Dsc complex resides in the Golgi (Figure 5). Fourth, Sre1 cleavage requires the E2 conjugating enzyme Ubc4, a functional Dsc1 RING domain, and the proteasome (Figure 6), suggesting that Dsc E3 ligase activity is essential for Sre1 cleavage. Finally, the Dsc complex binds to Sre2 when the Dsc1 RING domain is mutated (Figure 6). Collectively, these data support a direct role for the Dsc complex in targeting Sre1 and Sre2 for cleavage.
Model for Sre1 cleavage
One speculative model consistent with our data is that under low oxygen Scp1 transports Sre1 from the ER to the Golgi where Sre1 is ubiquitinylated by the E2-E3 pair Ubc4-Dsc1 and then targeted to the proteasome for cytosolic cleavage. Cleavage releases Sre1N from the membrane, activating transcription required for hypoxic growth. Sre2 does not bind Scp1 or require it for cleavage, suggesting that Sre2 transport to the Golgi is constitutive.
Precedent for limited proteolysis of transcription factors by the proteasome exists (Rape and Jentsch, 2004). In S. cerevisiae, the homologous ER membrane proteins Mga2 and Spt23 are ubiquitinylated by the E3 ligase Rsp5 and released from the ER membrane by proteasomal cleavage and the action of the AAA-ATPase Cdc48 (Hoppe et al., 2000; Rape et al., 2001; Shcherbik and Haines, 2007). Cleavage occurs at an internal site and complete degradation by the proteasome is prevented by tightly folded domains (Piwko and Jentsch, 2006). Likewise, generation of the p50 subunit of NF-κB occurs by a mechanism of incomplete degradation by the proteasome (Palombella et al., 1994).
Dsc-dependent processing of Sre1 or Sre2 leads to production of an N-terminal basic helix-loop-helix leucine zipper transcription factor that is a functional dimer (Figure 1B)(Parraga et al., 1998). Thus, proteasomal processing likely initiates at an internal site or from the C-terminus of the substrate. How the N-terminal domains are protected from complete degradation is unknown. Sre1 and Sre2 lack sequences required to protect other proteins, such as the glycine and alanine repeats of the viral protein EBNA1 (Levitskaya et al., 1997). Both Sre1 and Sre2 contain leucine zipper dimerization domains between the N-terminus and the site of processing. Perhaps these folded domains cooperate with downstream low complexity sequences to permit escape from proteasomal degradation in a mechanism similar to that shown for the transcription factors NF-κB and Cubitus interrupts (Tian et al., 2005). Future studies will be aimed at defining the sequence and structural requirements for Sre1 and Sre2 cleavage.
Sre1 cleavage requires its binding partner Scp1 (Hughes et al., 2005). Our results demonstrate that the Scp1 requirement is for Sre1 transport to the Golgi and not for Dsc complex recognition insomuch as Scp1 function can be bypassed by mixing Golgi and ER compartments (Figure 5) and Sre2 cleavage does not require Scp1 (Figure 4). Thus, the transport function of Scp1 and Scap appears to be conserved between yeast and mammalian cells (DeBose-Boyd et al., 1999). In addition, Dsc complex function does not require low oxygen since Sre2 is cleaved normally in the presence of oxygen as is Sre1 when cells are treated with brefeldin A (Figure 5).
Our data support a model in which dsc1–4 do not code for a protease, but rather the Dsc complex targets Sre1 and Sre2 for cleavage by the 26S proteasome. Consistent with this, deletion of each dsc resulted in reduced, but detectable, Sre2 cleavage (Figure 4). Interestingly, Sre2N did not localize to the nucleus in dsc mutants. Sre2 is a bHLH-leucine zipper transcription factor and presumably a dimer like SREBP (Parraga et al., 1998). Perhaps in the absence of Dsc function, inefficient cleavage by the proteasome leads to Sre2N retention on the membrane through binding to its uncleaved partner. Similarly, S. cerevisiae Mga2 requires the E3 ligase Rsp5 for membrane release, but not proteasomal cleavage (Shcherbik et al., 2003). Sre2 precursor does not accumulate in dsc mutants, possibly due to loss of positive feedback or Dsc-independent degradation (Figures 4B and 6B). Sre1 may also be cleaved at a low level in dsc mutants, but the short half-life of Sre1N may prevent detection of this residual cleavage (Hughes and Espenshade, 2008). Although Sre1 cleavage requires the proteasome, it is possible that the Dsc complex contains additional subunits that possess protease activity. In the future, detailed in vitro reconstitution experiments will be needed to demonstrate a direct role for the proteasome in Sre1 and Sre2 cleavage.
