Abstract
Important data obtained in mice raise the possibility that immunization against the saliva of sand flies could protect from leishmaniasis. Sand fly saliva stimulates the production of specific antibodies in individuals living in endemic areas of parasite transmission. To characterize the humoral immune response against the saliva of Phlebotomus papatasi in humans, we carried out a prospective study on 200 children living in areas of Leishmania major transmission. We showed that 83% of donors carried anti-saliva IgG antibodies, primarily of IgG4 isotype. Positive sera reacted differentially with seven salivary proteins. The protein PpSP30 was prominently recognized by all the sera. The salivary proteins triggered the production of various antibody isotypes. Interestingly, the immunodominant PpSP30 was recognized by all IgG subclasses, whereas PpSP12 was not by IgG4. Immunoproteomic analyses may help to identify the impact of each salivary protein on the L. major infection and to select potential vaccine candidates.
Introduction
Leishmaniasis affects millions of people worldwide. It includes a heterogeneous group of diseases that are caused by protozoan parasites of the genus Leishmania. Zoonotic cutaneous leishmaniasis (ZCL) is widespread in central Tunisia where it constitutes a public health problem with an annual incidence of ~5,000 cases.1 The clinical spectrum of the infection ranges from asymptomatic (sub-clinical) infection to self-limited cutaneous sores or more severe disease resulting in disfiguring scars. The etiological agent is an Old World Leishmania species, Leishmania major, which is transmitted by the sand fly vector, Phlebotomus papatasi. Transmission is greatest in the summer months (May to September) and the development of cutaneous lesions tends to appear in humans between October and March.2
While probing for a blood meal, the infected sand flies salivate into the host's skin. Sand fly saliva contains potent vasodilators, maxadilan and adenosine, described respectively in Lutzomyia longipalpis and P. papatasi, that prevent clotting at the biting site.3,4 Additionally, as clearly demonstrated by several investigators, sand fly saliva contains immunomodulatory molecules that have been shown to enhance disease progression.5–8
The possibility that leishmaniasis could be prevented by vaccination against sand fly saliva was supported by previous reports showing that pre-exposure to uninfected bites or pre-immunization with saliva abolished the enhancing effect of the saliva and/or prevented the disease in mice.8–10 Protection could be conferred by the host's antibodies that may neutralize a yet unidentified immunomodulatory component in the sand fly saliva.8 Alternatively, other data suggested that the protective effects of pre-exposure of saliva could be conferred in mice by a cell-mediated immune response and a delayed-type hypersensitivity to the salivary antigens.9,11,12
Little is known about the antibody response against sand fly saliva in humans. Individuals living in endemic areas for leishmaniasis develop antibodies against the saliva of the sand fly vectors13–15; yet, the protective role of these antibodies remains controversial. A correlation between the intensity of the antibody production against saliva of L. longipalpis and the appearance of a cell-mediated and a protective immunity against Leishmania chagasi has been shown.14 Contrastingly, the incidence of cutaneous leishmaniasis was still high in endemic areas even though the inhabitants were frequently bitten by uninfected sand flies arguing against a protective effect of pre-exposure to saliva in humans.15
To better understand the role of anti-sand fly saliva antibodies in humans, we characterized the features of the humoral immune response to the saliva of P. papatasi in 200 children who live in an endemic area of ZCL in Tunisia and monitored the antibody response throughout two seasons of Leishmania major transmission.
Material and Methods
Ethics statement.
All experiments were conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the ethic committee of the Institute Pasteur of Tunis. All participants provided written informed consent for the collection of blood samples and subsequent analyses.
Study population and samples.
Two hundred children, part from a previous study of ZCL in Tunisia, (age ranging from 6 to 12 years with a median of 8.2 years) were from endemic areas in Central regions (El Guettar and Souk Ejjdid). They were selected among 637 children on the basis of the availability of serum samples at different time points of the study. This cohort was followed up prospectively from April 2001 to October 2002 throughout two seasons of L. major transmission(Figure 1). Blood samples were collected from all donors at three different time points. The first sample was obtained at the beginning of the study (April–May 2001) before the ZCL transmission season, the second sample was collected after the first transmission season (April–May 2002), and the third sample was obtained in October 2002 immediately after the second transmission season. Several parameters such as leishmanin skin test (LST) reactivity and a complete clinical examination in search of the presence of typical scars or new active lesions were monitored during the study. Eighty-two individuals tested positive for the leishmanin skin test at enrollment and thirty five individuals developed ZCL after the first transmission season (data not shown).
Figure 1.
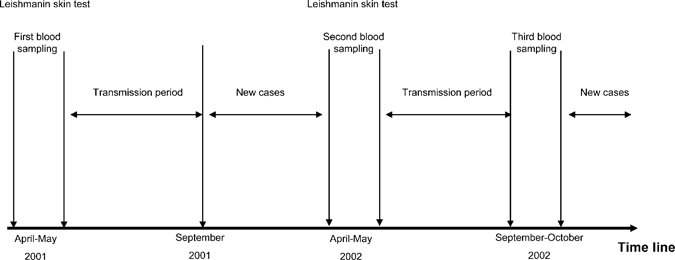
Timeline showing the temporal relationship between key events in the study and two seasons of Leishmania major transmission. Two hundred participants living in endemic areas of zoonotic cutaneous leishmaniasis (ZCL) in Central and Southwestern Tunisia (Gafsa and Sidi Bouzid) were followed up over 2 years throughout two seasons of L. major transmission. Several parameters such as leishmanin skin test or the presence of typical scars were monitored at the beginning of the study and after each transmission season and the triggering of new cases. Peripheral blood samples were obtained from each donor during these examinations.
Salivary glands extract preparation.
Sand fly salivary glands were kindly provided by Pr. E. Zhioua (Pasteur Institute of Tunis). They were obtained from a colony of P. papatasi that originated from El Felta, an endemic focus of ZCL located in the governorate of Sidi Bouzid in Central Tunisia.16 In some experiments, salivary gland extracts (SGE) derived from a colony of P. papatasi that originated from Anatolia, Turkey were used.16 The glands were dissected out in cold Tris buffer (20 mM Tris, 150 mM NaCl pH = 7.6), and then disrupted by three freezing and thawing cycles. After centrifugation, the supernatants were stored at −80°C with 10% glycerol. The salivary gland extracts were prepared just before use by dilution in phosphate saline buffer (Invitrogen, Cergy Pontoise, France).
Serum IgG anti-SGE antibody detection.
