Abstract
Regulation of the sumoylation system at the level of gene expression has not yet been explored. To begin to define transcriptional regulatory features, the promoter region for the SUMO1 gene was cloned from human genomic DNA and characterized. Initially, a 532 base pair fragment upstream of and including the predicted SUMO1 transcription start site (TSS) was cloned and shown to possess promoter activity. Subsequent deletion analysis showed that a smaller fragment containing 158 bp upstream of the TSS region exhibited basal promoter activity in both human and rodent cell lines. Within this basal promoter fragment, there were predicted binding sites for numerous transcription factors, including the nude mouse gene product, Whn (FoxN1). Electrophoretic mobility shift assays showed that Whn could bind to an ACGC motif adjacent to the TSR, and in transfection studies Whn stimulated a 3-fold increase in transcription from this cloned promoter in keratinocytes (HaCaT cells). Mutation of the ACGC motif abrogated both Whn binding and transcriptional activation, indicating that the Whn effect is likely due to direct interaction with this promoter element. Consistent with these observations on the cloned promoter region, Whn also modestly stimulated transcription from the endogenous, genomic SUMO1 promoter in HaCaT cells, consistent with Whn potentially playing a regulatory role for SUMO1 transcription in keratinocytes.
Keywords: Sumoylation, Differentiation, Keratinocytes
Introduction
Sumoylation is a dynamic post-translational modification process whereby a SUMO moiety is covalently attached to substrates via an isopeptide bond linking the carboxyl terminus of SUMO to the epsilon amino group of a lysine residue in the target protein [1]. In humans, there are four SUMO genes each encoding a polypeptide related to ubiquitin [2, 3]. SUMO addition to substrate proteins is catalyzed by a series of enzymatic steps utilizing the same biochemistry as ubiquitinylation, but involving a different and exclusive set of E1, E2, and E3 enzymes. Initially, SUMO is translated as a precursor that is proteolytically processed near the carboxyl terminal to reveal a functional diglycine motif [4]. This endoproteolytic cleavage is performed by SUMO proteases that can also function in removal of SUMO groups from substrates [5]. After maturation, SUMO undergoes an ATP-dependent activation step resulting in a thioester bond between SUMO and a cysteine residue in the heterodimeric activating E1 enzyme, SAE1/2. SUMO is subsequently transferred to the E2 conjugating enzyme, Ubc9, again forming a thioester linkage. Unlike the ubiquitin pathway where there are multiple E2 enzymes, Ubc9 is the sole conjugating enzyme for SUMOs. Lastly, SUMO is transferred from Ubc9 to the target protein. While this final transfer does not absolutely require an E3 ligase as in ubiquitinylation, several SUMO specific E3 enzymes have now been identified, including RanBP2 [6], PIAS proteins [7, 8], and Pc2 [9].
Sumoylation has now been linked to a variety of fundamental cellular processes, including transcriptional regulation, nucleocytoplasmic transport, DNA replication/ repair [10, 11], and differentiation [12]. Studies in yeast [13], C. elegans [14], and mice [15] also demonstrate an essential overall requirement for sumoylation in growth and development. Recent mammalian proteomics studies reveal that SUMO modification is widespread, and that hundreds of proteins are likely targeted by this modification, though the predominant class is nuclear transcription factors [16–18]. Given the emerging global importance of sumoylation for transcription, as well as for other critical cellular functions, there is surprisingly little known about the transcriptional regulation of the genes encoding the components of the sumoylation system. To facilitate characterization of the transcriptional control of SUMO component genes, we have begun to clone several of the relevant promoters. Here we report the cloning and initial characterization of the human SUMO1 promoter region.
Materials and methods
Cloning and truncation of the SUMO1 promoter
The region on human chromosome 2 (from contig NT 005403) containing the predicted SUMO1 promoter regions was identified using the Chip2Promoter software (Genomatix). Forward (5′-cagcagaattcgtttacaaatccttctcccttcc-3′) and reverse (5′-cagcagaattcttgtgttgggtcgttgcttt-3′) primers were chosen to amplify a 532 base pair fragment (designated SUMO1P for SUMO1 promoter) spanning the transcriptional start region (TSR) and to introduce exogenous EcoRI sites at both 5′ ends. Genomic DNA was extracted from a human keratinocyte line, HaCaT [19], using the DNeasy Kit (Qiagen) and was PCR amplified with the above primers. The amplification reaction had 50 ng genomic DNA in 25 μl containing 1× Taq polymerase buffer (Invitrogen), 2.0 mM MgCl2, 0.2 mM dNTPs, 1.6 mM of each primer, and 1 U of Taq polymerase (Invitrogen). Amplification was for 30 cycles with the following cycle conditions: 94°C for 45 s, 60° for 30 s, and 72°C for 60 s. The reaction product was directly ligated to the pCR2.1 TA cloning vector (Invitrogen) and was sub-cloned into the reporter vector, pSEAP2-BASIC (BD Biosciences), using the EcoRI sites to generate pSEAP-S1P. A mutant version of the promoter in which the central CG pair of the Whn binding site proximal to the transcription start site (see Fig. 1) was changed to TA was constructed with the Quik Change Site-Directed Mutagenesis kit (Stratagene). Truncations of the SUMO1P fragment were constructed by PCR amplification from pSEAP-S1P to generate four nested subfragments using the original forward primer and the following new primers: T1, 5′-gtgctagctagcggaagttactgcagcc-3′; T2, 5′-gtgctagcgaaggagctgacaaaactgc-3′; T3, 5′gtgctagccaatctaggttgtgagagtg-3′; and T4, 5′-gtgctagcctagagctagaagtactggc-3′. PCR products of each reaction were cloned directly into pCR2.1 and then subcloned into pSEAP-BASIC to generate final clones designated T1–T4. The identity and sequence fidelity of all clones and constructs were confirmed by DNA sequencing.