Other functions for the Dsc E3 ligase complex
The Dsc1 homolog, S. cerevisiae Tul1, is a Golgi E3 ligase that was isolated based on its ability to ubiquitinylate and target a mutant of the syntaxin-like SNARE Pep12 to the multivesicular body pathway (Reggiori and Pelham, 2002). In addition, Tul1 sorts the endosomal SNARE Tlg1 to the MVB pathway when it is unpalmitoylated (Valdez-Taubas and Pelham, 2005). The role of Tul1 in targeting normal cargo to the MVB pathway is less clear (Hettema et al., 2004; Katzmann et al., 2004). Given its action on mutant proteins, Tul1 has been proposed to function in protein quality control (Reggiori and Pelham, 2002).
Several lines of evidence demonstrate that the Dsc complex does not function in the MVB pathway but rather has a parallel or separate function in protein degradation and possibly quality control. First, dsc1–4 are not required to properly sort the MVB pathway protein Ub-GFP-Cps1 (Supplemental Figure S3). Second, E-MAP data indicate that dsc1–4 share a common function and that dsc mutants display aggravating interactions with ESCRT pathway mutants defective in MVB pathway sorting (Table 1). If the Dsc complex functioned to target proteins to the MVB, we would expect alleviating genetic interactions in E-MAPs. Third, these aggravating genetic interactions between dsc homologs and the ESCRT mutants are conserved in S. cerevisiae, which lacks SREBP, indicating that the Dsc complex functions outside of SREBP cleavage (Supplemental Table S4). Finally, structural parallels between the Dsc complex and the Hrd1 E3 ligase complex suggest a role for the Dsc complex in protein degradation by the proteasome.
ER-associated degradation (ERAD) is one component of a cellular protein quality control system that serves to maintain homeostasis (Ellgaard and Helenius, 2003). In yeast and mammalian cells, the Hrd1 E3 ligase complex plays a central role in ERAD to target misfolded ER luminal and membrane proteins for degradation by the proteasome (Hirsch et al., 2009). The overall structure of the Dsc complex resembles that of the Hrd1 E3 ligase complex with Dsc1, Dsc2, and Dsc3 corresponding to Hrd1, Der1, and Usa1, respectively. Dsc1 is a multi-transmembrane E3 ligase, Dsc2 is a Derlin family member, and Dsc3 is a Herp-like protein that bridges the interaction between Dsc1 and Dsc2 similar to Usa1 function in the Hrd1 ligase complex (Supplemental Figure S2)(Carvalho et al., 2006). Currently, the Dsc complex appears to lack an analog of Hrd3 that binds Hrd1 and permits recognition of glycan-based luminal ERAD substrates (Carvalho et al., 2006; Denic et al., 2006). Based on these parallels between the Dsc and Hrd1 E3 ligase complexes and the role of Tul1 in recognition of mutant Golgi proteins, it is tempting to speculate that the Dsc complex may function in Golgi protein quality control. Indeed, post-ER quality control mechanisms exist (Arvan et al., 2002), and current studies are focused on testing this hypothesis.
EXPERIMENTAL PROCEDURES
Materials and standard methods (cell culture, plasmid construction, antibody preparation, extract preparation, immunoprecipitation, fluorescence microscopy, tandem affinity purification and mass spectrometry) are described in Supplemental Experimental Procedures.
Yeast Strains
S. pombe strains were generated using homologous recombination and standard molecular biology and genetic techniques (Alfa et al., 1993; Bahler et al., 1998; Guthrie and Fink, 1991). Geneticin (100 µg/L, Gibco) and clonNAT (100 µg/L, Werner BioAgents, Germany) were used to select for the kanMX and natMX marker genes, respectively. Transformants were screened for homologous integration using PCR. The dsc1–1 mutant was isolated through a mutagenesis and selection for mutants that show reduced activity of an Sre1-dependent transcriptional reporter (details of the selection will be reported elsewhere). The dsc1–1 mutation Q673STOP is a single base change (CAA to TAA) that results in a premature stop codon and truncation of the terminal 23 amino acids of Dsc1. Supplemental Table S6 contains a complete list of strains used in this study.
S. pombe Deletion Collection Screen
Individual S. pombe deletion mutants (2,626 strains total) from the Bioneer Deletion Mutant Library v1.0 were manually streaked for singles onto YES and YES+CoCl2 medium (Kim et al., 2010). A YES plate and YES +CoCl2 plate were placed at 30°C in atmospheric oxygen. A second YES plate was wrapped in foil and placed in anaerobic conditions at 30°C. After 7 days, mutants were compared to control strains (wild-type, sre1Δ and scp1Δ) and scored for growth. Mutants that failed to grow both on cobalt medium and under anaerobic conditions were assayed for low oxygen Sre1 cleavage. Dsc mutants were the only strains that showed a complete block in Sre1 cleavage.
