Abstract
Infection with different strains of the same species of bacteria often results in vastly different clinical outcomes. Despite extensive investigation, the genetic basis of microbial strain-specific virulence remains poorly understood. Recent whole-genome sequencing has revealed that SNPs are the most prevalent form of genetic diversity among different strains of the same species of bacteria. For invasive serotype M3 group A streptococci (GAS) strains, the gene encoding regulator of proteinase B (RopB) has the highest frequency of SNPs. Here, we have determined that ropB polymorphisms alter RopB function and modulate GAS host-pathogen interactions. Sequencing of ropB in 171 invasive serotype M3 GAS strains identified 19 distinct ropB alleles. Inactivation of the ropB gene in strains producing distinct RopB variants had dramatically divergent effects on GAS global gene expression. Additionally, generation of isoallelic GAS strains differing only by a single amino acid in RopB confirmed that variant proteins affected transcript levels of the gene encoding streptococcal proteinase B, a major RopB-regulated virulence factor. Comparison of parental, RopB-inactivated, and RopB isoallelic strains in mouse infection models demonstrated that ropB polymorphisms influence GAS virulence and disease manifestations. These data detail a paradigm in which unbiased, whole-genome sequence analysis of populations of clinical bacterial isolates creates new avenues of productive investigation into the pathogenesis of common human infections.
Introduction
It has long been recognized that human infections with strains of the same bacterial species may have vastly different clinical outcomes that are not readily explained by host-specific differences (1–3). This observation has stimulated study of the genetic basis for differences in virulence among strains of the same bacterial species, which has generally focused on the effect of diversity in gene content (4–7). However, for many important bacterial pathogens it is clear that variation in gene content is insufficient to explain all virulence differences between strains (8–10). The recent development of low-cost, next-generation genome sequencing technologies has revealed that allelic variation, such as SNPs, are the most prevalent form of genetic diversity for several major bacterial pathogens (11–13). However, the role that SNPs and other minor genetic variants play in strain-specific virulence is only beginning to be dissected (14–16).
Group A streptococci are major Gram-positive pathogens that cause a broad array of disease manifestations in humans, ranging from simple colonization to uncomplicated pharyngitis or from skin infections to life-threatening bacteremia, pneumonia, and necrotizing fasciitis (17). In part because of its ability to cause such diverse human infections, group A streptococci have long served as a model pathogen for investigating the relationship between bacterial genetic variation and virulence (18). The fact that group A streptococci can infect distinct host sites strongly suggests that the ability of group A streptococci to alter gene expression in response to environmental cues is critical to its infectivity (19). Unlike many other pathogenic bacteria, group A streptococci do not appear to regulate virulence factor production by alternative sigma factors that influence transcription via interaction with core RNA polymerase, in response to different environment signals (20, 21). Thus, properly coordinated gene expression in group A streptococci depends on the activity of stand-alone transcriptional regulators and 2-component gene regulatory systems (TCS) (19, 22, 23).
Recent analysis of the genome sequences of 95 strains of invasive serotype M3 group A streptococci identified significant overabundance of SNPs in several genes, including genes encoding proteins contributing to pathogenesis, such as the anti-phagocytic M protein and the control of virulence (CovRS) TCS (13). Importantly, the highest frequency of SNPs was found in the gene encoding regulator of proteinase B (RopB, also known as Rgg) (13). RopB is a key positive transcriptional regulator of the gene encoding streptococcal proteinase B (SpeB), a secreted extracellular cysteine proteinase and proven virulence factor (24–26). RopB directly regulates speB and also appears to influence expression of several other genes encoding proteins involved in virulence and metabolic functions (27–29). A bioinformatic analysis of RopB-like proteins in various Gram-positive organisms identified 3 highly conserved amino acid residues in RopB, the substitution of any of which abolished group A streptococcal RopB function (30). Furthermore, a recent report demonstrated that an amino acid change in RopB (S103P) was found in a single group A streptococcal strain with decreased expression of speB (31). However, virulence studies performed to date involving RopB have used strains with insertional inactivation of ropB, and thus there is currently no information regarding the pathophysiologic consequences of naturally occurring single-amino-acid RopB variants (25, 32).
The strikingly high frequency of SNPs resulting in amino acid changes in RopB discovered through the aforementioned 95-genome sequencing study indicated that the ropB gene is under strong diversifying selective pressure in the host, suggesting a critical role in host-pathogen interactions (13). Here, we report on studies designed to test the hypothesis that allelic variation in ropB contributes to differences in group A streptococcal virulence. Our results demonstrate that single-amino-acid replacements in RopB significantly influence group A streptococcal gene expression and virulence, thereby expanding our understanding of how subtle changes in the bacterial genome influence pathogenesis.
Results
Analysis of ropB allelic variation and genetic background among invasive serotype M3 group A streptococcal strains.
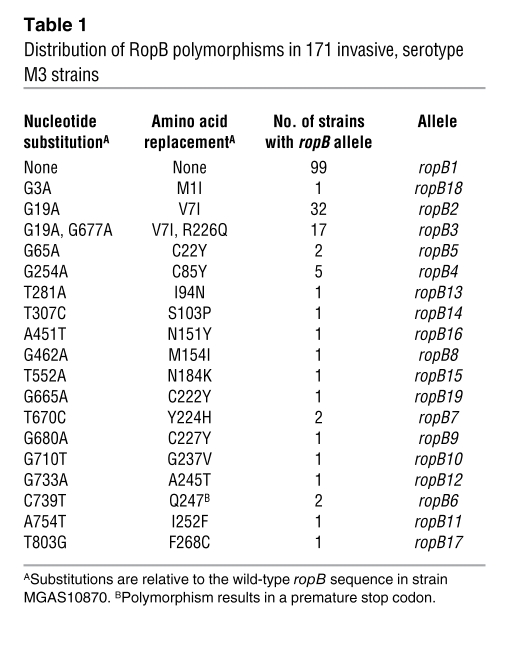
To obtain information about RopB variation in natural populations of group A streptococci, we sequenced the ropB gene from 171 unique patient isolates (strain details are provided in Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI45169DS1). These serotype M3 strains were obtained from patients with invasive group A streptococcal infections collected over 20 years in a population-based epidemiologic study in Ontario, Canada (13). We identified 19 distinct ropB alleles (Table 1). The most common ropB allele was present in 99 strains (57.9% of the strain cohort), and so we have designated this allele (ropB1) as wild-type. With one exception, all of the SNPs were non-synonymous; that is, they would result in an amino acid replacement in RopB (Table 1). The single exception was a C739T nucleotide polymorphism that generated a premature termination codon. Thus, despite the fact that ropB was the most polymorphic gene among invasive serotype M3 isolates, no synonymous or “silent” nucleotide changes in RopB were detected. This pattern of change is a strong signature of diversifying selection. All but 2 of the ropB polymorphic variants were observed in 5 or fewer strains, with most variants occurring in 2 or fewer strains (Table 1). Given that the vast majority of ropB SNPs result in amino acid replacements, we will refer to amino acid variants in this manuscript.
Table 1 .
Distribution of RopB polymorphisms in 171 invasive, serotype M3 strains
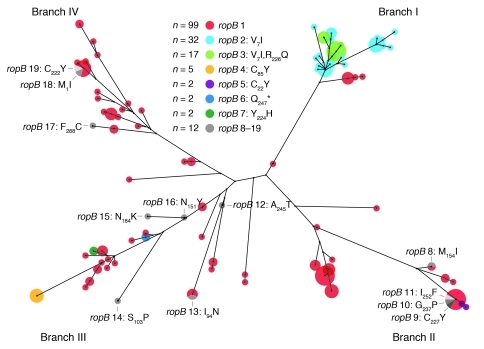
To test the hypothesis that ropB alleles occur in diverse serotype M3 genetic backgrounds, we assessed the distribution of the different ropB alleles within the context of the population genetic structure as defined by 280 core chromosomal biallelic SNPs (ref. 13 and Figure 1). This analysis revealed several findings. First, group A streptococcal strains comprising nodes located proximal on the branches of the phylogenetic tree tended to have the ropB1 allele (Figure 1). Second, polymorphisms generating new ropB alleles occurred on each of the 4 major branches of the tree (Figure 1). Finally, with the exception of ropB1, all of the ropB alleles were confined to only one branch of the tree — that is, to a single phylogenetic lineage. For example, the RopB C22Y variant (n = 2, ropB5) occurred in strains of branch II, whereas strains with the RopB C85Y polymorphism (n = 5, ropB4) were all of branch III. Similarly, the strains with the RopB V7I/R226Q variant (n = 17, ropB3) occurred only on branch I. These data indicate that although RopB polymorphisms have arisen in a diverse array of serotype M3 genetic backgrounds, particular RopB variants are found in strains of the same clonal lineage, suggesting identity by descent. In addition, the same ropB polymorphism can occur in clonally related group A streptococcal strains isolated from multiple patients, which suggests that strains with certain RopB variants can be successfully transmitted.
