Abstract
Human cancer cells frequently have regions of their DNA hypermethylated, which results in transcriptional silencing of affected genes and promotion of tumor formation. However, it is still unknown whether cancer-associated aberrant DNA methylation is targeted to specific genomic regions, whether this methylation also occurs in noncancerous cells, and whether these epigenetic events are maintained in the absence of the initiating cause. Here we have addressed some of these issues by demonstrating that transgenic expression of DNA methyltransferase 3b (Dnmt3b) in the mouse colon initiates de novo DNA methylation of genes that are similar to genes that become methylated in human colon cancer. This is consistent with the notion that aberrant methylation in cancer may be attributable to targeting of specific sequences by Dnmt3b rather than to random methylation followed by clonal selection. We also showed that Dnmt3b-induced aberrant DNA methylation was maintained in regenerating tissue, even in the absence of continuous Dnmt3b expression. This supports the concept that transient stressors can cause permanent epigenetic changes in somatic stem cells and that these accumulate over the lifetime of an organism in analogy to DNA mutations.
Introduction
Regional hypermethylation of DNA is frequently found in human cancer, causes transcriptional silencing of affected genes, and promotes tumor formation (1, 2). An unresolved question is whether aberrant DNA methylation of cancer is caused by stochastic DNA methylation followed by clonal selection, or whether specific sequences are targeted by the de novo methyltransferases (3). Furthermore, it is not known whether cancer-specific DNA methylation only occurs in neoplastic cells or whether it can also occur in normal cells. Another important question is whether aberrations in DNA methylation are maintained in the absence of the initiating cause.
Abundant evidence indicates that DNA methyltransferase 3b (Dnmt3b) is involved in de novo methylation of mammalian cells during development, is frequently activated in human tumors, and promotes tumor development in APCMin/+ mice (4, 5). To address whether Dnmt3b targets specific sequences for de novo methylation, we used transgenic mice that allow tetracycline-inducible expression of Dnmt3b in WT mice and in APCMin/+ mice, a model of intestinal tumorigenesis.
Results and Discussion
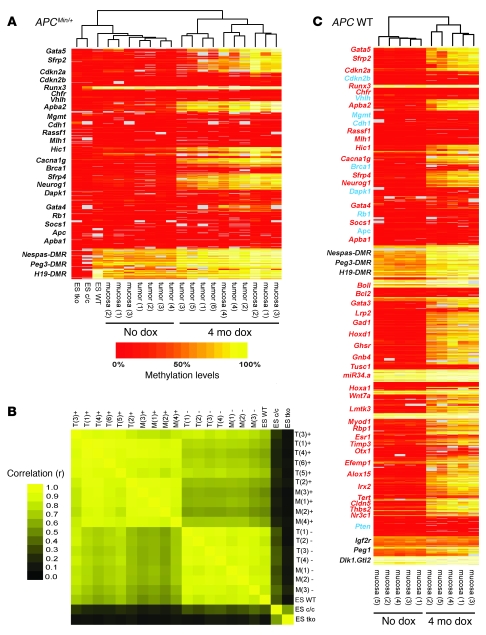
To determine whether aberrant de novo methylation is a stochastic or a targeted process, we compared DNA methylation of colon tumors, representing a mono-/oligoclonal cell population, with normal colon epithelial cells of APCMin/+ mice induced to express the Dnmt3b transgene by exposure to doxycycline (dox). We analyzed DNA methylation of 23 candidate genes that either are frequently methylated in human colon cancer or represent classical tumor suppressor genes (6). As control regions, 3 imprinted loci were included, and control samples included ES cells deficient for Dnmt1; triple-knockout ES cells deficient for Dnmt1, Dnmt3a, and Dnmt3b; and WT ES cells. For methylation analysis, we used gene-specific amplification of bisulfite-treated DNA followed by reverse transcription and MALDI-TOF analysis (7). An almost identical methylation pattern was seen in all tumor samples and nontumor cell samples from Dnmt3b-induced mice (Figure 1A). A significant increase in DNA methylation (defined as methylation increase of at least 0.1 and P ≤ 0.05 by t test) was detected in 11 of 26 candidate regions (including imprinted loci) in both tumor samples and nontumor samples, and pairwise analysis of methylation data confirmed a strong correlation of de novo methylation patterns among all Dnmt3b-expressing samples (correlation between induced sample pairs, 0.76–0.97; mean r, 0.90; Figure 1B). The observation that clonal and nonclonal cell populations showed the same de novo methylation pattern across multiple samples strongly suggests that de novo methylation by Dnmt3b is not random, but targets specific genetic loci. The data in Figure 1A also support the hypothesis that de novo methylation of targets such as Abpa2 (also known as Mint2), Hic1, Gata4, Gata5, Cacna1g, and Neurog1, considered to be hallmarks of methylated genes in cancer (6), can occur prior to tumor formation in normal cells (see also Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI43169DS1).