Specific anti-saliva IgG antibodies were measured by enzyme-linked immunosorbent assay (ELISA). The wells (NUNC, Maxisorp, Roskilde, Denmark) were coated overnight with SGE (0.5 glands per well) in 0.1 M carbonate-bicarbonate buffer (pH 9.6) at 4°C. The wells were then washed in phosphate buffer (PBS) with 0.1% Tween 20 and then incubated with 0.5% gelatin in PBS buffer with 0.1% Tween 20 for 1 hour at 37°C to block free binding sites. After washing, diluted sera (1:200) were incubated for 2 hours at 37°C. Antibody-antigen complexes were detected using peroxidase-conjugated anti-human IgG antibody diluted at 1:10,000 (Sigma, St. Louis, MO) for 1 hour at 37°C and visualized using orthophenylendiamine (OPD) in citrate buffer and hydrogen peroxide. The absorbance was measured using an automated ELISA reader (Awareness Technology Inc., Palm City, FL) at 492-nm wavelength. Because sand flies are present in almost all the country, 20 negative sera were obtained from healthy controls living outside Tunisia in sand fly-free regions. The cut-off value for the assays was the mean optical density (OD) of 20 negative controls plus three standard deviations. The relative OD was defined as the ratio of sample OD/mean OD of sera from 20 negative controls.
Detection of serum IgG subclasses (IgG1, 2, 3, and 4) and IgE anti-SGE antibody.
Specific anti-saliva IgG1, IgG2, IgG3, IgG4, and IgE antibodies were measured by ELISA. The optimal conditions of antigen concentration as well as sample, primary antibody and streptavidine-horseradish peroxidase dilutions were previously determined based on the differences obtained by positive and negative sera. The wells were coated with SGE and free binding sites were blocked as described previously. The wells were then incubated with diluted serum samples (1:200) for 2 hours at room temperature. After six washes, biotin-conjugated anti-human IgG isotypes (Sigma) or IgE (BD Biosciences, Le Pont de Claix, France) were incubated for 1 hour at 37°C at a dilution of 1:2,000; 1:20,000; 1:4,000; 1:20,000, or 1:250 for IgG1, IgG2, IgG3, IgG4 and IgE, respectively. After eight washes, streptavidine-horseradish peroxidase diluted at 1:6,000 (Amersham, Little Chalfont Buckinghamshire, UK) was added for 45 minutes at 37°C. Antibody-antigen complexes were visualized using OPD in citrate buffer and hydrogen peroxide. The absorbance was measured at 492 nm wavelength. The cut-off for the assays was the mean OD of 20 negative controls plus three standard deviations.
Western-blot analysis.
Salivary gland extracts were separated on a 12% or 15% sodium dodecyl sulfate- polyacrylamide gel electrophoresis (SDS-PAGE). The optimal conditions of antigen concentration as well as sample, primary antibody and streptavidine-horseradish peroxidase dilutions were previously determined. The equivalent of 40–60 salivary glands was loaded into a single long-well. The separated proteins were then transferred onto a nitrocellulose membrane. The membrane was incubated overnight at 4°C with a blocking buffer containing 5% nonfat milk and then cut into 8 to 10 strips. Each strip was incubated for 1 hour at room temperature with serum samples at different dilutions for each assay (1:200 for IgG1, IgG2, and IgG4, 1:20 for IgE and IgG3). After washing, the strips were incubated with horseradish peroxidase-linked anti-human IgG antibody (Sigma) at 1:10,000 or biotin-conjugated anti-human IgG1, IgG2, IgG3, IgG4 (Sigma) at different dilutions (1:1,000; 1:300; 1:500; 1:5,000, respectively) or IgE (BD Biosciences) at 1:250 for 1 hour at room temperature. A 45-minute incubation step at room temperature with streptavidine-horseradish peroxidase at 1:10,000 (Amersham) was performed when biotin-conjugated anti-human antibodies were used. After five washings, positive bands were visualized using enhanced chemiluminescence (Amersham). For all experiments, different molecular weight markers were used (RPN 800 or RPN 800E, Amersham).
Identification of salivary proteins by mass spectrometry (MS).
Salivary gland extracts of a Tunisian strain of P. papatasi were fractioned in a 15% SDS-PAGE gel. After Coomassie staining, gel bands corresponding to the major proteins that were recognized by positive sera were excised using the ProPic Investigator (Genomic Solutions, Ann Arbor, MI) and collected into a 96-well plate. Distaining, reduction, alkylation, and trypsin digestion of the proteins followed by peptide extraction were carried out with the ProGest Investigator (Genomic Solutions).
For MS and MS/MS analysis, peptides were eluted after the desalting step (C18-μZipTip, Millipore, Billerica, MA) directly using the ProMS Investigator, (Genomic Solutions) onto a 96-well stainless steel MALDI target plate (Applied Biosystems/MDS SCIEX, Framingham, MA) with 0.5 μL of CHCA matrix (5 mg/mL in 70% ACN/30% H2O/0.1% TFA).
Raw data for protein identification were obtained on the 4800 Proteomics Analyzer (Applied Biosystems) and analyzed with the GPS Explorer 3.6 software (Applied Biosystems). For positive-ion reflector mode spectra 3,000 laser shots were averaged. For MS calibration, autolysis peaks of trypsin ([M+H]+ = 842.5100 and 2211.1046) were used as internal calibrates. Monoisotopic peak masses were automatically determined within the mass range 800–4000 Da with a signal to noise ratio minimum set to 30. Up to 20 of the most intense ion signals were selected as precursors for MS/MS acquisition excluding common trypsin autolysis peaks and matrix ion signals. In MS/MS positive ion mode, 4,000 spectra were averaged, collision energy was 2 kV, collision gas was air, and default calibration was set using the Glu1-Fibrino-peptide B ([M+H]+ = 1570.6696) spotted onto 14 positions of the MALDI target. Combined peptide mass fingerprint (PMF) and MS/MS queries were performed using the MASCOT search engine 2.1 (Matrix Science Ltd., London, UK) embedded into GPS-Explorer Software 3.6 (Applied Biosystems) on the National Center for Biotechnology Information (NCBI) database (downloaded 2008 10 22; 7135729 sequences, 246233216 residues) with the following parameter settings: 50-ppm peptide mass accuracy, trypsin cleavage, one missed cleavage allowed, carbamidomethylation set as fixed modification, oxidation of methionines was allowed as variable modification. The MS/MS fragment tolerance was set to 0.3 Da. Only peptides with a MASCOT Ion score ≥ 51 (GPS Explorer confidence index ≥ 99%) were taken in account for protein identification. Protein hits with MASCOT Protein score ≥ 81 and a GPS Explorer Protein confidence index ≥ 99% were used for further manual validation.
Statistical analysis.