Figure 1. Genetic relationships among serotype M3 strains.
An unrooted neighbor-joining tree for 171 strains based on 280 biallelic SNPs located throughout the core chromosome is illustrated (13). Circles superimposed on nodes of the tree are proportionate in area to the number of strains having that haplotype. Circles are color coded by ropB allele. ropB alleles 8 to 19, represented by single strain, are shown in gray and individually labeled. Major branches of the tree are labeled I to IV.
Structural modeling of RopB suggests that observed amino acid replacements interfere with RopB oligomerization and/or DNA binding.
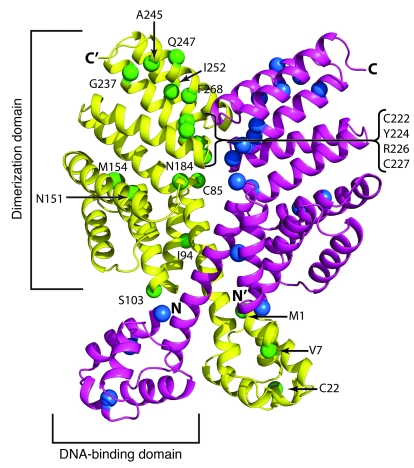
To gain insight into the potential consequences of the observed polymorphisms on RopB function, we modeled the structure of RopB using the structure of the related PlcR protein produced by Bacillus cereus (33). This comparison suggested that RopB functions as a homodimer, with DNA binding activity occurring in the aminoterminal domain and protein-protein interaction activity located in the carboxyterminal domain (Figure 2). The majority of observed RopB polymorphisms occur in the C-terminal domain, which is predicted to be involved in homodimer formation (33). The two exceptions were the V7I and C22Y polymorphisms, which are located in the predicted DNA binding domain. Clustering of amino acid polymorphisms was observed at sites predicted to affect dimerization (amino acids 222–227 and amino acids 245–268). Thus, the structure modeling data suggest that the majority of RopB polymorphisms occur in locations predicted to alter RopB function.
Figure 2. Mapping of the predicted location of amino acid replacements on RopB structural model.
The ribbon representation of the RopB model predicted by I-TASSER is shown. The monomer subunits of the RopB homodimer are shown in yellow and magenta. The areas of the putative DNA-binding and dimerization domains of 1 subunit are labeled. The amino- and carboxytermini of the molecule are denoted as N and C, respectively, and those of the second subunit are indicated by N′ and C′. The positions of the amino acid polymorphisms identified in the invasive serotype M3 strains are shown as colored spheres (blue for 1 subunit and green for the second subunit) and labeled.
Determination of kinetics of ropB and speB transcript levels and SpeB production in a serotype M3 strain.
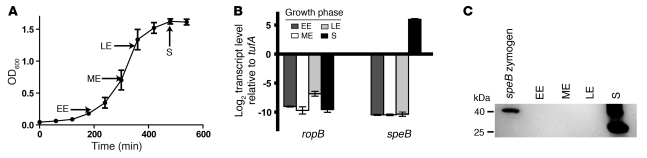
In previous studies using a serotype M5 group A streptococcal strain, ropB expression was reported to be maximal during late exponential growth, whereas speB expression and SpeB protein production peaked in stationary phase (34, 35). Given that considerable inter-serotype and even intraserotype variation in group A streptococcal regulatory patterns can exist (19), and given that an analogous study has not been conducted with a serotype M3 strain, we first determined the transcript levels of ropB and speB in strain MGAS10870 (ropB1 allele) during growth in a standard laboratory medium (Todd-Hewitt broth with yeast extract, THY) (Figure 3). We found that the level of ropB transcript peaked in late exponential growth, whereas the speB transcript level was highest during the stationary phase of growth (Figure 3B). These results are consistent with the serotype M5 findings that the RopB protein is needed for speB transcription (34). In accordance with the gene transcript data, we did not detect immunoreactive SpeB in the culture supernatant until the stationary growth phase (Figure 3C). These results indicate that the effect of RopB activity on speB peaks during the stationary phase of growth in strain MGAS10870. Thus studying RopB function in this phase is likely to be very informative.
Figure 3. Relationship of growth phase to ropB and speB transcript level and SpeB production.
(A) Growth curve of strain MGAS10870 in THY broth. Growth was done in triplicate on 3 separate occasions. Arrows indicate the points at which RNA and culture supernatants were harvested for analysis. EE, early exponential; ME, mid-exponential; LE, late exponential; S, stationary. (B) Transcript level of ropB and speB in strain MGAS10870 measured by TaqMan QRT-PCR and graphed relative to endogenous control gene tufA (71). Biologic replicates were performed in duplicate on 2 separate occasions and analyzed in duplicate. For A and B, the data shown are mean ± SD. (C) Western immunoblot using anti-SpeB polyclonal rabbit antibody and culture supernatants taken at the indicated time points. The zymogen form of SpeB is approximately 40 kDa. The multiple bands observed in the stationary-phase supernatant represent various SpeB maturation isoforms, with the majority of immunoreactive material being mature SpeB (~28 kDa) (72).
Strains with single-amino-acid replacements in RopB produce decreased amounts of functional SpeB.
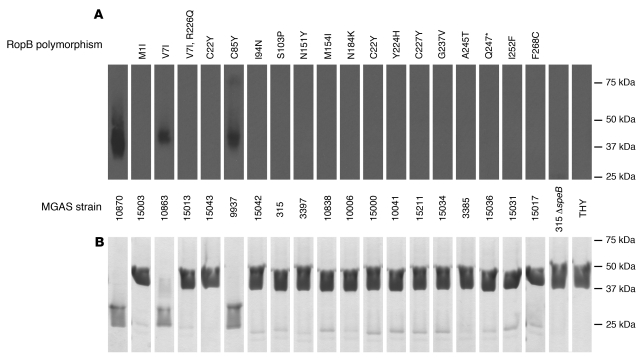
To begin to study the effect of RopB amino acid changes on RopB function, we compared the amount of immunoreactive SpeB produced during growth in THY by strains containing each of the RopB variants identified in the cohort of invasive strains. Immunoreactive SpeB was detected in the culture supernatants of only 3 of the 19 strains representing the spectrum of RopB variants (Figure 4A). The highest amount of immunoreactive SpeB was made by strain MGAS10870, which has a ropB1 allele (Figure 4A). Strains with the RopB V7I or C85Y variants also produced detectable immunoreactive SpeB (Figure 4A). To confirm that the immunoreactive SpeB was enzymatically functional, a laboratory-engineered mutant zymogen of SpeB that cannot autocatalyze its conversion to active SpeB due to a C192S substitution was incubated with culture supernatants from each group A streptococcal strain (36). In this assay, if enzymatically functional SpeB is present in the culture supernatant, the approximately 40-kDa SpeB zymogen will be processed to an approximately 28-kDa mature form. In support of the Western immunoblot results, functional SpeB activity was observed only in the culture supernatant from the group A streptococcal strains that produced immunoreactive SpeB (Figure 4B). These data indicate that the majority, but not all, of the amino acid replacements in RopB abrogate production of enzymatically active SpeB under the conditions tested.
Figure 4. Group A streptococcal strains with RopB single-amino-acid replacements produce different amounts of functional SpeB.