Figure 1. DNA methylation analysis of colon tumors and colon epithelial cells with or without Dnmt3b transgene expression.
4 mo dox, Dnmt3b induction; no dox, no Dnmt3b induction; ES c/c, Dnmt1-deficient ES cells; ES tko, Dnmt1/Dnmt3a/Dnmt3b triple-knockout ES cells; gray, no data. Heat map data were generated by mass array analysis and subjected to sample-based unsupervised hierarchical clustering. (A) Tumors and mucosa samples with Dnmt3b induction showed reproducible de novo DNA methylation of specific genomic loci. Imprinted regions (Nespas-, Peg3-, and H19-DMR) were partially methylated; ES tko and ES c/c cells showed loss of DNA methylation. (B) Pairwise correlation of methylation data confirmed similar de novo methylation in all samples with Dnmt3b induction (r, 0.76–0.97; mean r, 0.90). (C) Dnmt3b-mediated DNA methylation in colon mucosa of APC WT mice closely resembled de novo methylation reported for human colon cancer (Supplemental Table 1), which suggests that Dnmt3b can induce cancer-specific de novo methylation in normal cells (red, methylated; blue, unmethylated; black, control/imprinted regions).
To further compare DNA methylation in the mouse model with human colon cancer, we analyzed additional candidate genes that were previously reported to be methylated or unmethylated in human colon cancer (6). To confirm that de novo methylation of cancer-specific targets can occur in normal cells, the expanded candidate gene analysis was conducted using APC WT mice as opposed to the previously studied mutant APCMin/+ mice. Indeed, the Dnmt3b-induced DNA methylation pattern in WT mice fully reproduced the DNA methylation pattern detected in APCMin/+ mice (Figure 1C). This showed that the de novo methylation patterns obtained in Figure 1A do not require the presence of a mutant APC allele. Further, the expanded candidate gene analysis demonstrated that human cancer–related methylation targets were consistently methylated in mice with Dnmt3b expression: the target list (Figure 1C) included 31 genes that are frequently methylated in human colon cancer (6). Of these 31 human cancer methylation targets, 26 were also significantly methylated when Dnmt3b was induced in the colonic mucosa of WT mice (0.1 or greater absolute increase in DNA methylation and P ≤ 0.05 by t test; Supplemental Table 1). Conversely, 8 of 8 candidate genes reportedly unmethylated in human colon cancer also remained unmethylated in the Dnmt3b mouse model (0.1 or less de novo methylation and P ≥ 0.05; Supplemental Table 1).
