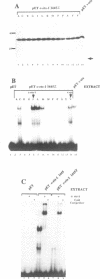
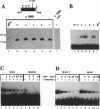
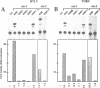
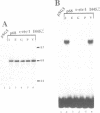
Abstract
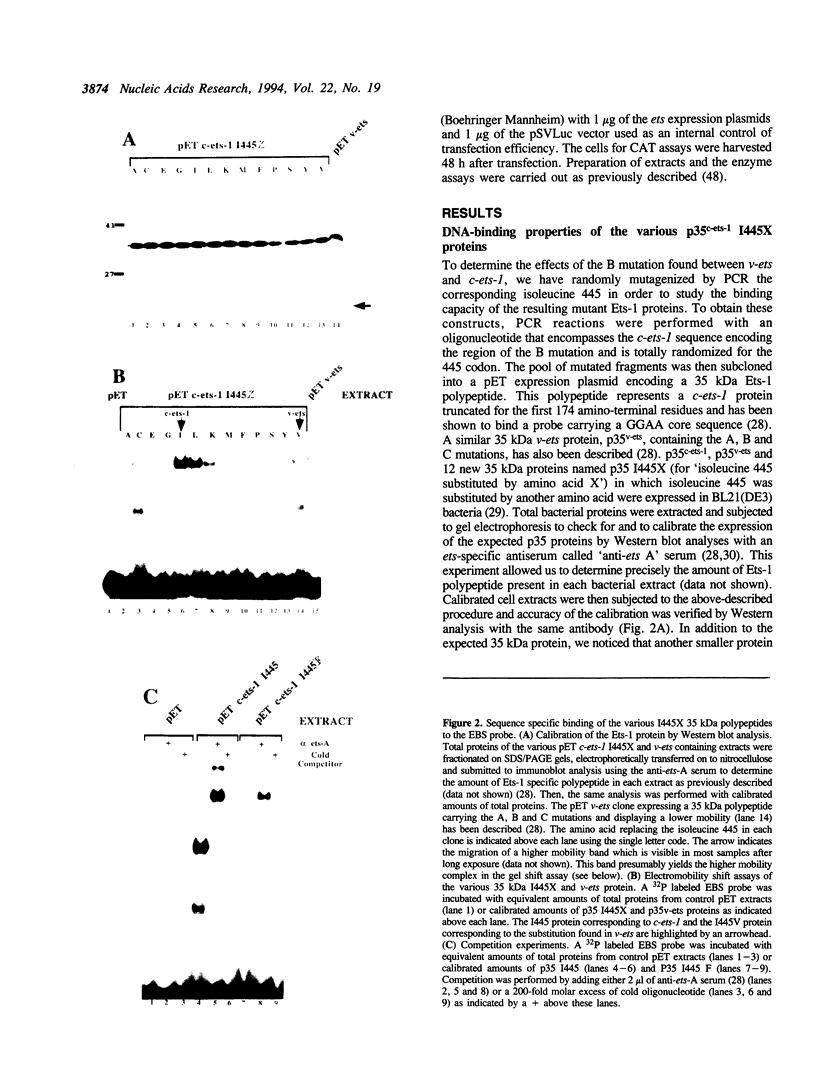
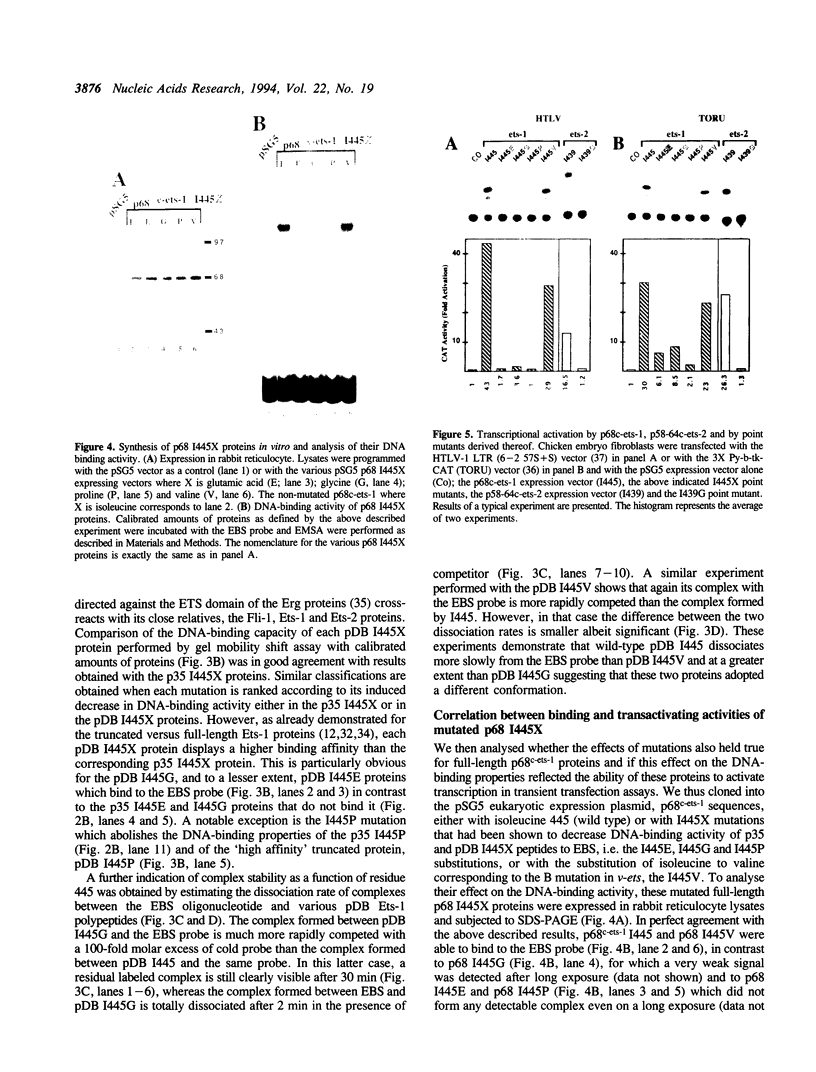
The c-ets-1 locus encodes two transcription factors, p54c-ets-1 and p68c-ets-1 that recognize purine-rich motifs. The v-ets oncogene of the avian retrovirus E26 differs from its cellular progenitor p68c-ets-1 by two amino acid substitutions (alanine 285 and isoleucine 445 in c-ets-1 both substituted by valine in v-ets, mutations A and B respectively) and its carboxy-terminal end (mutation C). The B mutation affects a well conserved residue in the carboxy-terminal 85 amino acids, ETS DNA-binding domain. To address the biological relevance of the B mutation found between v-ets and c-ets-1, we have randomly mutagenized isoleucine 445 of p68c-ets-1 by polymerase chain reaction. Using in vitro gel mobility shift assays, we show that this residue is crucial for the binding properties of c-ets-1 since the 12 mutations we have generated at this position, all diminish or even abolish the binding, to the 'optimized' Ets-1 binding site (EBS), of 35 kDa proteins corresponding to the 311 carboxy-terminal residues of c-ets-1. Among them, substitutions of isoleucine to glutamic acid, glycine or proline have the highest inhibitory effects. Similar results were obtained when the same mutations were introduced either in full-length p68c-ets-1 protein or into a carboxy-terminal polypeptide of 109 amino