Fig. 1.
Amplification of the SUMO1 promoter region from human genomic DNA. a Sequence of the SUMO1 promoter region derived from GenBank. The location of three mapped cDNA 5′ ends (GenBank Accession numbers BC006462, BC066306, and BC053528) are indicated with arrows, and the most 5′ position was designated the +1 nucleotide. Predicted binding sites for the Whn transcription factor and for Sp1 are as indicated; numerous other potential transcription factor binding sites are located in the region, but at not shown for simplicity. The 5′ endpoints of the truncations used in Fig. 3 are indicated as T2, T3, and T4. b Agarose gel of the PCR reaction use to amplify the SUMO1 promoter region. Lane 1 shows a 100-bp ladder and lane 2 is a sample of the PCR reaction using human genomic DNA and the primers described in “Materials and methods”
Transfections and SEAP assay
CHO or HaCaT cells were seeded at 4.0 × 105 cells per well in 6-well plates in DMEM Complete medium (Hy-clone) with 5% fetal bovine serum and were transfected the next day with Lipofectamine 2000 (Invitrogen) as suggested by the manufacturer. Each sample contained 3 μg of a pSEAP reporter vector construct plus 3 μg of either empty pCDNA3.1 or the pKSB51-Whn expression vector DNA (provided by T. Boehm). Cells were incubated at 37°C in a 5% CO2 incubator for 4 h, then the medium was removed and replaced with fresh medium. For assessment of alkaline phosphatase (AP) activity, 110 μl aliquots were removed at 0, 24, 48, and 72 h, with the 0 time sample taken immediately after the medium replacement. Samples were stored at −20°C until all time points were collected and were then assayed together. AP activity was determined with the Great EscAPe SEAP Chemiluminescent Assay (BD Biosciences) and a Lumi Count Plate luminometer (Packard Instrument Company). Luminometer readings for the transfected samples were converted to AP units using a standard curve generated with purified AP. All samples were analyzed in triplicate at each time point. Data reported in the figures are the average of 2–4 independent experiments. Statistical significance was determined by t-tests.
Electrophoretic mobility shift assay (EMSA)
The EMSA assay was performed with blunt-ended, double-stranded oligonucleotides and the his-tagged Whn DNA binding domain (WhnDBD) polypeptide as described by Schlake et al. [20], using their wild-type oligonucleotide (5′-atagggcgaattgggtaccaaagggacgctatcgagctccagcttt-3′) as our positive control. The oligonucleotide derived from the SUMO1 promoter region had the sequence 5′-ctcctccctgcgcgaagcggaagtgacgcgaggcgtagcggaagtt-3′ (putative Whn binding site underlined with the core motif in bold), and the mutant oligonucleotide had the CG in the core motif changed to TA. Oligonucleotides were end-labeled with T4 polynucleotide kinase and gamma-32P-ATP, and free radiolabel was removed by two passages through a Micro Bio-Spin P6 column (Bio-Rad Laboratories, Inc.). To purify the His-tagged WhnDBD protein, a 250 ml culture of E. coli M15 (Qiagen) expressing the pQE32-WhnDBD plasmid (kindly provided by T. Schlake and T. Boehm) was grown at 30°C to an OD600 of 1.0 then induced by adding IPTG to a final concentration of 0.2 mM. Incubation was continued for 4 h at room temperature, the cells were harvested by centrifugation, and the bacterial pellet was resuspended in lysis buffer (1× PBS, 10 mM β-mercaptoethanol, 10 mM imidazole, 860 mM NaCl, 5 mM PMSF and 1 mg/ml lysozyme). Resuspended bacteria were lysed by sonication, the lysate was cleared by centrifugation for 15 min at 13,000×g, and the supernatant was incubated with Ni-NTA agarose (Qiagen) for 3 h at 4°C on a circular rotor. After four washes with the wash buffer (1× PBS, 20 mM imidazole, 860 mM NaCl), the bound protein was eluted four times with 0.6 ml of elution buffer (1× PBS, 500 mM imidazole and 860 mM NaCl). The eluted fractions were dialyzed against 1× PBS and 10% glycerol, and were stored at −70°C. Protein quantitation was determined with a Bradford assay. For the binding reactions, 2.0 μg of the WhnDBD protein was incubated with 10 ng of labeled oligonucleotides for 30 min at 25°C. Binding reactions also contained 1 ug of poly DI-DC as a nonspecific competitor nucleic acid and 20 μg of BSA as a nonspecific protein competitor. After incubation for 30 min, 10× loading dye was added, samples were electrophoresed on 6% TBE gels, and the dried gels were visualized by autoradiography as previously described [21].