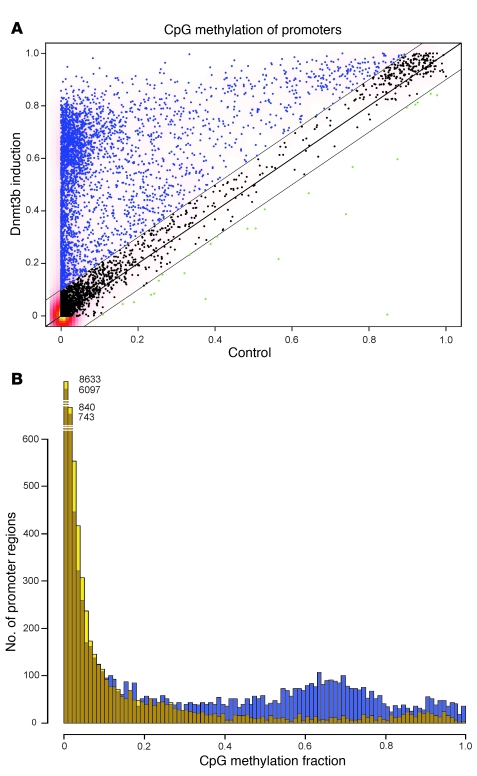
To complement this candidate gene approach and to further explore the similarities between de novo methylation in our transgenic model and aberrant DNA methylation of colon cancer, we next conducted genome-wide analysis of Dnmt3b-mediated de novo methylation using reduced representation bisulphite sequencing (RRBS; ref. 8). Using 3 colon mucosa samples with 3 months of Dnmt3b induction and 3 samples from age-matched control mice, we scored Dnmt3b-mediated de novo methylation in 13,361 promoter regions. Analogous to the above findings, we observed almost identical de novo methylation patterns among all 3 induced samples (pairwise correlation between induced sample pairs, r > 0.95; Supplemental Figure 2). This further supports the concept that Dnmt3b directly targets certain promoter regions. Interestingly, a discrete subset of genes (11.7%; 1,564 of 13,361) showed a strong methylation gain upon Dnmt3b expression (absolute methylation gain >0.5, false discovery rate [FDR] P < 0.01, complete methylation assigned as 1), whereas the majority of promoter regions remained unmethylated in both test and control DNA (Figure 2, A and B). This suggests that a subgroup of genes in the genome is inherently methylation sensitive. Importantly, in agreement with the results of the candidate gene analysis, we again found a strong concordance when comparing the murine Dnmt3b-mediated de novo methylation data with published data from human colon cancer: for the human cancer/mouse comparison, we selected human genes from the study of Widschwendter et al. (6) that were classified either as cancer-related methylation targets (absolute methylation gain >0.1 in tumors, PMR gain in tumors >10) or as nontargets (tumor methylation gain <0.1, excluding genes with baseline methylation in human control tissue >100% [PMR >100]). Mouse RRBS data were available for 44 human colon cancer methylation targets and for 71 nontargets (115 total). As shown in Supplemental Tables 2 and 3, 84% of genes (37 of 44) methylated in human colon cancer were also methylated by Dnmt3b (methylation gain >0.1, FDR P < 0.01); conversely, 92% of genes (65 of 71) not methylated in human colon cancer were also not methylated by Dnmt3b (methylation gain <0.1). This supports the concept that Dnmt3b targets the very same set of genes that are frequently methylated in human cancer. In addition, the similarity between murine and human data suggests conservation of DNA methylation sensitivity across species.
Figure 2. RRBS analysis of colon epithelial cells with or without Dnmt3b transgene expression in vivo.
RRBS data from colon epithelial cells with or without Dnmt3b transgene expression (n = 3 each) were used to calculate de novo methylation in 13,361 annotated promoter regions. (A) Scatter plot demonstrating Dnmt3b-mediated hypermethylation of a distinct subpopulation of promoter regions (11.7% of promoters showed >0.5 methylation gain in Dnmt3b-expressing tissue; lines parallel to the diagonal indicate the position of 0.1 methylation gain or loss). (B) Histogram further illustrating the appearance of a discrete subpopulation of hypermethylated promoters in Dnmt3b-expressing samples (blue), whereas the control samples (brown) contained few promoter regions with methylation scores >0.5.