(A and B) The quantity and activity of SpeB produced by strains with each of the RopB variants, analyzed by Western immunoblot (A) and SpeB zymogen (SpeBz) cleavage assay (B). MGAS strain numbers and variations in the RopB sequence are indicated for each sample. As shown in B, the SpeB present in culture supernatants of the wild-type RopB, V7I, and C85Y strains readily processed purified recombinant SpeBz C192S from the 40-kDa form to the 28-kDa form, indicating that the detected SpeB was enzymatically active (36). No SpeB or SpeB activity was detected in supernatants from the other strains. Strain MGAS315ΔspeB and sterile THY broth were used as negative controls. In both panels, all samples were run simultaneously on multiple gels and, following image acquisition, lanes were reordered such that RopB polymorphisms were ordered left to right with respect to the amino acid sequence.
Insertional inactivation of the ropB1 allele has a greater effect on global gene expression compared with inactivation of the ropB4 allele.
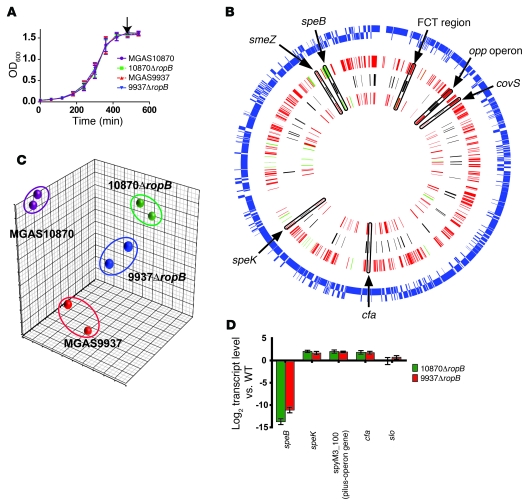
One key question regarding RopB function is why inactivation of ropB in different group A streptococci has markedly different effects on global gene expression. For example, study of 2 serotype M1 strains found that the RopB-controlled transcriptome consisted of 43 genes in 1 strain but only 3 genes in the other strain (37). To test the hypothesis that RopB amino acid polymorphisms influence the RopB transcriptome, we compared the RopB transcriptome in strain MGAS10870 (ropB1 allele) and strain MGAS9937 (ropB4 allele encoding the C85Y RopB variant). Both strains produced enzymatically active SpeB during growth in THY (Figure 4), indicating an active RopB protein. We used non-polar insertional mutagenesis to replace nearly the entire ropB gene in strain MGAS10870 and strain MGAS9937 with a spectinomycin resistance cassette to create strains 10870ΔropB and 9937ΔropB (see Supplemental Figure 1 for confirmatory Southern blot analysis). Consistent with other reports regarding insertional inactivation of ropB (29, 32, 37, 38), we found no significant difference in growth in THY between the parental and isogenic mutant strains (Figure 5A).
Figure 5. Analysis of RopB transcriptome in 2 serotype M3 strains.
(A) Growth curves of parental (MGAS10870, MGAS9937) and isogenic ropB-inactivated (10870ΔropB, 9937ΔropB) strains cultured in THY broth. Growth was done in triplicate on 3 separate occasions with data graphed as mean ± SD. Arrow depicts the point at which samples were taken for expression microarray analysis. (B) Summary of RopB transcriptomes. The outer circle (blue lines) represents the position of each of the 1,865 open reading frames in serotype M3 group A streptococcal strains with genes on the forward strand on the outside and genes on the reverse strand on the inside. The 3 inner circles represent (from the outside in) the RopB regulon in MGAS10870, genes found to be RopB regulated in both strains (in black), and the RopB regulon in MGAS9937 (innermost circle). Each gene is positioned according to its location in the genome. Genes colored red are RopB repressed, whereas genes colored green are activated by RopB. Specific examples of genes found to be RopB regulated in both strains are highlighted. FCT, fibronectin-binding, collagen-binding, T antigen. The FCT region contains the pilus operon genes. (C) Principal components analysis showing inter-sample variation for the expression microarray data set. (D) TaqMan QRT-PCR validation of expression microarray data. For D, samples were grown in duplicate on 2 separate occasions and analyzed in duplicate with data graphed as mean ± SD. For B and D, additional gene descriptions are provided in the text.
Next, we used a custom-made Affymetrix GeneChip containing all of the open reading frames of strains MGAS10870 and MGAS9937 to test the hypothesis that the RopB transcriptome differed between these strains and their RopB-inactivated isogenic derivatives. Duplicate cultures grown to early-stationary growth phase in THY were analyzed (see Figure 5A for depiction of growth phases). RopB inactivation affected the transcript level of 479 genes, or about 25% of the total genome of strain MGAS10870 (Figure 5B). In contrast, the RopB transcriptome in strain MGAS9937 was much smaller, consisting of only 159 genes or about 8% of the total genome. More than half of the genes whose transcript levels were affected by ropB inactivation in strain MGAS9937 also were affected by ropB inactivation in strain MGAS10870 (Figure 5B). For strains MGAS10870 and MGAS9937, genetic inactivation of ropB resulted in a similar transcriptome profile (note clustering of strains 10870ΔropB and 9937ΔropB on the principal components analysis plot shown in Figure 5C). These findings are consistent with the idea that the C85Y RopB amino acid replacement accounts for a substantial percentage of the transcriptome differences between strains MGAS10870 and MGAS9937.
For both strains MGAS10870 and MGAS9937, ropB inactivation mainly resulted in an increase in the number of gene transcripts (Figure 5B and Supplemental Table 2). Several genes that were previously identified as being influenced by RopB were affected by RopB inactivation in strains MGAS10870 and MGAS9937, including speB, covS, the opp operon that is involved in oligopeptide transport (39), and the smeZ gene, which encodes the superantigen streptococcal mitogenic exotoxin Z (40). There were also several genes not previously identified as being influenced by ropB whose transcript levels were affected by inactivation of ropB in both strains. These included the operon that encodes the pilus that is critical for group A streptococci adhesion to eukaryotic cells (41), the gene encoding the streptococcal pyrogenic exotoxin K (SpeK) superantigen (42), and the cfa gene encoding cAMP factor (43). Transcripts of genes encoding several key regulators were affected by ropB inactivation in strain MGAS10870 but not in strain MGAS9937, including those encoding the multigene activator (Mga) (44), catabolite control protein A (45), and the TCS Ihk/Irr (46). Similarly, there were many more genes putatively involved in translation, cellular replication, and cell wall biogenesis whose transcript levels were affected by RopB inactivation in strain MGAS10870 compared with strain MGAS9937 (Supplemental Figure 2). TaqMan quantitative real-time PCR (QRT-PCR) analysis confirmed the effect of ropB inactivation on genes analyzed (Figure 5D). Thus, ropB inactivation identified what we believe are novel RopB-regulated virulence factors in serotype M3 strains, and ropB inactivation in strain MGAS10870 (ropB1 allele) had a greater effect on global gene expression than did inactivation in strain MGAS9937, which contains the ropB4 allele encoding the C85Y variant.
Single-amino-acid replacements in RopB alter speB transcript level.