The variation in the percentage of positive donors and the median of the relative OD of anti-SGE antibodies throughout two transmission seasons was analyzed by McNemar's and Dunnet tests, respectively. The levels of serum IgG and IgE antibodies in donors with and without leishmaniasis were compared by using the non-parametric Mann-Whitney U test. The correlation between the levels of different isotypes of antibodies was assessed using Spearman's rank correlation. Statistical significance was assigned to a value of P < 0.05.
Results
Follow-up of IgG antibodies produced in response to the saliva of P. papatasi in a cohort of children living in an endemic area of L. major transmission.
The antibody response of 200 children was studied over 2 years corresponding to two transmission seasons of L. major. As shown in Figure 2, 83% of the participants tested positive for IgG antibodies against sand fly saliva at the beginning of the study. Although the median of the relative OD anti-SGE IgG antibodies varied throughout the seasons, the percentage of positive donors did not change significantly (Figure 2A and B). Among the 165 children who tested positive for IgG anti-saliva antibodies at the beginning of the study, 126 remained positive throughout the study and 39 donors became negative in the second and/or third sampling time (Figure 2C). Conversely, of the 35 donors who tested negative for anti-saliva IgG antibodies in the first blood sample, 27 developed specific anti-saliva antibodies at the second sampling and five tested positive at the third sampling. Only three children (9% of negative cases) remained unresponsive after two transmission seasons (Figure 2C). Altogether, these data suggest that nearly 90% of children living in an endemic area of ZCL who test negative for IgG anti-saliva antibodies develop specific antibodies within two transmission seasons.
Figure 2.
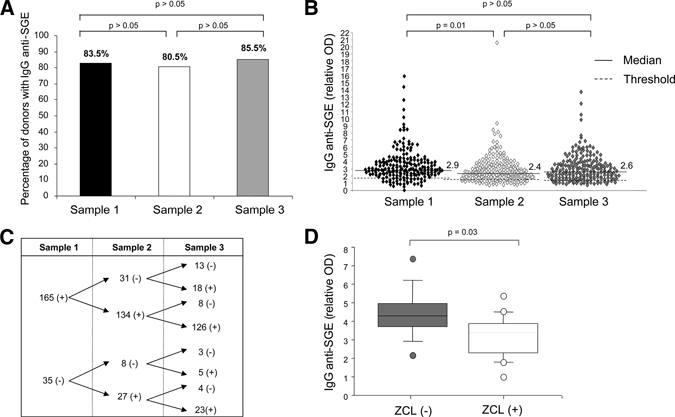
Serum levels of IgG antibodies to Phlebotomus papatasi saliva in people living in endemic areas of Leishmania major transmission. IgG antibodies to salivary gland extracts from P. papatasi were investigated in 200 children who live in endemic areas of zoonotic cutaneous leishmaniasis (ZCL). The IgG antibody levels were measured at three sampling points; at the beginning of the study before the transmission season (sample 1) and after one transmission season (sample 2) and two transmission seasons (sample 3). (A) The percentage of participants who had IgG antibodies to P. papatasi saliva did not vary significantly throughout the two transmission seasons (P > 0.05). (B) The results are expressed as the relative optical density (OD), which was the ratio of sample OD/mean OD of sera from 20 negative controls. The threshold (cut-off for the assays) was the mean optical density of sera from 20 negative controls plus three standard deviations. (C) Diagram showing the follow-up of the IgG anti-saliva antibodies throughout the three sampling points in the sera of all donors. Only three donors tested negative for specific IgG antibodies throughout the three sampling points. (D) Serum levels of IgG anti-saliva antibodies in 26 individuals with a naive immunological status against L. major. Of these 26 participants, 10 individuals developed ZCL after one transmission season (ZCL [+]) and 16 did not (ZCL [−]). The upper, middle, and lower box lines represent the 75th percentile, the median, and the 25th percentile of the OD values obtained in each group. The statistical significance between the two groups is based on the Mann-Whitney test.
We (H. Louzir, IPSOS V) and others17 formerly showed that the presence of salivary antibodies constitutes a risk factor for the development of ZCL. We, thus, focused our analysis on children who were negative for LST at the beginning of our study and who did not have any clinical evidence of previous ZCL.
During our follow-up, we found that the presence of IgG antibodies against saliva in this subgroup was associated with an increased risk of ZCL. Indeed, 9 of 20 (45%) children with anti-sand fly antibodies developed ZCL, whereas only 1 of 6 (16%) donors without antibodies contracted the disease (data not shown). Furthermore, the median level of the anti-saliva antibodies was significantly greater in patients who later developed ZCL (P = 0.03) (Figure 2D).
Characteristics of the antibodies to P. papatasi saliva in humans.
To better define the features of the IgG antibodies directed against the saliva of sand flies, we analyzed by an ELISA test the subclasses of these antibodies in 65 representative positive sera that cover the different range of positivity. Although IgG4 is a minor isotype among the IgG antibodies, anti-saliva antibodies were prominently of IgG4 subclass (76%) and at a lesser extent of IgG2 (50%) or IgG1 (39%) and IgG3 (39%) isotypes (Figure 3, panels A and B). Furthermore, the relative OD of IgG4 anti-saliva antibodies were significantly higher than those of the other isotypes (Figure 3A). Interestingly, the level of IgG4 anti-saliva antibodies correlated positively with the level of specific IgG1 and IgG2 antibodies (P = 0.01 and P = 0.09, respectively) but not with specific IgG3 antibodies (P > 0.05) (data not shown).
Figure 3.
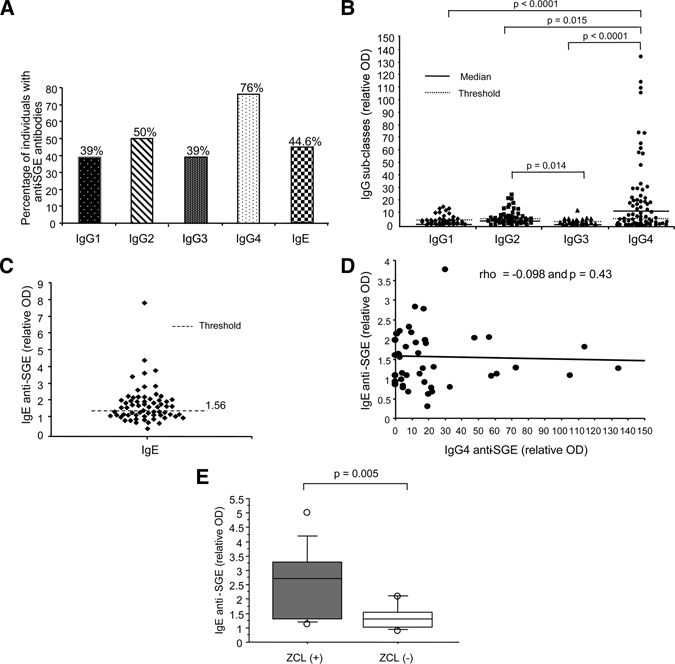
Quantification of IgG subclasses and IgE anti-saliva antibodies. Levels of IgG1, IgG2, IgG3, IgG4, and IgE antibodies directed against Phlebotomus papatasi saliva were studied in serum samples of 65 participants with specific IgG antibodies. (A) The percentage of positive donors was calculated for each assay. The results are expressed for (B) IgG subclass and (C) IgE antibodies as the relative optical density (OD), which corresponds to the ratio of sample OD/mean OD from the sera of 20 negative controls. The threshold corresponds to the mean OD obtained with the sera of 20 negative controls plus three standard deviations. (D) Regression line for pairs of OD values corresponding to IgG4 and IgE anti-saliva antibodies is shown for the 65 IgG-positive participants. No correlation between the levels of IgG4 and IgE anti-saliva antibodies was found. The Spearman's rank correlation coefficient (rho) and the P value are indicated. (E) The serum levels of IgE anti-saliva antibodies in 26 individuals with a naive immunological status against Leishmania major are shown. Of these 26 participants, 10 individuals developed ZCL after one transmission season (ZCL [+]) and 16 did not (ZCL [−]). The upper, middle, and lower box lines represent the 75th percentile, the median, and the 25th percentile of the OD values obtained in each group. The statistical significance between the two groups is based on the Mann-Whitney test.