Previous studies had shown a strong correlation between genes methylated in human cancer and genes targeted by the polycomb repressive complex 2 (PRC2) in ES cells (6, 9). We therefore used published data on mouse ES cells (10) to compare the PRC2 occupancy of genes targeted by Dnmt3b (methylation gain >0.5) with that of genes not targeted by Dnmt3b (methylation gain <0.1). Analogous to previous reports, we defined genes associated with either Suz12, Eed, or H3K27 methylation in mouse ES cells as PRC2 targets. Interestingly, ES cell PRC2 occupancy was 71% (721 of 1,015) for genes targeted by Dnmt3b and only 13% (716 of 5,564) for genes not targeted by Dnmt3b (Supplemental Tables 4 and 5). This strong enrichment of PRC2 targets (in ES cells) among genes methylated by Dnmt3b is in good agreement with the previously reported enrichment of PRC2 targets in genes methylated in human cancer (6) and further supports the concept that Dnmt3b targets the same genes that are methylated in cancer. Taken together, our results indicate that colon epithelial cell methylation in the Dnmt3b model predicts DNA methylation of human colon cancer with high confidence.
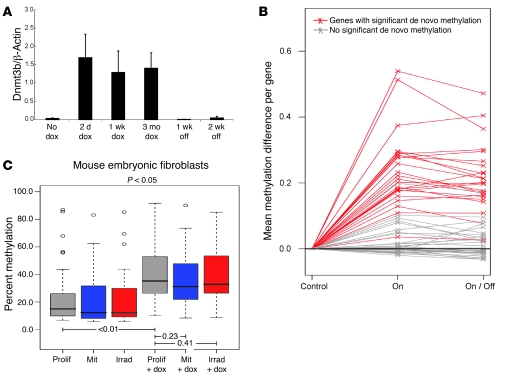
Having established that Dnmt3b can induce a cancer-specific signature of de novo methylation in healthy tissue, we next analyzed the stability of such alterations. Such information would help resolve the question whether altered DNA methylation is a potential initiating cause of disease, rather than only an effect of disease. We induced de novo methylation in the colon of transgenic mice by adding dox to the drinking water for 5 months and analyzed maintenance of these methylation marks after 4 months of dox withdrawal and silencing of transgene expression. The epithelial cell layer of the colon fully renews every 5 to 7 days (11) and is therefore ideal to test maintenance of epimutations in vivo. Dnmt3b expression in transgenic mice was rapidly activated by dox in the drinking water and quickly returned to baseline within 1 week of dox withdrawal (Figure 3A). Importantly, de novo methylation of individual genes was almost identical in mice on dox compared with mice that were maintained in the absence of dox for 4 months (Figure 3B). This supports the hypothesis that aberrant de novo DNA methylation is maintained in regenerating healthy tissue, even in the absence of the initiating cause. To test whether this may be caused by de novo methylation of the intestinal stem cell compartment, we used RNA FISH analysis to measure Dnmt3b transgene expression in colonic epithelial cells that express the intestinal stem cell marker Lgr5 (Supplemental Figure 1 and refs. 11, 12). Indeed, Dnmt3b expression in Lgr5-positive cells of a dox-induced transgenic mouse was approximately twice that of a littermate control (control, 0.0037 ± 0.00063 Dnmt3b particles/μm3; Dnmt3b transgene induction, 0.0076 ± 0.00124 Dnmt3b particles/μm3; P ≤ 0.001), supporting the concept that transgene-mediated de novo methylation of intestinal stem cells contributes to irreversible epigenetic alteration of the colonic mucosa.
Figure 3. Transient expression of Dnmt3b in vivo causes permanent epigenetic alterations of colonic mucosa.