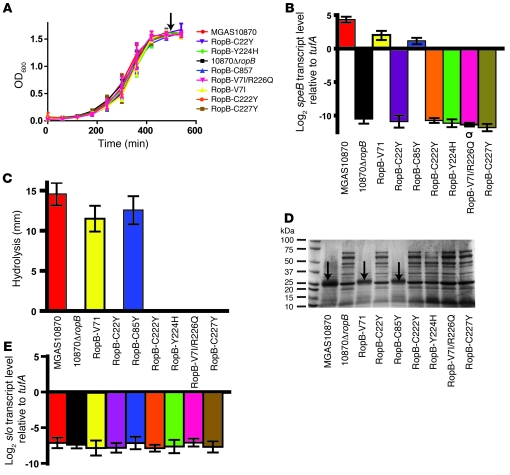
The above data demonstrated that 2 closely related strains with distinct ropB alleles differed considerably in transcriptomes (Figure 5). However, because strains MGAS10870 and MGAS9937 do not have identical genetic backgrounds (i.e., not all genes are isoallelic), the extent to which the observed expression differences are attributable to the different RopB variants is not known. To directly test the hypothesis that single-amino-acid replacements in RopB alter group A streptococci phenotypes, we used allelic exchange to create isoallelic strains with ropB-nonsynonymous SNPs in strain MGAS10870, which contains the ropB1 allele (see Table 2 for strain details and Supplemental Figure 3 for strain creation schematic). The strains constructed in this fashion differ from parental strain MGAS10870 only by the presence of the desired RopB amino acid change. We constructed 5 isoallelic strains containing ropB alleles present in strains that did not produce detectable SpeB under laboratory conditions. These included strains in which the RopB aminoterminal putative DNA-binding domain was altered (strain RopB-C22Y) and strains in which the carboxyterminal putative homodimerization domain was altered (strains RopB-C222Y, RopB-Y224H, RopB-V7I/R226Q, and RopB-C227Y). Two additional RopB isoallelic strains (RopB-V7I and RopB-C85Y) were made that contained the ropB2 and ropB4 alleles from the SpeB-producing strains MGAS15001 and MGAS9937, respectively. Each of the ropB isoallelic strains had growth characteristics in THY that were indistinguishable from parental strain MGAS10870 (Figure 6A). Strains RopB-C22Y, RopB-C222Y, RopB-Y224H, RopB-V7I/R226Q, and RopB-C227Y had a similar speB transcript level (Figure 6B), similar functional SpeB activity (Figure 6C), and a similar culture supernatant protein profile (Figure 6D) as strain 10870ΔropB, which contains an insertionally inactivated ropB gene. Conversely, strains RopB-V7I and RopB-C85Y had similar characteristics to the parental strain MGAS10870 (Figure 6). In some group A streptococci, RopB negatively influences the expression of slo, which encodes the streptolysin O cytotoxin (25, 32, 37, 47). In contrast to the differences observed in speB transcript level, there was no statistically significant difference in slo transcript between MGAS10870 and any of the RopB-altered strains (Figure 6E). Thus, we conclude that naturally occurring single nucleotide changes in ropB resulting in single-amino-acid replacements significantly alter speB transcript level and SpeB production and activity.
Table 2 .
Bacterial strains and plasmids used in this study
Figure 6. Characterization of ropB isoallelic strains.
(A) Growth curves of isoallelic ropB strains cultured in THY broth. Growth was done in triplicate on 3 separate occasions. Arrow indicates the point at which samples were taken for RNA isolation and protein analysis. (B) TaqMan QRT-PCR analysis of speB gene transcript level. Samples were grown in duplicate on 2 separate occasions and analyzed in duplicate. (C) Casein hydrolysis assay determining functional (proteolytically active) SpeB made by indicated strains. Samples were analyzed in triplicate on 3 separate occasions and the data graphed as mean ± SD. (D) SDS-PAGE analysis of culture supernatants from the indicated strains grown in THY. Arrows indicate production of SpeB only by strains MGAS10870, RopB-V7I, and RopB-C85Y, whereas a variety of other protein bands are visible in the SpeB– strains. (E) TaqMan QRT-PCR analysis of slo gene transcript level determined as in B. For C and E, strains are indicated in the area between the 2 panels. For A–C and E, data are graphed as mean ± SD.
Single-amino-acid replacements in RopB influence group A streptococcal virulence and gene transcript levels during infection.
We next tested the hypothesis that single-amino-acid replacements in RopB influence group A streptococcal infection at diverse infection sites. Because the C222Y, Y224H, R226Q, and C227Y amino acid replacements are all putatively located in very close proximity (Figure 2), we arbitrarily chose strain RopB-Y224H as representative of these 4 strains for the mouse virulence studies. The strains studied included MGAS10870, 10870ΔropB, RopB-C22Y, RopB-C85Y, and RopB-Y224H. We acknowledge that RopB influences genes other than SpeB, but for the sake of simplicity and brevity we will refer to strains MGAS10870 and RopB-C85Y as the SpeB+ strains and strains 10870ΔropB, RopB-C22Y, and RopB-Y224H as the SpeB– strains, based on the results shown in Figure 6.
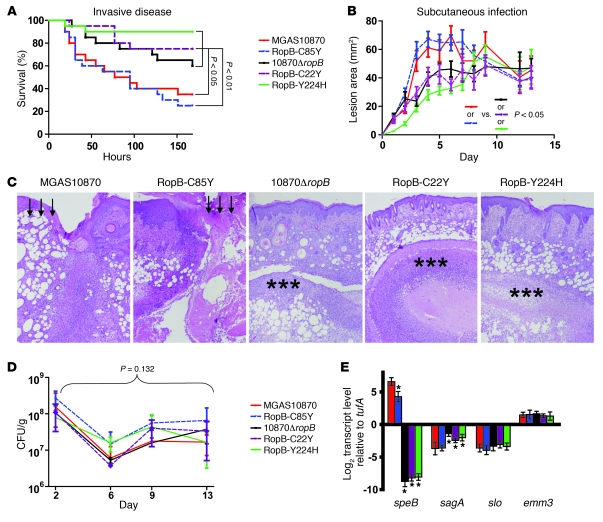
First, we studied the contribution of RopB polymorphisms to group A streptococcal virulence by challenging mice intraperitoneally with 1 × 107 CFUs of each strain. Consistent with each of the tested RopB variants occurring naturally in human invasive infection isolates, all of the strains were virulent and caused some extent of near-mortality in mice (Figure 7A). However, the survival curves of the mice infected with the 5 strains differed significantly (P < 0.0001 by log-rank test). Specifically, the survival curves of mice infected with SpeB+ strains were significantly different from mice infected with SpeB– strains (Figure 7A) (P < 0.05 for MGAS10870 and P < 0.01 for RopB-C85Y compared with each of the SpeB– strains). There was no significant difference in the survival curves between mice infected with the 2 SpeB+ strains (P = 0.78). Similarly, there was no significant difference in survival curves between mice infected with the various SpeB– strains (P = 0.33 for 10870ΔropB compared to RopB-C22Y; P = 0.06 for 10870ΔropB compared to RopB-Y224H; P = 0.29 for RopB-C22Y compared to RopB-Y224H).
Figure 7. RopB single-amino-acid changes influence group A streptococcal virulence and gene transcript levels during infection.
(A) Twenty outbred CD-1 mice were infected intraperitoneally with 1.0 × 107 CFUs of each indicated strain. Data shown are survival over time, with P values derived from Kaplan-Meier survival analysis. (B) Fifteen immunocompetent hairless mice were infected subcutaneously with 1.0 × 107 CFUs of each indicated strain. Lesion area was recorded daily and data graphed as mean ± SEM. P value refers to comparison of lesion size over time, performed using repeated measures analysis followed by Bonferroni’s adjustment for multiple comparisons. (C) Histologic analysis of lesions (original magnification, ×4) collected on day 9 following infection. Arrows indicate areas of ulceration and necrosis caused by the SpeB+ strains. Asterisks indicate focal abscess formation beneath an intact epithelium in mice infected with the SpeB– strains. (D) Bacterial density in skin/soft tissue over time for indicated strains. P value refers to repeated measures analysis of all strains. (E) In vivo gene expression of select group A streptococcal virulence factors was determined as described in Methods. For D and E, colors refer to strain inset, and data are graphed as mean ± SD of 3 biologic replicates. *P < 0.05 compared with parental strain MGAS10870. Gene names are referenced in the text.
Next, we performed a skin/soft tissue model of infection by challenging mice subcutaneously with the same set of wild-type and engineered isogenic RopB variant strains. Similar to the results from the intraperitoneal infection model, each of the strains caused disease in mice. Moreover, we found a statistically significant difference in lesion area among mice infected with the 5 strains (P < 0.0001 by repeated measures analysis) (Figure 7B). The lesion areas in mice infected with the SpeB+ strains were significantly larger than in mice infected with the SpeB– strains (P < 0.05 for MGAS10870 compared with each of the SpeB– strains and P < 0.05 for strain RopB-C85Y compared with each of the SpeB– strains). Similar to the intraperitoneal infection results, there were no statistically significant differences in lesion area among mice infected with the SpeB+ strains, nor were there statistically significant differences when comparing among mice infected with each of the SpeB– strains.