The IgE antibodies against mosquito saliva are commonly observed in the sera of people exposed to mosquito bites.18,19 They may indicate a classic type I hypersensitivity20 and are often associated with IgG4 antibodies.18 To test whether the presence of IgG4 antibodies against P. papatasi saliva antibodies was related or not to an allergic reaction to the saliva, the production of IgE antibodies against P. papatasi saliva was investigated and the correlation between IgE and IgG4 antibodies analyzed. Although 44% of children with positive IgG antibodies to saliva had also specific IgE antibodies (Figure 3C), there was no correlation (P = 0.43) between the presence of IgG4 and IgE antibodies (Figure 3D). This suggests that IgG4 production is rather a specific feature of the immune response against P. papatasi saliva and not an allergic phenomenon. Interestingly, the presence of IgE anti-saliva antibodies was associated with an enhanced risk of triggering ZCL among the 26 children who lacked clinical and biological features of previous ZCL at enrollment. Indeed, 7 of 10 (70%) of children with IgE antibodies developed ZCL, whereas only 3 of 16 (18%) of those without specific IgE antibodies acquired the disease. Furthermore, the median level of the IgE antibodies was significantly higher in patients who later developed ZCL (P = 0.005) (Figure 3E).
Identification of the target proteins in P. papatasi saliva.
The Western-blot analysis showed that seven salivary proteins of 12kDa, 15kDa, 21kDa, 28kDa, 30kDa, 36kDa, and 44kDa were commonly, but differentially, recognized by the tested positive sera (Figure 4A). The frequency and intensity of reactivity against these different proteins were evaluated in 20 positive sera selected randomly from donors with anti-saliva IgG antibodies (Table 1). To better distinguish between the reactivity against the 12kDa and 15kDa proteins, we reanalyzed some sera in blots after a 15% SDS-PAGE (data not shown). The 30kDa protein was prominently recognized by all these sera, whereas the other proteins were only moderately and inconsistently recognized (Table 1). Nearly 90% of the positive sera showed a moderate recognition of the 12kDa, 15kDa, and 36kDa proteins, whereas the reactivity against the 21kDa, 28kDa, and 44kDa proteins was only lightly detected in 60–70% of the positive sera (Table 1).
Figure 4.
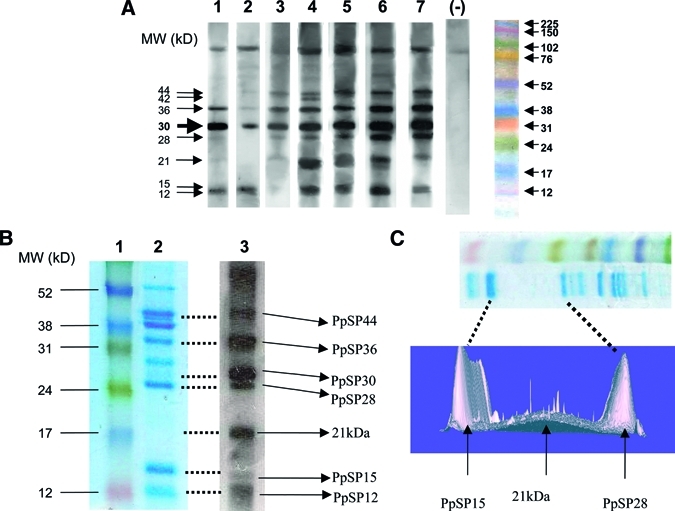
Identification of the target proteins in the saliva of Phlebotomus papatasi. Salivary gland extracts were fractionated with a (A) 12% or (B, C) 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). (A) Immunoblots profiles of salivary antigens from a Tunisian strain of P. papatasi with positive (1–7) and negative (−) human sera are shown. The molecular weight marker is shown. (B) A Coomassie-stained SDS-PAGE gel of 20 homogenized pairs of salivary glands from P. papatasi is shown in Lane 2. Lane 1 shows the molecular weight marker (RPN 800E, Amersham), whereas lane 3 shows the corresponding blot with a positive tested serum. (C) A three-dimensional analysis of the subregion of Coomassie-stained gel (shown in the upper panel) using PD Quest software.
Table 1.
Features of salivary protein recognition with positive sera
| Salivary proteins* | |||||||
|---|---|---|---|---|---|---|---|
| 12 kDA | 15 kDA | 21 kDA | 28 kDA | 30 kDA | 36 kDA | 44 kDA | |
| Percentage of positive sera | 94.1% | 88.1% | 70.5% | 60.5% | 100% | 94.1% | 64.7% |
| Intensity of reaction† | |||||||
| Range | 0–2 | 0–2 | 0–2 | 0–3 | 2–4 | 0–3 | 0–1 |
| Mean | 1.9 | 1.5 | 1.1 | 1.2 | 3.1 | 1.8 | 0.8 |
The percentage of positive sera and the intensity of the reaction were determined by western-blotting in 20 donors selected randomly from those with IgG anti-SGE.
The intensity of reaction was quoted 0 (absence), 1 (fair reactivity), 2 (moderate reactivity), 3 (strong reactivity) or 4 (very strong reactivity). The mean intensity represents the average intensity of reaction of each protein obtained in the 20 positive sera.