Quantitative real-time PCR
HaCaT cells plated in 6-well plates (4 × 105 cells/well) were transfected as above using 3 μg of a murine Whn (FoxN1) expression vector DNA (kindly provided by T. Boehm) or empty vector DNA (pCDNA3.1). Transfection efficiencies were ~30% as monitored by parallel transfection of pEGFP-C1 (BD Biosciences). RNA was harvested at 24-h intervals post-transfection using the RNAqueous kit (Ambion) and was stored at −80°C. The SUMO1 transcript was detected using custom LUX-primers (Invitrogen) with a FAM labeled forward primer (5′-gaatctcttggacaggatagcagtgaga[FAM]tc-3′) and an unlabeled reverse primer (5′- tcattggaacaccctgtctttg-3′). Each primer spans an exon-exon junction; therefore, amplification should be restricted to authentic transcript-derived cDNA and not genomic DNA. The reverse transcription and quantitative PCR reactions were performed in a single well of the real-time PCR plate using the one step RT-PCR Super Script III kit (Invitrogen). The RT step was performed for 20 min at 40°C followed by 90 s at 94°C to activate the Platinum Taq. For the quantitative real-time PCR, each 50 ll total reaction contained 50 ng of harvested RNA and 0.2 μM of each primer in addition to the RT-PCR Super Script III (Invitrogen) kit components. Subsequent amplification was performed for 40 cycles with the following steps: 30 s, 94°; 30 s, 60°C; 60 s, 72°C. The RT plate was read with an ABI 7500 real time PCR system machine, and readings of the FAM fluorescence were taken during the 72°C step. The LUX 18s rRNA primer set (Invitrogen) was used as the internal control. Average Ct values for the SUMO1 transcript were determined for each sample time (0–96 h), and the corresponding average Ct for the 18s rRNA was subtracted to generate the ΔCt. The ΔCCt for each of the 24–96 h samples was subtracted from the ΔCt for the 0 h sample to generate the ΔΔCt. Fold increase at each time point was determined as 2ΔΔCt. Each sample shown represents the average of RNAs collected from three independent transfections.
In vivo sumoylation
To evaluate the effect of exogenous Whn expression on sumoylation in HaCaT cells, the sumoylation state of a well-characterized SUMO substrate, RanGAP1 [22], was determined by immunoblotting. Transfections were as for the quantitative real-time PCR. At 24-h intervals post transfection, whole-cell extracts were prepared and analyzed by immunoblotting with either anti-RanGAP1 monoclonal antibody 19C7 (Zymed Laboratories) or anti-tubulin (Santa Cruz Biotechnology). Whole-cells extracts were prepared by direct extraction of plated cells with a 1:1 mixture of 1× RIPA buffer (50 mM Tris–Cl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 15 mM MgCl2, 1% NP40, 0.1% SDS, 1 mM DTT, 1:200 Protease Inhibitor Cocktail [Sigma], 10 mM N–ethylmaleimide) and 4× SDS-sample buffer (100 mM Tris–Cl, pH 6.8, 20% glycerol, 8% SDS, 0.02% bromophenol blue, 4% β-mercaptoethanol). Extracts were heated for 5 min at 95°C, electrophoresed on 10% SDS polyacrylamide gels, transferred to Immobilon-P membranes (Millipore), and blocked for 15–20 min with 3% nonfat milk in TTBS (150 mM NaCl, 50 mM Tris–Cl, pH 7.4, 0.005% Tween 20). Blocked membranes were incubated for 1 h with 1:5000 (anti-RanGAP1) or 1:15000 (anti-tubulin) dilutions of the primary antibodies in TTBS. After removal of the primary antibodies, the membranes were rinsed with TTBS and incubated for 1 hr with a 1:10000 dilution of the HRP-conjugated secondary antibody prior to detection with the Western Lightning Chemiluminescence reagent (PerkinElmer) as suggested by the manufacturer. The immunoblots were developed using horseradish peroxidaseconjugated secondary antibodies (Santa Cruz Biotechnology), the Western Lightning Chemiluminescence reagent (PerkinElmer), and X-ray film. RanGAP1 and sumoylated RanGAP1 bands were quantitated by densitometry using an Innotech Alphaimager and were normalized to the α-tubulin.
Results
Cloning the SUMO1 promoter
Changes in sumoylation profiles with varying cellular states are well documented, but little is known about control of the sumoylation-related genes at the transcriptional level. To perform a comprehensive exploration of the SUMO system gene regulation, the promoter regions for several of the major SUMO pathway genes are being cloned and characterized. Here we report the initial findings on the human SUMO1 gene promoter. The 5′ ends of several SUMO1-encoding cDNAs map within a 30 bp region on human chromosome 2 (Fig. 1a), and predictive algorithms suggested that the sequence ~500 bps upstream of this cluster would contain a functional promoter. Consequently, this region was amplified from human genomic DNA by high stringency PCR to yield a 532 bp fragment for analysis (Fig. 1b). The amplicon was cloned into a pCR2.1 TA vector and sequenced. The clone sequence was identical to the original genomic sequence shown in Fig. 1a. For reference purposes, the most 5′ transcriptional start site (TSS) was designated the +1 position.