To gain insight into pathophysiologic differences observed among the infecting strains, we performed histologic analysis of the skin/soft tissue lesions. The tissue from the 2 SpeB+ strains appeared similar, with extensive necrosis and skin ulceration (Figure 7C). Conversely, tissue from mice infected with the SpeB– strains had intact epithelium with well-demarcated abscesses (Figure 7C). There was no significant difference in the CFUs of group A streptococci recovered from the lesion sites among the 5 strains when measured over time (P = 0.132 by repeated measures analysis) (Figure 7D). This result suggested that alterations in gene expression, rather than organism density, are responsible for the observed differences in lesion size and histology. To test this hypothesis we used TaqMan QRT-PCR to measure select gene transcript levels in each of the 5 strains at the infection site. Consistent with the in vitro transcript level analysis, speB transcript level was significantly lower in the SpeB– strains compared with wild-type strain MGAS10870 and strain RopB-C85Y (Figure 7E). Conversely, a significant increase in the transcript level of sagA, the initial gene in the operon encoding the cytotoxin streptolysin S (48), was observed in the SpeB– strains compared with strain MGAS10870. No significant differences were observed in the transcript levels of the slo gene or the emm3 gene that encodes the anti-phagocytic M3 protein (ref. 49 and Figure 7E). Taken together, we conclude that RopB polymorphisms have a significant effect on group A streptococcal virulence in multiple mouse models of infection and that RopB amino acid replacements influence group A streptococcal virulence gene transcript levels in the host.
Lack of detection of covRS-inactiving mutations during infection.
Over the last several years it has been recognized that hypervirulent strains of group A streptococci can arise during pathogen-host interaction due to inactivating mutations in the CovRS 2-component system, a negative regulator of many virulence genes (50–52). These mutations usually result in a SpeB– phenotype (53, 54). It is thought that the abrogation of SpeB production may be a key factor driving the emergence of such strains (50, 51). Thus, one explanation for the observed differences in virulence among strains with the various RopB amino acid replacements is that RopB polymorphisms influence the development of CovRS polymorphisms. To test this possibility, we sequenced the entire covRS operon of group A streptococci obtained from the infection site and from mouse spleen (disseminated infection site) at 2, 6, 9, and 13 days after infection. We studied more than 100 isolates from skin and spleen, but none of the strains had polymorphisms in the covR or covS genes. Similarly, visual inspection of more than 10,000 colonies isolated from mice over the course of infection detected no hypermucoid strains, a phenotype that can indicate the presence of CovRS–inactivating polymorphisms (50–52). Thus, we conclude that differential emergence of CovRS polymorphic strains is unlikely to have accounted for the variation in virulence observed among strains with RopB amino acid changes.
Discussion
In recent years there has been a marked increase in our understanding of how SNPs in the human genome contribute to many diseases, including susceptibility to particular infections such as tuberculosis and leprosy (55, 56). However, the role that microbial SNPs play in human infection is only beginning to be appreciated (14). Herein we used the results of a recent 95-strain whole-genome sequencing study as the stimulus to examine how single-amino-acid replacements in the RopB regulatory protein contribute to group A streptococcal virulence. Importantly, we discovered that naturally occurring ropB SNPs identified in strains causing human invasive infections result in single-amino-acid replacements that profoundly influence RopB function and significantly affect virulence in mouse models of infection.
Among 171 invasive, serotype M3 strains, we identified 19 different RopB variants (Table 1). Virtually every nucleotide change identified resulted in an amino acid replacement. Thus, we concluded that group A streptococci are under strong evolutionary pressure to alter RopB composition and, presumably, function. These data stand in contrast to those observed for the covS gene, in which deletions or insertions resulting in premature protein termination, rather than single-amino-acid substitutions, predominate in many invasive group A streptococci (32, 50, 57). Inasmuch as the only known role of RopB is as a regulatory protein, it stands to reason that the evolutionary pressure to modify RopB function occurs because of consequent alterations in RopB-regulated virulence factors. There are several key virulence factors influenced by RopB, including the secreted cysteine protease SpeB (Figure 5 and refs. 29, 37). Our phylogenetic analysis indicated that group A streptococci isolated from the highest numbers of patients are those with a wild-type RopB and those with RopB variants that reduce but do not eliminate SpeB production (Table 1 and Figure 4). Thus, a RopB protein capable of activating speB transcription may facilitate person-to-person transmission of group A streptococci and/or establishment of clinically apparent infection.
Our animal infection model data indicate that single-amino-acid replacements in RopB that abrogate its ability to activate speB transcription under the conditions studied result in decreased virulence, raising the question of how variance in the RopB amino acid sequence provides an evolutionary benefit to group A streptococci. It may be that amino acid replacements in RopB actually increase bacterial fitness under distinct conditions commonly encountered by group A streptococci, such as during the initial establishment of oropharyngeal colonization. It is clear from studies of Pseudomonas aeruginosa during long-term infection in the lungs of patients with cystic fibrosis (58) that increased virulence does not necessarily provide evolutionary benefit to a microbe.
Previous studies of the effect of RopB inactivation on group A streptococcal virulence have resulted in conflicting conclusions. Similar to the results of our study, ropB inactivation in a serotype M1 strain resulted in decreased mortality in a bacteremic mouse infection model (25). However, recent data from Japan showed that RopB inactivation in a serotype M3 strain increased mouse lethality in a bacteremia model of infection and increased lesion area in a subcutaneous infection model (32). A potential explanation for the variation in results in the various RopB studies is that RopB inactivation increased slo transcript levels only in the Japanese study, whereas no effect on slo transcript level was noted in our study or in the previously mentioned serotype M1 study (25, 32). Thus, the function of RopB in the various parental strains in which it has been inactivated may be quite distinct. The importance of the choice of the parental strain to study RopB inactivation was clearly demonstrated by the markedly different size of the RopB transcriptome in strains MGAS10870 and MGAS9937 (Figure 5). Our finding that a SNP can be associated with significant differences in the breadth of the RopB transcriptome provides a potential explanation for previous observations regarding marked strain-to-strain variation in the effect of RopB inactivation on group A streptococcal global gene expression (37).
Historically, in the absence of a crystal structure, the contribution of amino acid residues to protein function has been studied using site-specific mutagenesis or by identifying highly conserved amino acid residues by bioinformatic analysis of orthologous proteins from diverse bacterial species (30, 59). The studies described herein provide a different strategy for probing the contribution of particular amino acids to protein function, namely by comparative study of naturally occurring variants found in clinical bacterial isolates. A previous bioinformatic-based study of group A streptococcal RopB using sequences from diverse Gram-positive organisms identified 3 invariant amino acid residues among RopB-like regulatory proteins (30). Interestingly, none of these 3 amino acids (G4, R11, or W142) was altered in the strains from our invasive cohort. Similarly, many of the polymorphisms that we found altered group A streptococcal RopB function were present in amino acids that are highly varied among RopB orthologs (e.g., C22, C85, N184, and C222) (30). Thus, through the use of sequence data obtained from a large sample of clinical infection strains, we have markedly expanded the understanding of the amino acids that are critical to group A streptococcal RopB function.
In summary, we have exploited the results obtained in a large multi-genome sequencing project to design productive studies bearing on how single-amino-acid replacements in a regulatory protein influence streptococcal pathogenesis. Given the ability to rapidly and relatively economically sequence large microbial strain collections, we anticipate that similar approaches will be applied to a broad array of major microbial pathogens, thereby expanding our understanding of how bacteria cause disease in humans and potentially providing new avenues for therapeutic intervention.
Methods
Bacterial strains, culture media, and growth experiments.
Bacterial strains and plasmids used in this study are listed in Table 1 and Supplemental Table 1. Invasive serotype M3 strains were recovered from an active, population-based surveillance of group A streptococcal infection in Ontario, Canada, that has been previously described (13, 60). Group A streptococci were grown on Trypticase soy agar containing 5% sheep blood (BSA; Becton Dickinson) or in Todd-Hewitt broth containing 0.2% (wt/vol) yeast extract (THY; Difco). Spectinomycin or chloramphenicol (Sigma-Aldrich) was added when appropriate to THY agar or broth at a concentration of 150 μg/ml and 10 μg/ml, respectively. All growth experiments were done in triplicate on 3 separate occasions, for a total of 9 replicates. Overnight cultures were inoculated into fresh medium to achieve a uniform starting OD600 of 0.03. Growth was monitored by measuring OD at 600 nm.
Analysis of ropB and covRS gene sequences.
The ropB gene and the covRS operon were amplified by PCR from chromosomal DNA using primers listed in Supplemental Table 3. Sanger sequencing of the target sequences was done with a 3730xl DNA analyzer (Applied Biosystems).
Mass spectroscopy SNP analysis and phylogenetic reconstruction.