The target proteins in the saliva of P. papatasi were identified by MS after trypsin digestion and elution (Table 2). As seen in Figure 4B, six of the target proteins correspond to previously identified salivary proteins in P. papatasi—PpSP12 (SL1 protein, gi|15963505); PpSP15 (SL1 protein, gi|15963509); PpSP28 (D7 protein, gi|15963513); PpSP30(D7 protein, gi|15963513); PpSP36 (Salivary apyrase, gi|10443907); PpSP44 (yellow protein, gi|15963519). The 21kDa protein was difficult to identify because it was present at such a low level. However, a three-dimensional analysis of the Coomassie-stained SDS gel revealed a light expression of this protein in a wide area between PpSP15 and PpSP28 (Figure 4C). We subsequently enlarged the size of the band cut and a new MS analysis revealed that the 21kDa target protein matched the salivary protein PpSP42 (yellow protein, gi|15963517).
Table 2.
Protein identification of antibody target proteins by mass spectrometry
| Protein bands (kDa) | Protein name | NCBI accession no. | Calculated MW (kDa) | Peptide count | Sequence coverage (%) | Peptide sequences identified by MS/MS experiment | MASCOT ion score | MASCOT protein score |
|---|---|---|---|---|---|---|---|---|
| 44 | 44kDa salivary protein precursor | gi|15963519 | 45.9 | 20 | 42 | LYFGVPR | 65 | 1060 |
| DGHRPAYYIAGSSTK | 87 | |||||||
| YSNIPYTLAEIDTR | 102 | |||||||
| EFTSIYQPVIDDCR | 71 | |||||||
| NPEIIAFDLNQEGNPEVHR | 139 | |||||||
| HGNEYPTKNPEIIAFDLNQEGNPEVHR | 207 | |||||||
| 36 | salivary apyrase (Phlebotomus papatasi) | gi|10443907 | 38.3 | 18 | 61 | NQWVFLPR | 74 | 747 |
| TATYITVIDITGR | 74 | |||||||
| LYTFDDKSGIVFR | 93 | |||||||
| SGTIYNFAIIADLDKK | 86 | |||||||
| IPNGFIWHEAVNWSK | 108 | |||||||
| 30 | 30kDa salivary protein precursor | gi|15963513 | 30.5 | 12 | 41 | IPNGFIWHEAVNWSK | 87 | 458 |
| NGDQTYWAFNTCQR | 123 | |||||||
| 28 | 28kDa salivary protein precursor | gi|15963511 | 29.7 | 8 | 36 | HACSAYYYR | 77 | 320 |
| NADQTLWAFR | 86 | |||||||
| TISFFNNNVADLR | 104 | |||||||
| 21 | 42kDa salivary protein precursor | gi|15963517 | 44.7 | 8 | 21 | VPITFAQLSTR | 68 | 188 |
| AGIFGIALGDR | 51 | |||||||
| 15 | 15kDa salivary protein precursor | gi|15963509 | 16.9 | 9 | 56 | EDSHWLNCR | 72 | 415 |
| EDSHWLNCR | 82 | |||||||
| YQYYGFVAMDNNIAKPEIR | 108 | |||||||
| 12 | 12kDa salivary protein precursor | gi|15963505 | 16.3 | 9 | 56 | QKLENLLR | 79 | 474 |
| FVGAIIAYDKK | 70 | |||||||
| LGCHTSIDYYR | 76 | |||||||
| KQIDQFVDVLINGK | 120 |
NCBI = National Center for Biotechnology Information; MW = molecular weight; MS = mass spectrometry; MASCOT = search engine.
Finally, we showed that the immunodominant PpSP30 protein was recognized by all sub-classes of IgG antibodies (Figure 5). The PpSP15 and PpSP36 were recognized mostly by IgG1, IgG2, and IgG4 antibodies, whereas PpSP12 protein was targeted mainly by IgG1 and more inconsistently by IgG2 antibodies but not by the other isotypes. In positive sera, IgE antibodies against SGE were directed mainly against PpSP30 and the 21kDa protein.
Figure 5.
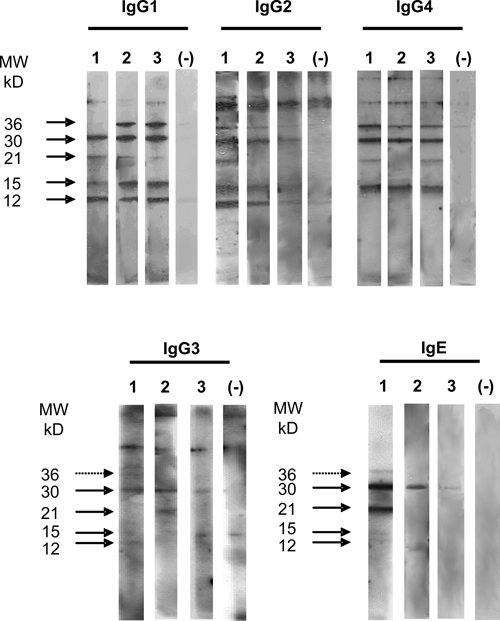
IgG subclasses and IgE immunoblots against salivary proteins of Phlebotomus papatasi. Salivary gland proteins of P. papatasi were fractioned with a 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). IgG subclasses and IgE antibodies react against salivary antigens from P. papatasi in three positive participants (1–3). Immunoblots of one negative human serum is also shown. The results were representative of 6 positive sera for IgG subclasses.
Discussion
Phlebotomus papatasi is one of the major sand fly vectors of Leishmania in the Old World. A spatial correlation between the abundance of P. papatasi in Tunisia and the incidence of ZCL showed that the disease is endemic in the arid and Saharan bioclimatic areas corresponding to Central and Southwestern Tunisia.21 Little is known about the human antibody response to the bites of sand flies from the Old World. In Sanliurfa, Turkey (a focus of Leishmania tropica), Rohousova and others15 showed that ~40% of the people carried IgG antibodies to the saliva of P. papatasi. In this study, nearly 80% of our participants tested positive, a result that may suggest a higher exposure to P. papatasi bites in the selected areas. Because there was no information about the age of the participants tested by Rohousova and others, such discrepancy could also be explained by a difference in the median age of the two cohorts.
Monitoring the antibody response to sand fly saliva revealed that nearly 90% of children who tested negative for IgG antibodies against P. papatasi saliva developed specific antibodies after two transmission seasons. However, the overall percentage of positive children did not vary significantly throughout the time. Interestingly, most of the IgG antibodies were of the IgG4 isotype and to a lesser extent, of the IgG2 subclass. One published study analyzed the IgG isotypes developed by humans in response to the sand fly saliva. It reported that individuals who were experimentally exposed to the bites of uninfected L. longipalpis developed anti-saliva antibodies restricted mostly to IgG1 and IgG4 subclasses.22 Such results were close to those obtained in individuals exposed to the bites of Aedes.23–26 In all these studies, the presence of IgG4 antibodies was correlated to the production of IgE antibodies suggesting that IgG4 was the result of an allergic reaction to mosquitoe bites. Contrasting with such data, we did not find any correlation between the presence of specific IgG4 and IgE antibodies in our cohort suggesting that the prominence of IgG4 anti-saliva antibodies did not reflect an allergic response (hypersensitivity type I) to sand fly bites. Moreover, the IgE antibodies were restricted to two salivary antigens, whereas the IgG4 antibodies recognized all the target proteins.