Characterization of SUMO1 promoter activity
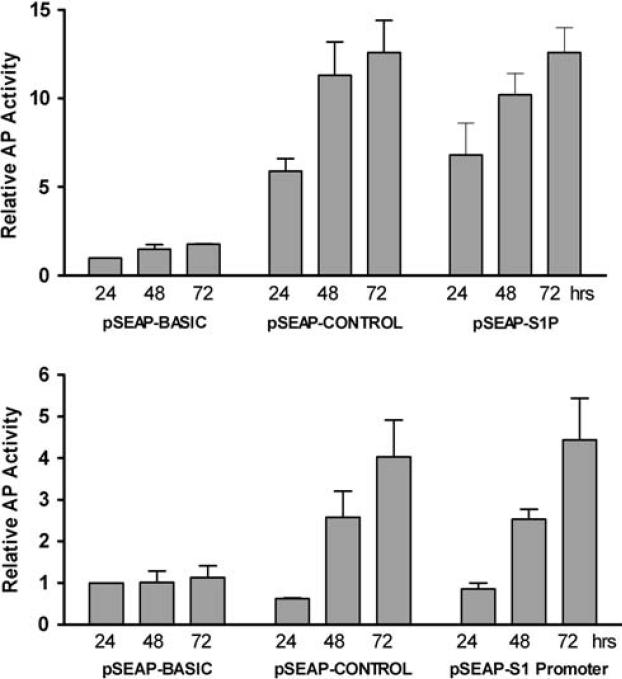
To assess promoter activity, the cloned fragment was transferred from the TA plasmid to the promoterless pSEAP-BASIC reporter vector to generate the pSEAP-S1Promoter construct. Transient transfections were performed into both human (HaCaT) and rodent (CHO) cell lines, and reporter activity was assessed from 1–3 days post-transfection (Fig. 2). In both cell types the pSEAP-S1Promoter construct produced reporter activity comparable to the pSEAP-CONTROL vector which contains an SV40 early promoter. In CHO cells, the relative activity values at all time points for pSEAP-S1Promoter and pSEAP-CONTROL were significantly greater than those for pSEAP-BASIC (P < 0.05), while in HaCaT cells only the 48 and 72 h values were significant. Note that the relative reporter activity for both pSEAP-S1P and pSEAP-CONTROL was also much greater in the CHO cells compared to the HaCaT cell, likely reflecting the higher transfection efficiency for the CHO cells (not shown). For both the pSEAP-S1Promoter and pSEAP-CONTROL plasmids, reporter activity increased during the time course examined, in contrast to the promoterless control vector (pSEAP-BASIC) which showed relatively constant and low basal reporter activity throughout this time period. Observed reporter activity for pSEAP-S1Promoter was also dependent upon the amount of DNA used in the transfections (data not shown). Overall, these results indicate that the cloned genomic fragment in the pSEAP-S1Promoter construct contains intrinsic promoter activity.
Fig. 2.
Promoter activity of the pSEAP-S1Promoter construct. The pSEAP-S1Promoter vector, the positive control (pSEAP-CONTROL), and the negative control (pSEAP-BASIC) plasmids were each individually transfected into CHO (a) or HaCaT (b) cells. Culture supernatants were collected at 24, 48, and 72 h post-transfection and analyzed for alkaline phosphatase (AP) activity. Values for all samples were normalized to the 24 h pSEAP-BASIC sample which was set at 1.0. Values shown at each time point are the average of 2–3 independent experiments. Values for the 24, 48, and 72 h pSEAP-CONTROL and pSEAP-S1Promoter samples in CHO cells (a) were all statistically significant compared to the pSEAP-BASIC samples (P < 0.05). In HaCaT cells (b), only the 48 and 72 h samples achieved statistical significance
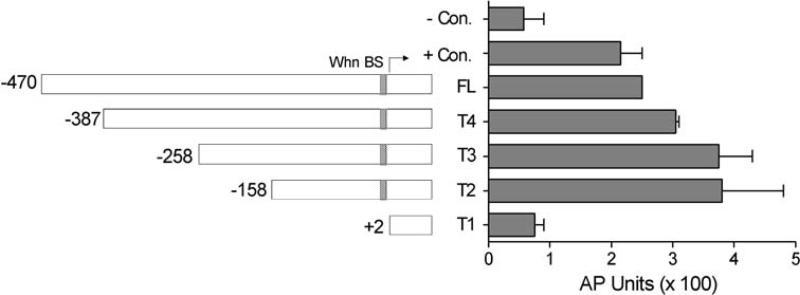
To determine the location of sequences critical for basal SUMO1 promoter activity, a series of truncations (T1–T4) of the 470 base pair region upstream of the TSS was constructed into the pSEAP-BASIC reporter vector. After transfection, each truncation was assayed for its ability to express the alkaline phosphatase reporter enzyme (Fig. 3). Removal of upstream sequences from −470 to −158 did not decrease promoter activity, indicating that all the requisite basal elements were present in the T2 clone. Subsequent truncation to +2 (T1 clone) totally abrogated promoter activity, confirming that critical basal promoter elements reside in the region from −158 to +2. While not statistically significant, there was a slight increase in promoter activity for the T4, T3, and T2 truncations compared to the parental clone, suggesting that there might be a weak negative regulatory element in the distal portion of the cloned region.
Fig. 3.