Genetic relationships among serotype M3 strains were determined using ClustalX and SplitsTree v4 based on assessment of 280 core genome polymorphic loci, using a mass spectroscopy–based method as previously described (13).
Modeling of RopB polymorphisms.
The structure of RopB was modeled using the I-TASSER structure prediction server ( http://zhanglab.ccmb.med.umich.edu/I-TASSER). The top template identified by I-TASSER to model RopB was PlcR, a virulence gene regulator protein made by Bacillus cereus (33). The algorithm for structure prediction by I-TASSER involves template identification by threading alignments, assembly by iterative simulations, and refinement of the resulting model (61, 62). The accuracy of the predicted model is validated by two parameters: the confidence score (c-score) and the template modeling score (TM-score) (61, 62). The predicted RopB model has a high c-score of 0.44, indicating the accuracy of the prediction, and a very reliable TM-score of 0.77, pointing to the topological match of the model to the template. The resulting model covers 98% of all RopB residues. The RopB dimer was generated by superimposition of individual subunits of RopB to the PlcR dimer using COOT (63). The RopB model (Figure 2) was created using the program Pymol (64).
RNA isolation and transcript level analysis.
For transcript level analysis in laboratory media, strains were grown to indicated growth phases, and RNA was isolated and purified with an RNeasy kit (Qiagen) (59). The quality and the concentration of RNA were assessed with an Agilent 2100 Bioanalyzer and by analysis of the A260/A280 ratio. cDNA was reverse transcribed from RNA using Superscript III (Invitrogen) according to the manufacturer’s instructions. TaqMan QRT-PCR was performed with an ABI Thermocycler 7500 Fast System (Applied Biosystems) using the ΔCT method of analysis (65). The primers and probes used are listed in Supplemental Table 3.
Western immunoblot analysis of SpeB present in culture supernatants.
Following overnight growth in THY, 2 ml of culture was removed, bacterial cells were pelleted by centrifugation, and the culture supernatant was sterilized with a 0.22-μm membrane filter. Samples were then concentrated approximately 10-fold using centrifugation filter units with a 10-kDa MWCO (Milipore). Total protein concentration was determined using the Bradford assay (Bio-Rad) and volumes of condensed culture supernatant containing equal quantities of protein were analyzed on a 15% SDS-PAGE gel, transferred to nitrocellulose membrane, and probed with anti-SpeB polyclonal rabbit antiserum at a dilution of 1:100,000. Detection and visualization of immunoreactive SpeB was carried out with SuperSignal West Pico Rabbit IgG detection kit (Thermo Scientific). To generate Figure 4A, all strains were tested simultaneously using multiple gels and, following image acquisition, lanes corresponding to each strain were reordered such that RopB polymorphisms were ordered left-to-right with respect to the amino acid sequence.
Assay of SpeB protease activity in culture supernatants.
The proteolytic activity of SpeB in culture supernatants was determined using a SpeB zymogen cleavage assay as described previously (14). Aliquots of each culture supernatant were incubated with 5 μg of purified recombinant SpeB zymogen containing a C192S substitution (SpeBz C192S). SpeBz C192S is proteolytically inactive but is readily processed from the 40-kDa to the 28-kDa form by proteolytically active SpeB (36). A reducing environment is required for maximal SpeB activity; therefore, reactions were performed in the presence of 10 mM β-mercaptoethanol at 37°C for 3 hours. Following incubation, samples were separated on three 15% SDS-PAGE gels and stained with Coomassie Brilliant Blue. After image acquisition, individual lanes corresponding to different strains were reordered to align with results from Western blot analysis (Figure 4A).
Insertional inactivation of ropB.
Strains 10870ΔropB and 9937ΔropB, in which nearly the entire ropB open reading frame was replaced with a spectinomycin resistance cassette, were created using the PCR splicing overlap extension method with the spectinomycin resistance cassette being amplified from plasmid pSL60 (66, 67). The isogenic mutant strains were analyzed by Southern hybridization and DNA sequencing to confirm that the proper genetic construct was obtained (Supplemental Figure 1).
Expression microarray analysis.
Expression microarray analysis was performed using biologic replicates of indicated strains grown to early stationary growth phase (Figure 5A). A custom-made Affymetrix GeneChip that contains probes for 100% of the genes of strain MGAS10870 was used for expression microarray (transcriptome) studies (45). Principal component analysis was performed using the Partek Genomics Suite (Figure 5C). Expression microarray data were deposited in the GEO database (GSE26655).
Creation of ropB isoallelic strains.
A schematic explaining isoallelic strain construction is provided in Supplemental Figure 3. For each of the indicated strains, primers were designed to amplify a DNA fragment that consisted of approximately 600 bp on either side of the ropB SNP plus restriction digest sites for PstI and XbaI. The resultant PCR products were then cloned into the multi-cloning site of the temperature-sensitive plasmid pJL1055 (gift of D. Kasper, Channing Laboratory, Boston, Massachusetts, USA). (68). The resultant plasmids were electroporated into group A streptococci as described (69). Incorporation of the vector into the group A streptococcal chromosome was accomplished by growing the cells in THY with chloramphenicol at 28°C (permissive temperature), whereas insertion of the vector into the chromosome was accomplished by shifting the culture temperature to 39°C (non-permissive temperature) and continuing to grow the cells in THY with chloramphenicol. Plasmid curing was performed by growing the cells at 28°C in the absence of chloramphenicol. After 3 cycles of growth, cells were plated onto BSA, and individual colonies were checked for sensitivity to chloramphenicol. Sequencing of 1,000 bp on either side of the desired polymorphism was then performed to ensure the correct genetic construct was obtained. Additional sequencing of the covRS operon, mga, and emm gene were performed to check for the presence of spurious mutations that may have developed during the strain construction process (none were found).
Casein hydrolysis assays.
Functional SpeB protease activity was determined using casein hydrolysis (26).
One-dimensional PAGE analysis of culture supernatants.
Cells from 5 ml of group A streptococcal culture grown overnight in THY were pelleted by centrifugation. The resultant supernatant was sterilized by passage through a 0.22 μM filter and mixed with 2 volumes of 100% methanol for 1 h at room temperature. The precipitated proteins were collected via centrifugation, suspended in 200 μl sterile phosphate buffered saline, and 10 μl separated on a 4%- to 15%-gradient denaturing polyacrylamide gel, followed by staining with Coomassie Brilliant Blue.
Animal virulence studies.
Mouse experiments were performed according to protocols approved by the Methodist Hospital Research Institute’s Institutional Animal Care and Use Committee. For the bacteremia model of infection, 20 female outbred CD-1 Swiss mice (Harlan-Sprague-Dawley) were injected intraperitoneally with 1.0 × 107 CFUs of each strain, and mice were monitored at least daily for near-mortality. For the skin/soft tissue infection model, 15 immunocompetent hairless SKH1-hrbr female mice (Charles River BRF) per strain were injected subcutaneously with 1.0 × 107 CFUs, and lesion size was measured daily. For determination of inoculation site CFUs, 3 mice per strain were euthanized at the time points indicated in the figures and skin lesions were excised, homogenized in phosphate buffered saline, plated onto BSA, and incubated for 24 hours, and CFUs were counted. To detect spontaneous covRS mutations in group A streptococcal infection isolates, individual group A streptococci colonies from the skin lesions and spleens of mice were randomly selected for sequencing of the entire covRS operon using PCR and sequencing primers listed in Supplemental Table 3.
Microscopic pathologic analysis.
On day 9 following subcutaneous inoculation, mouse lesions were excised and tissue was fixed in 10% phosphate-buffered formalin, serially sectioned, and embedded in paraffin using automated standard instruments. H&E- and Gram’s-stained sections were examined with an Olympus BX5 microscope and photographed with an Olympus DP70 camera. Micrographs of tissue taken from the inoculation sites that showed skin and soft tissue pathology characteristic of each strain were selected for publication.
Measurement of cDNA levels during infection.