E-MAPs and Bioinformatic Methods
Genetic screening of dsc mutants was performed using the PEM (Pombe Epistatic Mapper) system (Roguev et al., 2007). NAT-marked mutant queries in the h− PEM2 background (P392, Supplemental Table S6) were mated against the Bioneer G418-resistant deletion set (h+, ura4-D18, leu1–32, ade6-M21X, yfg∷kanMX6)(Kim et al., 2010). Mating, haploid selection, data acquisition and analysis were as described previously (Roguev et al., 2007). Pair correlation coefficients (CCs) to determine the relationship between genetic screens (471 screens in total) in Figure 2B and Supplemental Table S2 were calculated by the CORREL function (Excel). Predicted transmembrane segments and signal peptides were identified using full protein sequences and the Phobius prediction server (http://phobius.sbc.su.se/) at the Stockholm Bioinformatics Center.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health HL077588 (PJE), GM084279 (NJK), DK176743 (DWP); Searle Foundation (NJK) and Keck Foundation (NJK). PJE is an Established Investigator of the American Heart Association; CCN is an Isaac Morris Hay and Lucille Elizabeth Hay Graduate Fellow. We thank M. Shales for technical assistance and members of the Espenshade lab: J. Burg for computational assistance, S. Zhao for technical assistance, and A. Hughes and B. Hughes for preliminary experiments. We are grateful to these investigators for sharing strains and plasmids: Shelley Sazer (Baylor College of Medicine), Snezhana Oliferenko (TLL, Singapore), Hiroaki Seino (Osaka City U., Japan), Ben Glick (U. of Chicago), Colin Gordon (Medical Research Council, UK), Kaoru Takegawa (Kagawa U., Japan).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with Fission Yeast: A Laboratory Course Manual. New York: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Antonny B, Schekman R. ER export: public transportation by the COPII coach. Current Opinion in Cell Biology. 2001;13:438–443. doi: 10.1016/s0955-0674(00)00234-9. [DOI] [PubMed] [Google Scholar]
- Arvan P, Zhao X, Ramos-Castaneda J, Chang A. Secretory pathway quality control operating in Golgi, plasmalemmal, and endosomal systems. Traffic. 2002;3:771–780. doi: 10.1034/j.1600-0854.2002.31102.x. [DOI] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, Steever AB, III, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Beltrao P, Cagney G, Krogan NJ. Quantitative genetic interactions reveal biological modularity. Cell. 2010;141:739–745. doi: 10.1016/j.cell.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien CM, Chang YC, Nes WD, Kwon-Chung KJ, Espenshade PJ. Cryptococcus neoformans Site-2 protease is required for virulence and survival in the presence of azole drugs. Molecular Microbiology. 2009;74:672–690. doi: 10.1111/j.1365-2958.2009.06895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien CM, Espenshade PJ. Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis. Eukaryot Cell. 2010;9:352–359. doi: 10.1128/EC.00358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Burg JS, Powell DW, Chai R, Hughes AL, Link AJ, Espenshade PJ. Insig regulates HMG-CoA reductase by controlling enzyme phosphorylation in fission yeast. Cell Metab. 2008;8:522–531. doi: 10.1016/j.cmet.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBose-Boyd RA, Brown MS, Li WP, Nohturfft A, Goldstein JL, Espenshade PJ. Transport-dependent proteolysis of SREBP: Relocation of Site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Espenshade PJ, Hughes AL. Regulation of sterol synthesis in eukaryotes. Annu Rev Genet. 2007;41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- Gordon C, McGurk G, Wallace M, Hastie ND. A conditional lethal mutant in the fission yeast 26 S protease subunit mts3+ is defective in metaphase to anaphase transition. J Biol Chem. 1996;271:5704–5711. doi: 10.1074/jbc.271.10.5704. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. Vol 194. San Diego: Academic Press, Inc.; 1991. [Google Scholar]
- Hampton RY, Garza RM. Protein quality control as a strategy for cellular regulation: lessons from ubiquitin-mediated regulation of the sterol pathway. Chem Rev. 2009;109:1561–1574. doi: 10.1021/cr800544v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann-Petersen R, Semple CA, Ponting CP, Hendil KB, Gordon C. UBA domain containing proteins in fission yeast. Int J Biochem Cell Biol. 2003;35:629–636. doi: 10.1016/s1357-2725(02)00393-x. [DOI] [PubMed] [Google Scholar]