Truncation analysis of the SUMO1 promoter region. The full-length SUMO1 promoter (the pSEAP-S1Promoter construct designated FL in the figure) and truncated constructs (T4–T1) are diagramed with the position of the 5′ end of each fragment indicated relative to the transcription start site (marked with arrow). The position of the predicted Whn binding site is also indicated. The measured activity of each truncation is shown in the graph. Each construct was transfected into HaCaT cells and supernatants were collected at 48 h post-transfection for analysis of AP activity. Values are reported as actual AP units without normalization and are the average of two independent experiments (error bars for the FL sample are too small to be visible in this figure). The negative (−) control was the pSEAP-BASIC vector and the positive (+) control was the pSEAP-CONTROL vector. The values for the T4, T3, and T2 samples were not significantly different from the FL sample (P > 0.05) while the reduction in the T1 sample was statistically significant (P < 0.05)
Functional evaluation of a predicted Whn binding site
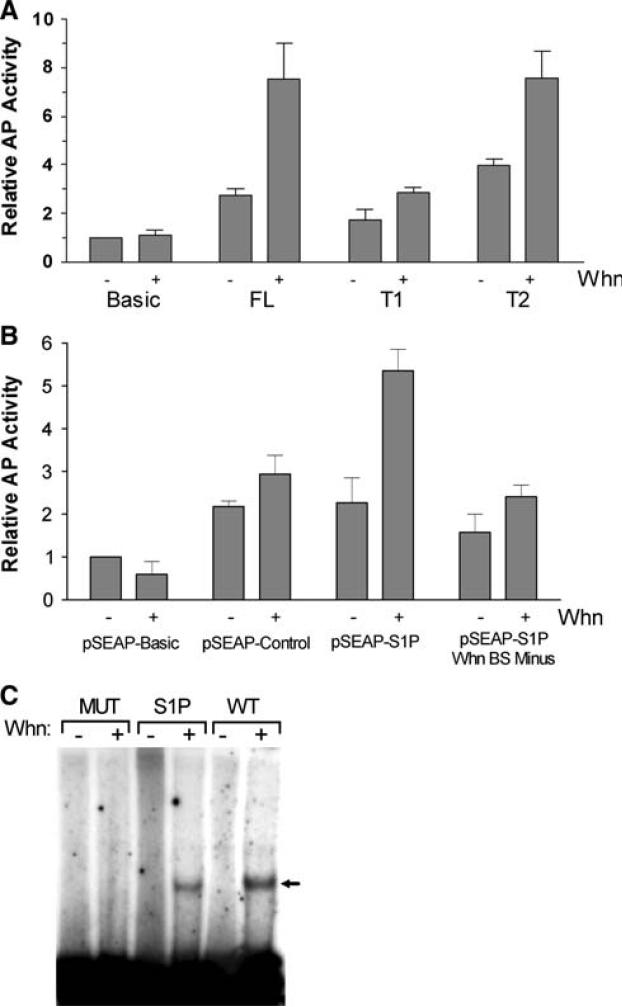
Interrogation of the −158 to +2 region with the MatIn-spector software (Genomatix) revealed over 20 potential transcription factor binding sites upstream of the TSS, including a possible site for the Whn (FoxN1) protein (see Figs. 1a, 3). A critical feature for Whn binding is a central core ACGC motif [20], and this core is present in this predicted binding site. To evaluate the responsiveness of the SUMO1 promoter to this transcription factor, cotransfection experiments were performed with various promoter constructs and an expression vector for Whn. Endogenous Whn in HaCaT cells could not be detected by immunoblotting, but expression of exogenous Whn was confirmed (data not shown). Exogenous expression of Whn consistently produced a statistically significant (P < 0.05) 2–4 fold stimulation of AP activity for the full-length SUMO1 promoter clone (FL) (Fig. 4a and data not shown). A similar transcriptional enhancement was observed for the T2 clone (P < 0.05), suggesting that sequences upstream of −158 were not required for this effect. Deletion of the predicted Whn binding site in the T1 clone eliminated activation by Whn.
Fig. 4.
Whn effects on the cloned SUMO1 promoter. a The full-length pSEAP-SUMO1 promoter (FL) and two truncations (T1 and T2) were co-transfected into HaCaT cells with either 3 μg of pCDNA (−lanes) or 3 μg of the Whn expression vector (+lanes). Supernatants were collected at 48 h post-transfections and analyzed for AP activity. Activity results were normalized to the value for the negative control (pSEAP-BASIC) in the absence of Whn. The increase in the FL and T2 samples in the presence of Whn was statistically significant (P < 0.05), while the increase for T1 was not. b The various plasmids indicated were transfected in HaCaT cells with pCDNA (−lanes) or with the Whn plasmid (+lanes) as in part a. The pSEAP-S1P Whn BS Minus plasmid was constructed from the pSEAP-S1Promoter and contains a CG to TA change in the core motif of the Whn binding site. Samples were collected and analyzed as in part a and represent the average of 2–3 independent experiments. Only the increase seen with pSEAP-S1P in the presence of Whn was statistically significant (P < 0.05). c EMSA analysis of the purified Whn DNA binding domain (WhnDBD) protein. WhnDBD binding to three different oligonucleotides was determined by EMSA as described in “Material and methods”. The WT oligonucleotide was as described previously [20] and contains a strong Whn binding site. The SUMO1P (S1P) oligonucleotide is derived from the SUMO1 promoter region and contains the predicted Whn binding site. The MUT oligonucleotide has the CG from the critical ACGC core motif of the Whn binding site changed to TA, a change which was previously shown to eliminate WhnDBD binding [20]. Minus (−) lanes contained a comparable amount of the bovine papillomavirus E2 DNA binding protein as a specificity control. No E2 binding site was present in the oligo substrates
To further localize the Whn-responsive element, the predicted Whn binding site was mutated in the context of the full-length SUMO1 promoter. Mutation of the CG dinucleotide in the predicted core motif to TA abrogated Whn responsiveness without significantly impairing basal activity in the absence of Whn (Fig. 4b). The very slight activation by Whn seen with the mutant promoter and the pSEAP-CONTROL vector, as well as seen with the T1 clone in Fig. 4a, likely reflects nonspecific promoter activation as previously described and was not statistically significant [23]. The combined results of the point mutations and the truncations strongly support the presence of a Whn-responsive promoter element located adjacent to the TSR.