For in vivo transcript levels, 3 mouse skin lesions per strain were obtained at day 9, immediately placed into RNAlater (Qiagen), and then snap-frozen with liquid nitrogen. Group A streptococcal RNA was isolated from mouse limbs as previously described (70). In brief, the frozen limbs were subjected to vigorous mechanical lysis with a series of sharp blows using a 3-pound drill hammer and FastPrep Lysing Matrix B (MP Biomedicals). RNA was isolated using a Qiagen RNeasy kit and treated vigorously with Turbo DNAse (Ambion). RNA integrity was assessed with an Agilent 2100 Bioanalyzer, and cDNAs were prepared with and without reverse transcriptase to ensure that TaqMan QRT-PCR signal amplification did not reflect DNA contamination. Mouse lesions inoculated with PBS were also included in the analysis to ensure that the observed signal did not arise from eukaryotic RNA.
Statistics.
A 2-sided Student’s t test was employed to compare differences in the QRT-PCR and casein hydrolysis assays. For the animal studies, comparison of mouse near-mortality rates following intraperitoneal inoculation was performed using a Kaplan-Meier survival analysis, whereas comparison of lesion size following subcutaneous inoculation was done using a repeated measures analysis. Comparison of CFU number was performed as described for lesion size. All differences were considered significant at P < 0.05 after applying Bonferroni’s test to account for multiple comparisons. For expression microarray analysis, a 2-sided Student’s t test (unequal variance) was applied, followed by a false discovery rate correction (Q < 0.05) to account for multiple testing with transcript levels being considered significantly different when the corrected P value was less than 0.05 and the mean difference was at least 2-fold.
Supplementary Material
Acknowledgments
This work was supported by American Heart Association grant 0765115Y (to S.A. Shelburne III) and National Institute Allergy and Infectious Diseases K08 Career Development Award AI-064564 (to S.A. Shelburne III).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(5):1956–1968. doi:10.1172/JCI45169.
Ronan Carroll’s present address is: Department of Clinical Microbiology, Trinity College Dublin, St. James’s Hospital, Dublin, Ireland.
References
- 1.Davies HD, et al. Invasive group A streptococcal infections in Ontario, Canada. Ontario Group A Streptococcal Study Group. N Engl J Med. 1996;335(8):547–554. doi: 10.1056/NEJM199608223350803. [DOI] [PubMed] [Google Scholar]
- 2.Fridkin SK, et al. Methicillin-resistant Staphylococcus aureus disease in three communities. . N Engl J Med. 2005;352(14):1436–1444. doi: 10.1056/NEJMoa043252. [DOI] [PubMed] [Google Scholar]
- 3.Laupland KB, Gregson DB, Church DL, Ross T, Pitout JD. Incidence, risk factors and outcomes of Escherichia coli bloodstream infections in a large Canadian region. . Clin Microbiol Infect. 2008;14(11):1041–1047. doi: 10.1111/j.1469-0691.2008.02089.x. [DOI] [PubMed] [Google Scholar]
- 4.Bae IG, et al. Presence of genes encoding the Panton-Valentine leukocidin exotoxin is not the primary determinant of outcome in patients with complicated skin and skin structure infections due to methicillin-resistant Staphylococcus aureus: results of a multinational trial. . J Clin Microbiol. 2009;47(12):3952–3957. doi: 10.1128/JCM.01643-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Afset JE, et al. Identification of virulence genes linked with diarrhea due to atypical enteropathogenic Escherichia coli by DNA microarray analysis and PCR. . J Clin Microbiol. 2006;44(10):3703–3711. doi: 10.1128/JCM.00429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lintges M, et al. Superantigen genes are more important than the emm type for the invasiveness of group A Streptococcus infection. . J Infect Dis. 2010;202(1):20–28. doi: 10.1086/653082. [DOI] [PubMed] [Google Scholar]
- 7.Fierer J, Guiney DG. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. . J Clin Invest. 2001;107(7):775–780. doi: 10.1172/JCI12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pirnay JP, et al. Pseudomonas aeruginosa population structure revisited. . PLoS One. 2009;4(11):e7740. doi: 10.1371/journal.pone.0007740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lalani T, et al. Associations between the genotypes of Staphylococcus aureus bloodstream isolates and clinical characteristics and outcomes of bacteremic patients. . J Clin Microbiol. 2008;46(9):2890–2896. doi: 10.1128/JCM.00905-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinberger DM, et al. Association of serotype with risk of death due to pneumococcal pneumonia: a meta-analysis. Clin Infect Dis. 2010;51(6):692–699. doi: 10.1086/655828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327(5964):469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He M, et al. Evolutionary dynamics of Clostridium difficile over short and long time scales. . Proc Natl Acad Sci U S A. 2010;107(16):7527–7532. doi: 10.1073/pnas.0914322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beres SB, et al. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc Natl Acad Sci U S A. 2010;107(9):4371–4376. doi: 10.1073/pnas.0911295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsen RJ, et al. Decreased necrotizing fasciitis capacity caused by a single nucleotide mutation that alters a multiple gene virulence axis. Proc Natl Acad Sci U S A. 2010;107(2):888–893. doi: 10.1073/pnas.0911811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy AD, et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. . Proc Natl Acad Sci U S A. 2008;105(4):1327–1332. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen SL, et al. Positive selection identifies an in vivo role for FimH during urinary tract infection in addition to mannose binding. Proc Natl Acad Sci U S A. 2009;106(52):22439–22444. doi: 10.1073/pnas.0902179106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musser JM, Shelburne SA., 3rd A decade of molecular pathogenomic analysis of group A Streptococcus. . J Clin Invest. 2009;119(9):2455–2463. doi: 10.1172/JCI38095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olsen RJ, Shelburne SA, Musser JM. Molecular mechanisms underlying group A streptococcal pathogenesis. Cell Microbiol. 2009;11(1):1–12. doi: 10.1111/j.1462-5822.2008.01225.x. [DOI] [PubMed] [Google Scholar]
- 19.McIver KS. Stand-alone response regulators controlling global virulence networks in Streptococcus pyogenes. . Contrib Microbiol. 2009;16:103–119. doi: 10.1159/000219375. [DOI] [PubMed] [Google Scholar]
- 20.Cho BK, et al. The transcription unit architecture of the Escherichia coli genome. . Nat Biotechnol. 2009;27(11):1043–1049. doi: 10.1038/nbt.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown RN, Gulig PA. Roles of RseB, sigmaE, and DegP in virulence and phase variation of colony morphotype of Vibrio vulnificus. . Infect Immun. 2009;77(9):3768–3781. doi: 10.1128/IAI.00205-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kreikemeyer B, McIver KS, Podbielski A. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. . Trends Microbiol. 2003;11(5):224–232. doi: 10.1016/s0966-842x(03)00098-2. [DOI] [PubMed] [Google Scholar]
- 23.Shelburne SA, et al. A combination of independent transcriptional regulators shapes bacterial virulence gene expression during infection. PLoS Pathog. 2010;6(3):e1000817. doi: 10.1371/journal.ppat.1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyon WR, Gibson CM, Caparon MG. A role for trigger factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. . EMBO J. 1998;17(21):6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollands A, Aziz RK, Kansal R, Kotb M, Nizet V, Walker MJ. A naturally occurring mutation in ropB suppresses SpeB expression and reduces M1T1 group A streptococcal systemic virulence. . PLoS ONE. 2008;3(12):e4102. doi: 10.1371/journal.pone.0004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukomski S, et al. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. . J Clin Invest. 1997;99(11):2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dmitriev AV, McDowell EJ, Kappeler KV, Chaussee MA, Rieck LD, Chaussee MS. The Rgg regulator of Streptococcus pyogenes influences utilization of nonglucose carbohydrates, prophage induction, and expression of the NAD-glycohydrolase virulence operon. . J Bacteriol. 2006;188(20):7230–7241. doi: 10.1128/JB.00877-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaussee MA, Callegari EA, Chaussee MS. Rgg regulates growth phase-dependent expression of proteins associated with secondary metabolism and stress in Streptococcus pyogenes. . J Bacteriol. 2004;186(21):7091–7099. doi: 10.1128/JB.186.21.7091-7099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaussee MS, Somerville GA, Reitzer L, Musser JM. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. . J Bacteriol. 2003;185(20):6016–6024. doi: 10.1128/JB.185.20.6016-6024.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loughman JA, Caparon MG. Contribution of invariant residues to the function of Rgg family transcription regulators. J Bacteriol. 