In light of these results, one might expect that the recall of a memory immune response in previously sensitized individuals would impair the host response and lead to the commitment of the anti-Leishmania immunity toward a Th2 response. This response may support an exacerbating effect of pre-exposure to sand fly saliva in humans. People living in endemic areas are normally exposed to bites of wild populations of P. papatasi. In Tunisia, the highest prevalence of infection of P. papatasi with L. major during the transmission peak is 7.9%.2,27 The high seroprevalence observed among our study population is the result of a high degree of contact between people and wild populations of P. papatasi. Therefore, despite that pre-exposure to uninfected bites of P. papatasi are more frequent than infected ones, people are still succumbing to ZCL in endemic areas. This finding is corroborated by the results of a recent study showing that pre-immunization of mice with salivary gland proteins of wild-caught Tunisian strain of P. papatasi did not provide any protection against L. major infection compared with a significant protection obtained with salivary gland proteins of long-term colonized ones.28 The absence of protection is probably caused by the polymorphism of salivary gland proteins of the wild population of sand flies to avoid host immune systems.28,29 Interestingly, when we followed up the participants with a naive immunological status against L. major, we showed that children with anti-saliva antibodies were more likely to develop ZCL. Accordingly, the levels of anti-saliva IgG antibodies were significantly higher in patients who developed ZCL. Our results are consistent with the data from two previous reports on leishmaniaisis from either the New World16 or the Old Word (Louzi H, personal communication). More generally, a correlation between the level of specific IgG anti-saliva antibodies and the risk of disease has been demonstrated for different vector-host models.30–32 We also found that the enhanced risk of developing ZCL was associated with the production of anti-saliva antibodies of IgE isotype. One may suggest that individuals who have allergies are more prone to develop a Th2 immune response and therefore have an increased risk of developing leishmaniasis.
Interestingly, our parallel work on the cellular immune response developed against P. papatasi saliva further confirms that this immune response is dominated by a Th2 profile. Indeed, we showed that specific memory cells are mainly CD8 + T lymphocytes that produce high levels of Interleukin-10 (IL-10) and IL-4 (Abdeladhim and others, manuscript in preparation). Nevertheless, we also showed that specific IFN-γ-producing CD4 + T cells could be activated after blocking IL-10 production (Abdeladhim and others, manuscript in preparation). This fact is of great interest as it provides a new rationale for immunological approaches that target the salivary components inducing a Th1 immune response. This strategy could be used to create a combined vaccine that contains Leishmania antigens and salivary components. For that purpose, we further investigated the proteins from P. papatasi saliva that are targeted by the antibody response. In accordance with previous results obtained by Rohousova and others,15 we found that all positive sera from naturally exposed children reacted strongly with a 30kDa protein, suggesting that this protein is the major immunodominant antigen of P. papatasi saliva in humans. Clements and others33 recently highlighted the utility of measuring the antibody response to the saliva of Phlebotomus argentipes to assess the exposure among humans living in endemic areas for leishmaniasis and subsequently to evaluate vector control programs. However, these authors showed the possibility of cross-reactive reactions between different species of sand flies. Thus, developing serological tests with the recombinant form of the immunodominant protein may overcome such limits.
Other salivary antigens such as 12 kDa, 15 kDa, and 36 kDa proteins were also frequently recognized by positive sera but to a lesser degree. Noticeably, the 12 kDa and 15 kDa proteins have not been demonstrated by Rohousova and others15 as target antigens. Previous studies showed that the composition and antigenicity of sand fly saliva may vary with the geographical origin of the fly.34 Because the Anatolian strain differs from the Tunisian strain in that it can only reproduce anautogenously, needs a previous blood meal to produce fertile eggs,16 we tested the reactivity of our positive sera against the saliva of a P. papatasi strain that originated from Anatolia in Turkey but no significant difference in the reactivity pattern between the two tested strains of P. papatasi was found (data not shown).
Mass spectrometry analyses showed that the immunodominant salivary protein recognized by all the tested sera was related to PpSP30 and the sequence obtained from the corresponding band was different from that obtained from the bands of 28 kDa and 32 kDa. Similarly, five of the other six target proteins have been previously described and correspond to PpSP12, PpSP15, PpSP28, PpSP36, and PpSP44.12 Regarding the target protein of a 21 kDa antigen, which has not been previously described, further analyses revealed that it was related to the yellow protein PpSP42, corresponding possibly to a minor isoform of this protein. However, we cannot exclude that 21 kDa antigen might be a product of degradation or proteolytic cleavage of the PpSP42. Remarkably, PpSP42 exhibits much homology with PpSP44, another yellow protein that we identified as a target protein of antibodies in humans. Further analysis of the identity of the salivary proteins of P. papatasi using proteomic approaches are in progress and may bring some clarification to this issue.
Finally, our data showed that the subclasses of IgG antibodies to the saliva of P. papatasi may differ according to the target protein. Although PpSP15, PpSP30, and PpSP36 were recognized by all IgG isotypes, PpSP12 was targeted only by IgG1 and IgG2 antibodies and not by IgG4 and IgE antibodies. This suggests that the immune response mediated by this antigen may not be polarized toward a Th2 phenotype. We are currently investigating the immune profiles induced by the different salivary gland proteins and attempting to predict their specific effects on the outcome of leishmaniasis in a large cohort of naturally exposed individuals. These investigations could help us to define the salivary proteins that might be useful for vaccination against leishmaniasis.
In conclusion, our data suggest that antibody response developed by naturally exposed individuals against saliva of P. papatasi is mainly polarized toward a Th2 phenotype. This antibody response is mainly directed toward an immunodominant protein, PpSP30. The production of the related recombinant protein and the confirmation of its recognition by the positive sera are currently in progress. Such experiments will serve ultimately to develop a serological test that could be useful for monitoring exposure of humans to sand fly bites and for predicting the risk of triggering leishmaniasis. Additionally, immunoproteomic analyses of target proteins may help us to better define the impact of each protein on Leishmania infection and possibly to define potential candidates for vaccination.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Zhioua and I. Chelbi for the critical review of the manuscript. We also thank the International Clinical Sciences Support Center (ICSSC) at Family Health International for editorial support as we prepared the manuscript.