As further evidence for the functionality of the TSR proximal Whn binding site, direct binding of Whn protein to this sequence was tested by EMSA (Fig. 4c). The DNA binding domain of Whn (WhnDBD) was expressed in E. coli and purified for in vitro testing. The purified WhnDBD protein bound both the positive control oligonucleotide (WT) and the oligonucleotide containing the wild-type Whn binding site sequence derived from the SUMO1 promoter (S1P). In contrast, WhnDBD did not bind to a mutant oligonucleotide (MUT) whose central core motif CG pair was changed to TA, thus confirming the binding specificity. The in vitro binding of WhnDBD to sequences adjacent to the TSR in the SUMO1 promoter supports the conclusion that the in vivo transactivation by Whn is a direct effect on the SUMO1 promoter.
Functional evaluation of the endogenous SUMO1 promoter
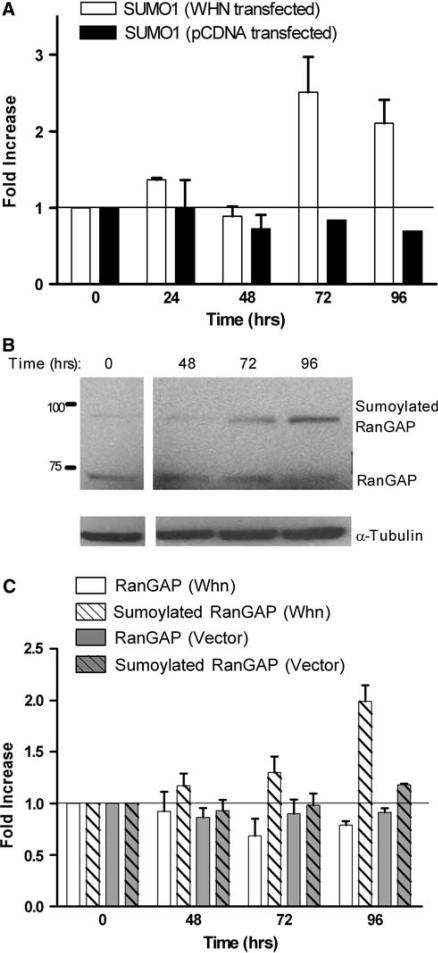
To verify that the endogenous SUMO1 promoter in the chromosomal context is activated by Whn, the Whn expression construct was transfected into HaCaT cells and the endogenous SUMO1 transcript was monitored by quantitative real-time PCR. By 72 h post-transfection there was a 2–3 fold increase in the amount of SUMO1 transcript in the Whn-transfected culture while the empty vector-transfected cells showed no change in amount of SUMO1 transcript (Fig. 5a). The observed increase in the Whn transfected samples at 72 and 96 h was statistically significant (P < 0.05). These results confirm that Whn can regulate the authentic SUMO1 promoter in its chromosomal context. Also, note that the actual transcriptional enhancement by Whn is likely greater as the transfection efficiency is only about 30% for these cells.
Fig. 5.
Effect of Whn on the endogenous SUMO1 promoter and overall sumoylation. a HaCaT cells were transfected with the Whn expression vector (WHN transfected) or the empty parental vector (pCDNA transfected) as indicated. RNA was collected at 24 h intervals from 0 to 96 h post-transfection, and the SUMO1 transcript was measured by quantitative real-time PCR. SUMO1 transcript values were normalized to the 18S RNA value for each sample and then compared to the 0 time samples. The increase in the SUMO1 transcripts in the presence of Whn at 72 and 96 h was statistically significant (P < 0.05). b HaCaT cells were transfected with the Whn expression vector, and total protein extracts were prepared at times 0, 48, 72, and 96 h post-transfection. Samples were electrophoresed on a 10% SDS polyacrylamide gel and immunoblotted with anti-RanGAP1 and anti-tubulin. The positions of the molecular weight markers in kDa are indicated on the left. c The relative levels of RanGAP1 and sumoylated RanGAP1 were quantitated by densitometric analysis of immunoblots such as shown in b. HaCaT cells were transfected with either the Whn expression vector (Whn samples) or the empty parental pCDNA plasmid (Vector samples), and extracts were prepared and immunoblotted as in b. The quantitation represents the average of three experiments. Compared to the 0 time sample, the increase in sumoylated RanGAP seen in the presence of Whn at 96 h was statistically significant (P < 0.05). None of the other samples showed any significant change
The increase in expression from the SUMO1 promoter suggests that overall sumoylation might increase due to more abundant free SUMO1. To evaluate this possibility, we first assessed the extent of total sumoylation following Whn transfection. However, the total pattern of sumoylated proteins was complex and no consistent increase could be observed, again likely reflecting low transfection efficiency and expression of Whn in only a small subset of the total cell population (data not shown). As an alternative, the extent of modification of a prominent SUMO1 substrate, RanGAP1, was examined. In parallel with the Whn-dependent increase in transcription of the SUMO1 promoter, by 96 h we observed a statistically significant (P < 0.05) increase in sumoylated RanGAP1 coupled with a slight decrease in the unsumoylated form of RanGAP1 (Fig. 5b). Quantitation of multiple independent experiments confirmed a 2-fold increase in sumoylated RanGAP1 following Whn transfection, while no effect on RanGAP1 sumoylation was detected in parallel transfections with empty vector (Fig. 5c). Other SUMO1 substrates have not yet been examined; however, the findings with RanGAP1 imply that the Whn overexpression leads to a modest functional upregulation of cellular sumoylation in keratinocytes.