(A) Dnmt3b transgene expression in colon epithelial cells in vivo was activated within 2 days of dox administration and returned to baseline within 1 week of dox withdrawal (n = 2 each; quantitative PCR analysis, mean ± SD). (B) Dnmt3b was induced in transgenic mice for 5 months (On, n = 5), and a subset was followed up for an additional period of 4 months after dox withdrawal (On/Off, n = 5). Controls were age-matched to the On/Off group (n = 4). Almost all genes that underwent significant de novo methylation (P < 0.01, t test) were equally methylated in the On and On/Off groups. (C) Dnmt3b-mediated de novo methylation was analyzed in dividing (prolif, n = 5) and nondividing mouse embryonic fibroblasts (mit, mitomycin C, n = 3; irrad, γ-irradiated, n = 2) in the absence of dox or after 14 days of dox-induced Dnmt3b transgene expression. Box plot shows combined methylation data of genes with significant de novo methylation in proliferating cells (P < 0.05, paired t test; n = 25). Box denotes interquartile range; line within box denotes median; whiskers denote 1.5× interquartile range; symbols denote outliers. Significant Dnmt3b-mediated de novo methylation was detected in both proliferating and growth-arrested cells, which suggests that cell division is not required for this process. P values between groups are shown by brackets.
Several previous studies have shown a strong correlation between cell proliferation and acquisition of aberrant de novo DNA methylation (8, 13), which suggests that infrequently dividing cells, such as somatic stem cells, are possibly less sensitive to Dnmt3b-mediated de novo methylation than more rapidly dividing cells. To address this question, we analyzed Dnmt3b-mediated de novo methylation of proliferating and growth-arrested (mitomycin C–treated or irradiated) mouse embryonic fibroblasts after 14 days of dox-induced transgene expression. We did not detect a significant difference between de novo methylation of actively dividing cells and growth-arrested cells (mitomycin C, P = 0.23; γ-irradiated, P = 0.41; paired t test versus proliferating cells; Figure 3C). This further supports the concept that Dnmt3b activation in the slow-cycling stem cell compartment can contribute to permanent epigenetic alterations in regenerating tissues.
In summary, our results show that Dnmt3b induction in the mouse colon generates de novo methylation that closely resembles aberrant DNA methylation of human colon cancer. We demonstrated that cancer-specific DNA methylation can be a targeted process and may not be the result of stochastic methylation followed by clonal selection in the incipient tumor cell population. Importantly, our results showed that de novo methylation can occur in normal cells, which suggests that regional hypermethylation may contribute to the earliest stages of tumor formation. Finally, we demonstrated that transient induction of aberrant DNA methylation, most likely in intestinal stem cells, caused permanent epigenetic alterations of the intestinal mucosa. These observations support the concept that transient stressors can cause permanent epigenetic changes in somatic stem cells that accumulate over the lifetime of an organism, analogous to DNA mutations.
Methods
Transgenic mice.
All mice used in this study and the genotyping protocol were described previously (5). Transgene induction was achieved by administering 0.5 g/l dox in the drinking water. All animal studies were reviewed and approved by the Committee on Animal Care (protocol no. 1107-088-10; Department of Comparative Medicine, MIT).
Tissue harvesting.
Colon tumors from APCMin/+ mice were harvested under the dissecting microscope. Colon epithelial cells from tumor-free colon samples of APCMin/+ mice and WT mice were harvested as described previously (14).
Mouse embryonic fibroblasts.
Double-homozygous embryos (Dnmt3b+/+ and Rosa-rtTA+/+) were harvested 12.5 days after conception. Growth arrest of cells was achieved by 24 Gy irradiation or mitomycin C treatment and verified using BrdU staining. After 14 days of dox treatment, cells were harvested for DNA methylation analysis. See Supplemental Methods for details.
Lgr5 and Dnmt3b mRNA FISH.