Footnotes
Financial support: This work was supported by the NIH/NIAID grant no. SP50AI074178.
Authors' addresses: Soumaya Marzouki, Mélika Ben Ahmed, Maha Abdeladhim, and Hechmi Louzir, Laboratoire d'Immunologie clinique, Institut Pasteur de Tunis, Tunisia, E-mails: marzouki_sou@yahoo.fr, melika.benahmed@pasteur.rns.tn, maha_et_abdel@yahoo.fr, and hechmi.louzir@pasteur.rns.tn. Thouraya Boussoffara, Laboratoire d'Immunologie, de Vaccinologie et de Génétique Moléculaire, Institut Pasteur de Tunis, Tunisia, E-mail: thouraya.boussoffara@pasteur.rns.tn. Nissaf Ben Aleya-Bouafif, Nabil Belhaj Hamida, and Afif Ben Salah, Laboratoire d'Épidémiologie Médicale, Institut Pasteur de Tunis, Tunisia, E-mails: nissaf.bouafif@rns.tn, nabil.belhadjhmida@pasteur.rns.tn, and afif.bensalah@pasteur.rns.tn. Abdelkader Namane, Plateforme de Protéomique, Institut Pasteur de Paris, Paris, France, E-mail: abdelkader.namane@pasteur.fr.
References
- 1.Aoun K, Amri F, Chouihi E, Haouas N, Bedoui K, Benikhlef R, Ghrab J, Babba H, Chahed MK, Harrat Z, Bouratbine A. Epidemiology of Leishmania (L.) infantum, L. major and L. killicki in Tunisia: results and analysis of the identification of 226 human and canine isolates. Bull Soc Pathol Exot. 2008;101:323–328. doi: 10.3185/pathexo3201. [DOI] [PubMed] [Google Scholar]
- 2.Chelbi I, Derbali M, Al-Ahmadi Z, Zaafouri B, El Fahem A, Zhioua E. Phenology of Phlebotomus papatasi (Diptera: Psychodidae) relative to the seasonal prevalence of zoonotic cutaneous leishmaniasis in central Tunisia. J Med Entomol. 2007;44:385–388. doi: 10.1603/0022-2585(2007)44[385:poppdp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 3.Lerner EA, Ribeiro JM, Nelson RJ, Lerner MR. Isolation of maxadilan, a potent vasodilatory peptide from the salivary glands of the sand fly Lutzomyia longipalpis. J Biol Chem. 1991;266:11234–11236. [PubMed] [Google Scholar]
- 4.Ribeiro JM, Katz O, Pannell LK, Waitumbi J, Warburg A. Salivary glands of the sand fly Phlebotomus papatasi contain pharmacologically active amounts of adenosine and 5′-AMP. J Exp Biol. 1999;202:1551–1559. doi: 10.1242/jeb.202.11.1551. [DOI] [PubMed] [Google Scholar]
- 5.Rogers KA, Titus RG. Immunomodulatory effects of Maxadilan and Phlebotomus papatasi sand fly salivary gland lysates on human primary in vitro immune responses. Parasite Immunol. 2003;25:127–134. doi: 10.1046/j.1365-3024.2003.00623.x. [DOI] [PubMed] [Google Scholar]
- 6.Titus RG, Ribeiro JM. Salivary gland lysates from the sand fly Lutzomyia longipalpis enhance Leishmania infectivity. Science. 1988;239:1306–1308. doi: 10.1126/science.3344436. [DOI] [PubMed] [Google Scholar]
- 7.Theodos CM, Ribeiro JM, Titus RG. Analysis of enhancing effect of sand fly saliva on Leishmania infection in mice. Infect Immun. 1991;59:1592–1598. doi: 10.1128/iai.59.5.1592-1598.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belkaid Y, Kamhawi S, Modi G, Valenzuela J, Noben-Trauth N, Rowton E, Ribeiro J, Sacks DL. Development of a natural model of cutaneous leishmaniasis: powerful effects of vector saliva and saliva preexposure on the long-term outcome of Leishmania major infection in the mouse ear dermis. J Exp Med. 1998;188:1941–1953. doi: 10.1084/jem.188.10.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamhawi S, Belkaid Y, Modi G, Rowton E, Sacks D. Protection against cutaneous leishmaniasis resulting from bites of uninfected sand flies. Science. 2000;290:1351–1354. doi: 10.1126/science.290.5495.1351. [DOI] [PubMed] [Google Scholar]
- 10.Morris RV, Shoemaker CB, David JR, Lanzaro GC, Titus RG. Sandfly maxadilan exacerbates infection with Leishmania major and vaccinating against it protects against L. major infection. J Immunol. 2001;167:5226–5230. doi: 10.4049/jimmunol.167.9.5226. [DOI] [PubMed] [Google Scholar]
- 11.Belkaid Y, Valenzuela JG, Kamhawi S, Rowton E, Sacks DL, Ribeiro JM. Delayed-type hypersensitivity to Phlebotomus papatasi sand fly bite: an adaptive response induced by the fly? Proc Natl Acad Sci USA. 2000;97:6704–6709. doi: 10.1073/pnas.97.12.6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valenzuela JG, Belkaid Y, Garfield MK, Mendez S, Kamhawi S, Rowton ED, Sacks DL, Ribeiro JM. Toward a defined anti-Leishmania vaccine targeting vector antigens: characterization of a protective salivary protein. J Exp Med. 2001;194:331–342. doi: 10.1084/jem.194.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barral A, Honda E, Caldas A, Costa J, Vinhas V, Rowton ED, Valenzuela JG, Charlab R, Barral-Netto M, Ribeiro JM. Human immune response to sand fly salivary gland antigens: a useful epidemiological marker? Am J Trop Med Hyg. 2000;62:740–745. doi: 10.4269/ajtmh.2000.62.740. [DOI] [PubMed] [Google Scholar]
- 14.Gomes RB, Brodskyn C, de Oliveira CI, Costa J, Miranda JC, Caldas A, Valenzuela JG, Barral-Netto M, Barral A. Seroconversion against Lutzomyia longipalpis saliva concurrent with the development of anti-Leishmania chagasi delayed-type hypersensitivity. J Infect Dis. 2002;186:1530–1534. doi: 10.1086/344733. [DOI] [PubMed] [Google Scholar]
- 15.Rohousova I, Ozensoy S, Ozbel Y, Volf P. Detection of species-specific antibody response of humans and mice bitten by sand flies. Parasitology. 2005;130:493–499. doi: 10.1017/s003118200400681x. [DOI] [PubMed] [Google Scholar]