Discussion
Changes in the levels of SUMO proteins and in the overall sumoylation pattern are known to occur in response to various signals [18], but little is known about control of this process at the level of transcription of the sumoylation components. Identification of effectors and their response elements affecting SUMO system genes will be necessary to understand global regulation of the sumoylation process in response to biologically relevant changes in growth conditions. To begin investigation of the regulation of SUMO1, an active promoter was cloned from the genomic region containing the major transcription start sites (TSS) for the human SUMO1 gene. The initial cloned fragment extended from 470 bp upstream of the most 5′ TSS to 62 bp downstream. This fragment exhibited significant promoter activity in human and rodent cells, indicating that it contained a functional basal promoter element. When the cloned region was truncated to −158 there was no loss of promoter function. Further truncation to +2 completely abrogated promoter activity, indicating that essential basal promoter elements are located between −158 and +2. This finding is consistent with a recent comparative genomics study which found that conserved transcriptional regulatory motifs tend to cluster within 100 base pairs upstream of the TSS [24]. Whether or not there are additional regulatory elements further upstream or downstream of the cloned fragment has not been examined.
Inspection of the functional −158 to +2 region revealed the absence of a TATA element as was previously noted from an examination of the genomic DNA sequence upstream of the human SUMO1 gene [25]. Like other TATA-less promoters [26], the SUMO1 basal promoter region is GC rich and has multiple predicted Sp1 binding sites. In addition to the possible Sp1 sites, there are a number of other predicted transcription factor binding motifs in the −158 to + 2 regions, including NF1, WT1, E2F, and Whn (FoxN1). Whn was chosen as an example to demonstrate responsiveness of the cloned promoter to potential regulatory transcription factors. The results presented here clearly indicate that Whn can activate the SUMO1 promoter, at least under transfection conditions, but the contributions of the other transcription factors to SUMO1 promoter activity have not yet been tested. None of the other predicted transcription factor binding sites overlap the Whn site, so it is unlikely the loss of transcriptional activity observed with the Whn binding site mutants in Fig. 4 are due to effects on other factors.
Whn, a product of the Nude gene [reviewed in 27], is a member of the forkhead/winged helix family of transcription factors, and its expression is restricted to epithelial cells, including keratinocytes [28]. Whn expression is low in basal keratinocytes and increases rapidly upon induction of differentiation, suggesting an early role for Whn in the differentiation process [29]. However, only a few genes have been identified as targets for Whn activation [30, 31], and none of these are related to the sumoylation system or to transcription factors that might regulate the SUMO system genes. The biology of Whn implies that it would not be a general regulator of the SUMO1 promoter in most cell types, and instead would have at best a more limited role confined to certain epithelial cells. The results presented here suggest that Whn can stimulate the SUMO1 promoter, but additional studies will be required to establish whether or not Whn is a functionally significant transcription factor under normal cellular conditions.
Acknowledgments
We gratefully thank the following for providing valuable reagents for this work: Dr. T. Boehm (pKSB51-Whn), Dr. Schlake (pQE32-WhnDBD), Dr. S. Safe (pCDNA3.1-Sp1), Dr. L. Bundy (pCDNA3.1-C/EBP beta 1), and Dr. C. Lima (pET-Ulp1403–621). This work was supported by CA089289 from the National Cancer Institute.
Abbreviations
- TSS
Transcriptional start site
- AP
Alkaline phosphatase
- S1P
SUMO 1 promoter
- PMSF
Phenylmethylsulfonylflouride
References
- 1.Desterro JM, Thomson J, Hay RT. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. doi:10.1016/S0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 2.Su HL, Li SSL. Molecular features of human ubiquitin-like SUMO genes and their encoded proteins. Gene. 2002;296:65–73. doi: 10.1016/s0378-1119(02)00843-0. doi: 10.1016/S0378-1119(02)00843-0. [DOI] [PubMed] [Google Scholar]
- 3.Bohren KM, Nadkarni V, Song JH, Gabbay KH, Owerbach D. A M55 V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J Biol Chem. 2004;279:27233–27238. doi: 10.1074/jbc.M402273200. doi:10.1074/jbc.M402273200. [DOI] [PubMed] [Google Scholar]
- 4.Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. doi: 10.1038/18457. doi:10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 5.Watts F. SUMO Proteases. In: Wilson VG, editor. Sumoylation: molecular biology, biochemistry. Horizon Bioscience; Norfolk: 2004. pp. 113–130. [Google Scholar]
- 6.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. doi:10.1016/S0092-8674(01)00633-X. [DOI] [PubMed] [Google Scholar]
- 7.Rogers RS, Horvath CM, Matunis MJ. SUMO modification of STAT1 and its role in PIAS-mediated inhibition of gene activation. J Biol Chem. 2003;278:30091–30097. doi: 10.1074/jbc.M301344200. doi:10.1074/jbc.M301344200. [DOI] [PubMed] [Google Scholar]
- 8.Ungureanu D, Vanhatupa S, Kotaja N, Yang J, Aittomaki S, Janne OA, Palvimo JJ, Silvennoinen O. PIAS proteins promote SUMO-1 conjugation to STAT1. Blood. 2003;102:3311–3313. doi: 10.1182/blood-2002-12-3816. doi:10.1182/blood-2002-12-3816. [DOI] [PubMed] [Google Scholar]