Frozen sections of colon samples were hybridized with labeled probes targeting mouse Lgr5 and Dnmt3b mRNA (12). Individual mRNA molecules were located using the semiautomated method described previously (12). Lgr5-positive cells were manually identified to calculate Dmnt3b mRNA density in intestinal stem cells. See Supplemental Methods for details and probe sequences.
cDNA and quantitative PCR.
cDNA production and quantitative PCR was conducted as described previously (5). See Supplemental Methods for details and primer sequences.
DNA methylation analysis (mass array platform).
Genomic DNA was sodium bisulfite converted and PCR amplified. When feasible, amplicons were designed to cover CpG islands in the same region as the 5′ untranslated region. MassCLEAVE biochemistry, mass spectra acquisition, and methylation ratio analysis was performed as previously described (7). Primers are listed in Supplemental Methods.
DNA methylation analysis (reduced representation bisulfite sequencing).
RRBS methylation analysis was conducted as described previously (8). Briefly, mouse genomic DNA was digested with MspI, and resulting fragments between 40 and 220 bp were subjected to sequencing (Illumina genome Analyzer II) (8). Sequence reads that mapped to a promoter region (defined as 2 kb up- and downstream of the transcription start site) were then used to calculate promoter methylation. Raw data and processed RRBS data were deposited in GEO ( http://www.ncbi.nlm.nih.gov/geo; accession no. GSE26758). See Supplemental Methods for details.
Statistics.
All correlations were performed using the Pearson correlation. Significant difference in methylation fraction for the RRBS data was assayed with the moderated 2-tailed t test, corrected for FDR, with significance defined as FDR P value less than 0.01. Significant difference for colon mass array methylation data was assayed by 2-tailed t test and for cell culture mass array data by 2-tailed paired t test. For RNA FISH, we obtained errors by bootstrap resampling; for P values, we computed the fraction of events that the null hypothesis would be satisfied given a random resampling of the data set. In all cases, P values less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Jessica Dausman, Ruth Flannery, and Qing Gao for help with maintenance of the mouse colony and Tom DiCesare for support in preparation of figures. E.J. Steine is supported by Prins Bernhard Cultuurfonds “Banning-de Jong Fonds”; H.G. Linhart is supported by a fellowship from the Fritz-Thyssen Stiftung. Research described in this article was supported by grants to R. Jaenisch by Philip Morris International and by NIH grant RO1-CA087869. A. Raj was supported by a Burroughs-Wellcome Fund Career Award at the Scientific Interface. A. Raj was supported by a Burroughs-Wellcome Fund Career Award at the Scientific Interface. A. van Oudenaarden is supported by an NIH Director’s Pioneer award and award U54CA143874 from the National Cancer Institute.
Footnotes
Conflict of interest: M. Ehrich is employed by Sequenom Inc. S. Reddy is employed by Oral Cancer Prevention International.
Citation for this article: J Clin Invest. 2011;121(5):1748–1752. doi:10.1172/JCI43169.
References
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447(7143):433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 3.Opavsky R, et al. CpG island methylation in a mouse model of lymphoma is driven by the genetic configuration of tumor cells. PLoS Genet. 2007;3(9):1757–1769. doi: 10.1371/journal.pgen.0030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nosho K, et al. DNMT3B expression might contribute to CpG island methylator phenotype in colorectal cancer. Clin Cancer Res. 2009;15(11):3663–3671. doi: 10.1158/1078-0432.CCR-08-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linhart HG, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21(23):3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39(2):157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 7.Ehrich M, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102(44):15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39(2):232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 10.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 11.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 12.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5(10):877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velicescu M, Weisenberger DJ, Gonzales FA, Tsai YC, Nguyen CT, Jones PA. Cell division is required for de novo methylation of CpG islands in bladder cancer cells. Cancer Res. 2002;62(8):2378–2384. [PubMed] [Google Scholar]
- 14.Fujimoto K, Beauchamp RD, Whitehead RH. Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology. 2002;123(6):1941–1948. doi: 10.1053/gast.2002.37065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.