2007;189(2):650–655. doi: 10.1128/JB.01437-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kappeler KV, Anbalagan S, Dmitriev AV, McDowell EJ, Neely MN, Chaussee MS. A naturally occurring Rgg variant in serotype M3 Streptococcus pyogenes does not activate speB expression due to altered specificity of DNA binding. . Infect Immun. 2009;77(12):5411–5417. doi: 10.1128/IAI.00373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikebe T, et al. Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS Pathog. 2010;6(4):e1000832. doi: 10.1371/journal.ppat.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Declerck N, et al. Structure of PlcR: Insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria. Proc Natl Acad Sci U S A. 2007;104(47):18490–18495. doi: 10.1073/pnas.0704501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neely MN, Lyon WR, Runft DL, Caparon M. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. . J Bacteriol. 2003;185(17):5166–5174. doi: 10.1128/JB.185.17.5166-5174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loughman JA, Caparon M. Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: a model for in vivo gene expression. . J Bacteriol. 2006;188(2):399–408. doi: 10.1128/JB.188.2.399-408.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Musser JM, Stockbauer K, Kapur V, Rudgers GW. Substitution of cysteine 192 in a highly conserved Streptococcus pyogenes extracellular cysteine protease (interleukin 1beta convertase) alters proteolytic activity and ablates zymogen processing. . Infect Immun. 1996;64(6):1913–1917. doi: 10.1128/iai.64.6.1913-1917.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dmitriev AV, McDowell EJ, Chaussee MS. Inter- and intraserotypic variation in the Streptococcus pyogenes Rgg regulon. . FEMS Microbiol Lett. 2008;284(1):43–51. doi: 10.1111/j.1574-6968.2008.01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaussee MS, Watson RO, Smoot JC, Musser JM. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. . Infect Immun. 2001;69(2):822–831. doi: 10.1128/IAI.69.2.822-831.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang CH, et al. Effects of oligopeptide permease in group A streptococcal infection. Infect Immun. 2005;73(5):2881–2890. doi: 10.1128/IAI.73.5.2881-2890.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unnikrishnan M, et al. The bacterial superantigen streptococcal mitogenic exotoxin Z is the major immunoactive agent of Streptococcus pyogenes. . J Immunol. 2002;169(5):2561–2569. doi: 10.4049/jimmunol.169.5.2561. [DOI] [PubMed] [Google Scholar]
- 41.Zähner D, Scott JR. SipA is required for pilus formation in Streptococcus pyogenes serotype M3. . J Bacteriol. 2008;190(2):527–535. doi: 10.1128/JB.01520-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banks DJ, Lei B, Musser JM. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. . Infect Immun. 2003;71(12):7079–7086. doi: 10.1128/IAI.71.12.7079-7086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gase K, Ferretti JJ, Primeaux C, McShan WM. Identification, cloning, and expression of the CAMP factor gene (cfa) of group A streptococci. . Infect Immun. 1999;67(9):4725–4731. doi: 10.1128/iai.67.9.4725-4731.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ribardo DA, McIver KS. Defining the Mga regulon: Comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A Streptococcus. . Mol Microbiol. 2006;62(2):491–508. doi: 10.1111/j.1365-2958.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- 45.Shelburne SA, 3rd, et al. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. . Proc Natl Acad Sci U S A. 2008;105(5):1698–1703. doi: 10.1073/pnas.0711767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voyich JM, et al. Engagement of the pathogen survival response used by group A Streptococcus to avert destruction by innate host defense. . J Immunol. 2004;173(2):1194–1201. doi: 10.4049/jimmunol.173.2.1194. [DOI] [PubMed] [Google Scholar]
- 47.Chaussee MS, et al. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. . Infect Immun. 2002;70(2):762–770. doi: 10.1128/IAI.70.2.762-770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nizet V, et al. Genetic locus for streptolysin S production by group A Streptococcus. . Infect Immun. 2000;68(7):4245–4254. doi: 10.1128/IAI.68.7.4245-4254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlsson F, Berggard K, Stalhammar-Carlemalm M, Lindahl G. Evasion of phagocytosis through cooperation between two ligand-binding regions in Streptococcus pyogenes M protein. . J Exp Med. 2003;198(7):1057–1068. doi: 10.1084/jem.20030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2(1):e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker MJ, et al. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat Med. 2007;13(8):981–985. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
- 52.Engleberg NC, Heath A, Miller A, Rivera C, DiRita VJ. Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. . J Infect Dis. 2001;183(7):1043–1054. doi: 10.1086/319291. [DOI] [PubMed] [Google Scholar]
- 53.Aziz RK, et al. Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. . Mol Microbiol. 2004;51(1):123–134. doi: 10.1046/j.1365-2958.2003.03797.x. [DOI] [PubMed] [Google Scholar]
- 54.Kansal RG, Datta V, Aziz RK, Abdeltawab NF, Rowe S, Kotb M. Dissection of the molecular basis for hypervirulence of an in vivo-selected phenotype of the widely disseminated M1T1 strain of group A Streptococcus bacteria. J Infect Dis. 2010;201(6):855–865. doi: 10.1086/651019. [DOI] [PubMed] [Google Scholar]
- 55.Khor CC, et al. CISH and susceptibility to infectious diseases. N Engl J Med. 2010;362(22):2092–2101. doi: 10.1056/NEJMoa0905606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang FR, et al. Genomewide association study of leprosy. N Engl J Med. 2009;361(27):2609–2618. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 57.Turner CE, Kurupati P, Jones MD, Edwards RJ, Sriskandan S. Emerging role of the interleukin-8 cleaving enzyme SpyCEP in clinical Streptococcus pyogenes infection. . J Infect Dis. 2009;200(4):555–563. doi: 10.1086/603541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith EE, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A. 2006;103(22):8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu Y, Inouye M. The role of the G2 box, a conserved motif in the histidine kinase superfamily, in modulating the function of EnvZ. Mol Microbiol. 2002;45(3):653–663. doi: 10.1046/j.1365-2958.2002.03061.x. [DOI] [PubMed] [Google Scholar]
- 60.Sharkawy A, et al. Severe group A streptococcal soft-tissue infections in Ontario:1992–1996. Clin Infect Dis. 2002;34(4):454–460. doi: 10.1086/338466. [DOI] [PubMed] [Google Scholar]
- 61.Wu S, Skolnick J, Zhang Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 2007;5:17. doi: 10.1186/1741-7007-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(12 pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64. DeLano WL.The PyMOL Molecular Graphics System . Palo Alto California, USA: DeLano Scientific; 2002. [Google Scholar]
- 65.Shelburne SA, 3rd, et al. Maltodextrin utilization plays a key role in the ability of group A Streptococcus to colonize the oropharynx. . Infect Immun. 2006;74(8):4605–4614. doi: 10.1128/IAI.00477-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuwayama H, Obara S, Morio T, Katoh M, Urushihara H, Tanaka Y. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 2002;30(2):E2. doi: 10.1093/nar/30.2.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lukomski S, et al. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. . Infect Immun. 2000;68(2):535–542. doi: 10.1128/IAI.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J, Kasper DL, Ausubel FM, Rosner B, Michel JL. Inactivation of the alpha C protein antigen gene, bca, by a novel shuttle/suicide vector results in attenuation of virulence and immunity in group B Streptococcus. . Proc Natl Acad Sci U S A. 1997;94(24):13251–13256. doi: 10.1073/pnas.94.24.13251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perez-Casal J, Price JA, Maguin E, Scott JR. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. . Mol Microbiol. 1993;8(5):809–819. doi: 10.1111/j.1365-2958.1993.tb01628.x. [DOI] [PubMed] [Google Scholar]
- 70.Graham MR, et al. Analysis of the transcriptome of group A Streptococcus in mouse soft tissue infection. . Am J Pathol. 2006;169(3):927–942. doi: 10.2353/ajpath.2006.060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Virtaneva K, et al. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. . Proc Natl Acad Sci U S A. 2005;102(25):9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lyon WR, Caparon MG. Trigger factor-mediated prolyl isomerization influences maturation of the Streptococcus pyogenes cysteine protease. . J Bacteriol. 2003;185(12):3661–3667. doi: 10.1128/JB.185.12.3661-3667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lei B, Mackie S, Lukomski S, Musser JM. Identification and immunogenicity of group A Streptococcus culture supernatant proteins. . Infect Immun. 2000;68(12):6807–6818. doi: 10.1128/iai.68.12.6807-6818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.