- 9.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. doi:10.1016/S0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 10.Girdwood DWH, Tatham MH, Hay RT. SUMO and transcriptional regulation. Semin Cell Dev Biol. 2004;15:201–210. doi: 10.1016/j.semcdb.2003.12.001. doi: 10.1016/j.semcdb.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Muller S, Ledl A, Schmidt D. SUMO: a regulator of gene expression and genome integrity. Oncogene. 2004;23:1998–2008. doi: 10.1038/sj.onc.1207415. doi: 10.1038/sj.onc.1207415. [DOI] [PubMed] [Google Scholar]
- 12.Deyrieux AF, Rosas-Acosta G, Ozbun MA, Wilson VG. Sumoylation dynamics during keratinocyte differentiation. J Cell Sci. 2007;120:125–136. doi: 10.1242/jcs.03317. doi:10.1242/jcs.03317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dohmen RJ, Stappen R, McGrath JP, Forrova H, Kolarov J, Goffeau A, Varshavsky A. An essential yeast gene encoding a homolog of ubiquitin activating enzyme. J Biol Chem. 1995;270:18099–18109. doi: 10.1074/jbc.270.30.18099. doi:10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- 14.Jones D, Crowe E, Stevens TA, Candido EPM. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: ubiquitin- conjugating enzymes, ubiquitin- activating enzymes, and ubiquitin-like proteins. Genome Biol. 2001;3:2.1–2.15. doi: 10.1186/gb-2001-3-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi T, Sharma P, Athanasiou M, Kumar A, Yamada S, Kuehn MR. Mutation of SENP1/SuPr-2 reveals an essential role for desumoylation in mouse development. Mol Cell Biol. 2005;25:5171–5182. doi: 10.1128/MCB.25.12.5171-5182.2005. doi:10.1128/MCB.25.12.5171-5182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao YM, Kwon SW, Anselmo A, Kaur K, White MA. Broad spectrum identification of cellular small ubiquitin-related modifier (SUMO) substrate proteins. J Biol Chem. 2004;279:20999–21002. doi: 10.1074/jbc.M401541200. doi:10.1074/jbc.M401541200. [DOI] [PubMed] [Google Scholar]
- 17.Vertegaal ACO, Ogg SC, Jaffray E, Rodriguez MS, Hay RT, Andersen JS, Mann M, Lamond AI. A proteomic study of SUMO-2 target proteins. J Biol Chem. 2004;279:33791–33798. doi: 10.1074/jbc.M404201200. doi: 10.1074/jbc.M404201200. [DOI] [PubMed] [Google Scholar]
- 18.Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol Cell Proteomics. 2005;4:56–72. doi: 10.1074/mcp.M400149-MCP200. doi:10.1074/mcp.M400149-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. doi:10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlake T, Schorpp M, Nehls M, Boehm T. The nude gene encodes a sequence-specific DNA binding protein with homologs in organisms that lack an anticipatory immune system. Proc Natl Acad Sci USA. 1997;94:3842–3847. doi: 10.1073/pnas.94.8.3842. doi:10.1073/pnas.94.8.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez A, Bazaldua-Hernandez C, West M, Woytek K, Wilson VG. Identification of a short, hydrophilic amino acid sequence critical for origin recognition by the bovine papillomavirus E1 protein. J Virol. 2000;74:245–253. doi: 10.1128/jvi.74.1.245-253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin- related polypeptide involved in targeting Ran-GAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. doi:10.1016/S0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 23.Brissette JL, Li J, Kamimura J, Lee D, Dotto GP. The product of the mouse nude locus, Whn, regulates the balance between epithelial cell growth and differentiation. Genes Dev. 1996;10:2212–2221. doi: 10.1101/gad.10.17.2212. doi:10.1101/gad.10.17.2212. [DOI] [PubMed] [Google Scholar]
- 24.Xie XH, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3 `UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. doi:10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howe K, Williamson J, Boddy N, Sheer D, Freemont P, Solomon E. The ubiquitin-homology gene PIC1: characterization of mouse (Pic1) and human (UBL1) genes and pseudogenes. Genomics. 1998;47:92–100. doi: 10.1006/geno.1997.5091. doi:10.1006/geno.1997.5091. [DOI] [PubMed] [Google Scholar]
- 26.Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 27.Schlake T. The nude gene and the skin. Exp Derm. 2001;10:293–304. doi: 10.1034/j.1600-0625.2001.100501.x. doi:10.1034/j.1600-0625.2001.100501.x. [DOI] [PubMed] [Google Scholar]
- 28.Lee D, Prowse DM, Brissette JL. Association between mouse nude gene expression and the initiation of epithelial terminal differentiation. Dev Biol. 1999;208:362–374. doi: 10.1006/dbio.1999.9221. doi:10.1006/dbio.1999.9221. [DOI] [PubMed] [Google Scholar]
- 29.Baxter RM, Brissette JL. Role of the nude gene in epithelial terminal differentiation. J Invest Dermatol. 2002;118:303–309. doi: 10.1046/j.0022-202x.2001.01662.x. doi:10.1046/j.0022-202x.2001.01662.x. [DOI] [PubMed] [Google Scholar]
- 30.Schlake T, Boehm T. Expression domains in the skin of genes affected by the nude mutation and identified by gene expression profiling. Mech Dev. 2001;109:419–422. doi: 10.1016/s0925-4773(01)00538-x. doi:10.1016/S0925-4773(01)00538-X. [DOI] [PubMed] [Google Scholar]
- 31.Schlake T, Schorpp M, Maul-Pavicic A, Malashenko AM, Boehm T. Forkhead/winged-helix transcription factor Whn regulates hair keratin gene expression: Molecular analysis of the nude skin phenotype. Dev Dyn. 2000;217:368–376. doi: 10.1002/(SICI)1097-0177(200004)217:4<368::AID-DVDY4>3.0.CO;2-Z. doi:10.1002/(SICI) 1097-0177(200004)217:4<368::AID-DVDY4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]