- 16.Chelbi I, Zhioua E. Biology of Phlebotomus papatasi (Diptera: Psychodidae) in the laboratory. J Med Entomol. 2007;44:597–600. doi: 10.1603/0022-2585(2007)44[597:boppdp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 17.de Moura TR, Oliveira F, Novais FO, Miranda JC, Clarencio J, Follador I, Carvalho EM, Valenzuela JG, Barral-Netto M, Barral A, Brodskyn C, de Oliveira CI. Enhanced Leishmania braziliensis infection following pre-exposure to sandfly saliva. PLoS Negl Trop Dis. 2007;1:e84. doi: 10.1371/journal.pntd.0000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reunala T, Brummer-Korvenkontio H, Palosuo T. Are we really allergic to mosquito bites? Ann Med. 1994;26:301–306. doi: 10.3109/07853899409147906. [DOI] [PubMed] [Google Scholar]
- 19.Peng Z, Rasic N, Liu Y, Simons FE. Mosquito saliva-specific IgE and IgG antibodies in 1059 blood donors. J Allergy Clin Immunol. 2002;110:816–817. doi: 10.1067/mai.2002.128736. [DOI] [PubMed] [Google Scholar]
- 20.Shan EZ, Taniguchi Y, Shimizu M, Ando K, Chinzei Y, Suto C, Ohtaki T, Ohtaki N. Immunoglobulins specific to mosquito salivary gland proteins in the sera of persons with common or hypersensitive reactions to mosquito bites. J Dermatol. 1995;22:411–418. doi: 10.1111/j.1346-8138.1995.tb03415.x. [DOI] [PubMed] [Google Scholar]
- 21.Chelbi I, Kaabi B, Bejaoui M, Derbali M, Zhioua E. Spatial correlation between Phlebotomus papatasi Scopoli (Diptera: Psychodidae) and incidence of zoonotic cutaneous leishmaniasis in Tunisia. J Med Entomol. 2009;46:400–402. doi: 10.1603/033.046.0229. [DOI] [PubMed] [Google Scholar]
- 22.Vinhas V, Andrade BB, Paes F, Bomura A, Clarencio J, Miranda JC, Bafica A, Barral A, Barral-Netto M. Human anti-saliva immune response following experimental exposure to the visceral leishmaniasis vector, Lutzomyia longipalpis. Eur J Immunol. 2007;37:3111–3121. doi: 10.1002/eji.200737431. [DOI] [PubMed] [Google Scholar]
- 23.Brummer-Korvenkontio H, Lappalainen P, Reunala T, Palosuo T. Detection of mosquito saliva-specific IgE and IgG4 antibodies by immunoblotting. J Allergy Clin Immunol. 1994;93:551–555. doi: 10.1016/s0091-6749(94)70066-4. [DOI] [PubMed] [Google Scholar]
- 24.Reunala T, Brummer-Korvenkontio H, Palosuo K, Miyanij M, Ruiz-Maldonado R, Love A, Francois G, Palosuo T. Frequent occurrence of IgE and IgG4 antibodies against saliva of Aedes communis and Aedes aegypti mosquitoes in children. Int Arch Allergy Immunol. 1994;104:366–371. doi: 10.1159/000236693. [DOI] [PubMed] [Google Scholar]
- 25.Palosuo K, Brummer-Korvenkontio H, Mikkola J, Sahi T, Reunala T. Seasonal increase in human IgE and IgG4 antisaliva antibodies to Aedes mosquito bites. Int Arch Allergy Immunol. 1997;114:367–372. doi: 10.1159/000237696. [DOI] [PubMed] [Google Scholar]
- 26.Remoue F, Alix E, Cornelie S, Sokhna C, Cisse B, Doucoure S, Mouchet F, Boulanger D, Simondon F. IgE and IgG4 antibody responses to Aedes saliva in African children. Acta Trop. 2007;104:108–115. doi: 10.1016/j.actatropica.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 27.Esseghir S, Ftaiti A, Ready PD, Khadraoui B, Zaafouri B, Dellagi K, Ben Ismail R. The squash blot technique and the detection of Leishmania major in Phlebotomus papatasi in Tunisia. Arch Inst Pasteur Tunis. 1993;70:493–496. [PubMed] [Google Scholar]
- 28.Ben Hadj Ahmed S, Chelbi I, Kaabi B, Cherni S, Derbali M, Zhioua E. Differences in the salivary effects of wild-caught versus colonized Phlebotomus papatasi (Diptera: Psychodidae) on the development of zoonotic cutaneous leishmaniasis in BALB/c mice. J Med Entomol. 2010;47:74–79. doi: 10.1603/033.047.0110. [DOI] [PubMed] [Google Scholar]
- 29.Milleron RS, Mutebi JP, Valle S, Montoya A, Yin H, Soong L, Lanzaro GC. Antigenic diversity in maxadilan, a salivary protein from the sand fly vector of American visceral leishmaniasis. Am J Trop Med Hyg. 2004;70:286–293. [PubMed] [Google Scholar]
- 30.Poinsignon A, Remoue F, Rossignol M, Cornelie S, Courtin D, Grebaut P, Garcia A, Simondon F. Human IgG antibody response to Glossina saliva: an epidemiologic marker of exposure to Glossina bites. Am J Trop Med Hyg. 2008;78:750–753. [PubMed] [Google Scholar]
- 31.Schwartz BS, Nadelman RB, Fish D, Childs JE, Forseter G, Wormser GP. Entomologic and demographic correlates of anti-tick saliva antibody in a prospective study of tick bite subjects in Westchester County, New York. Am J Trop Med Hyg. 1993;48:50–57. doi: 10.4269/ajtmh.1993.48.50. [DOI] [PubMed] [Google Scholar]
- 32.Remoue F, Cisse B, Ba F, Sokhna C, Herve JP, Boulanger D, Simondon F. Evaluation of the antibody response to Anopheles salivary antigens as a potential marker of risk of malaria. Trans R Soc Trop Med Hyg. 2006;100:363–370. doi: 10.1016/j.trstmh.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 33.Clements MF, Gidwani K, Kumar R, Hostomska J, Dinesh DS, Kumar V, Das P, Muller I, Hamilton G, Volfova V, Boelaert M, Das M, Rijal S, Picado A, Volf P, Sundar S, Davies CR, Rogers ME. Measurement of recent exposure to Phlebotomus argentipes, the vector of Indian visceral leishmaniasis, by using human antibody responses to sand fly saliva. Am J Trop Med Hyg. 2010;82:801–807. doi: 10.4269/ajtmh.2010.09-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volf P, Tesarova P, Nohynkova EN. Salivary proteins and glycoproteins in phlebotomine sandflies of various species, sex and age. Med Vet Entomol. 2000;14:251–256. doi: 10.1046/j.1365-2915.2000.00240.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