- Hettema EH, Valdez-Taubas J, Pelham HR. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 2004;23:1279–1288. doi: 10.1038/sj.emboj.7600137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 2005;120:831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Hughes BT, Espenshade PJ. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J. 2008;27:1491–1501. doi: 10.1038/emboj.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes BT, Nwosu CC, Espenshade PJ. Degradation of SREBP precursor requires the ERAD components UBC7 and HRD1 in fission yeast. J Biol Chem. 2009;284:20512–20521. doi: 10.1074/jbc.M109.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki T, Onishi M, Ikeuchi M, Kita A, Sugiura R, Giga-Hama Y, Fukui Y, Takegawa K. Essential roles of class E Vps proteins for sorting into multivesicular bodies in Schizosaccharomyces pombe. Microbiology. 2007;153:2753–2764. doi: 10.1099/mic.0.2007/006072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Sarkar S, Chu T, Audhya A, Emr SD. Multivesicular body sorting: ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S. Mol Biol Cell. 2004;15:468–480. doi: 10.1091/mbc.E03-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Bien CM, Hughes AL, Espenshade PJ, Kwon-Chung KJ, Chang YC. Cobalt chloride, a hypoxia-mimicking agent, targets sterol synthesis in the pathogenic fungus Cryptococcus neoformans. Mol Microbiol. 2007;65:1018–1033. doi: 10.1111/j.1365-2958.2007.05844.x. [DOI] [PubMed] [Google Scholar]
- Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci MG. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc Natl Acad Sci U S A. 1997;94:12616–12621. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- Parraga A, Bellsolell L, Ferre-D'Amare AR, Burley SK. Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution. Structure. 1998;6:661–672. doi: 10.1016/s0969-2126(98)00067-7. [DOI] [PubMed] [Google Scholar]
- Piwko W, Jentsch S. Proteasome-mediated protein processing by bidirectional degradation initiated from an internal site. Nat Struct Mol Biol. 2006;13:691–697. doi: 10.1038/nsmb1122. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- Rape M, Jentsch S. Productive RUPture: activation of transcription factors by proteasomal processing. Biochim Biophys Acta. 2004;1695:209–213. doi: 10.1016/j.bbamcr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Pelham HR. A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat Cell Biol. 2002;4:117–123. doi: 10.1038/ncb743. [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Roguev A, Wiren M, Weissman JS, Krogan NJ. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat Methods. 2007;4:861–866. doi: 10.1038/nmeth1098. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Seino H, Kishi T, Nishitani H, Yamao F. Two ubiquitin-conjugating enzymes, UbcP1/Ubc4 and UbcP4/Ubc11, have distinct functions for ubiquitination of mitotic cyclin. Mol Cell Biol. 2003;23:3497–3505. doi: 10.1128/MCB.23.10.3497-3505.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbik N, Haines DS. Cdc48p(Npl4p/Ufd1p) binds and segregates membrane-anchored/tethered complexes via a polyubiquitin signal present on the anchors. Mol Cell. 2007;25:385–397. doi: 10.1016/j.molcel.2007.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbik N, Zoladek T, Nickels JT, Haines DS. Rsp5p is required for ER bound Mga2p120 polyubiquitination and release of the processed/tethered transactivator Mga2p90. Curr Biol. 2003;13:1227–1233. doi: 10.1016/s0960-9822(03)00457-3. [DOI] [PubMed] [Google Scholar]
- Tian L, Holmgren RA, Matouschek A. A conserved processing mechanism regulates the activity of transcription factors Cubitus interruptus and NF-kappaB. Nat Struct Mol Biol. 2005;12:1045–1053. doi: 10.1038/nsmb1018. [DOI] [PubMed] [Google Scholar]
- Todd BL, Stewart EV, Burg JS, Hughes AL, Espenshade PJ. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol Cell Biol. 2006;26:2817–2831. doi: 10.1128/MCB.26.7.2817-2831.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez-Taubas J, Pelham H. Swf1-dependent palmitoylation of the SNARE Tlg1 prevents its ubiquitination and degradation. EMBO J. 2005;24:2524–2532. doi: 10.1038/sj.emboj.7600724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vjestica A, Tang XZ, Oliferenko S. The actomyosin ring recruits early secretory compartments to the division site in fission yeast. Mol Biol Cell. 2008;19:1125–1138. doi: 10.1091/mbc.E07-07-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaal KJ, Smith CL, Polishchuk RS, Altan N, Cole NB, Ellenberg J, Hirschberg K, Presley JF, Roberts TH, Siggia E, et al. Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell. 1999;99:589–601. doi: 10.1016/s0092-8674(00)81548-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.