acids encompassing the ETS-domain which has previously been shown to display a very high binding activity as compared with the full-length protein. Consistent with the in vitro results, point mutations in p68c-ets-1 that decrease binding activity to EBS abrogate its ability to transactivate reporter plasmids carrying either the TPA Oncogene Response Unit of the Polyoma virus enhancer (TORU) or a sequence derived from the HTLV-1 LTR. Furthermore, as this isoleucine residue is rather well-conserved within the ETS gene family, we show that mutation of the corresponding isoleucine of c-ets-2 into glycine also abrogates its DNA-binding and hence, transactivating properties. Thus, the v-ets B mutation highlights the isoleucine 445 as an essential amino acid of the c-ets-1 and c-ets-2 DNA-binding domains.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anton I. A., Frampton J. Tryptophans in myb proteins. Nature. 1988 Dec 22;336(6201):719–719. doi: 10.1038/336719a0. [DOI] [PubMed] [Google Scholar]
- Bosselut R., Duvall J. F., Gégonne A., Bailly M., Hémar A., Brady J., Ghysdael J. The product of the c-ets-1 proto-oncogene and the related Ets2 protein act as transcriptional activators of the long terminal repeat of human T cell leukemia virus HTLV-1. EMBO J. 1990 Oct;9(10):3137–3144. doi: 10.1002/j.1460-2075.1990.tb07511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosselut R., Levin J., Adjadj E., Ghysdael J. A single amino-acid substitution in the Ets domain alters core DNA binding specificity of Ets1 to that of the related transcription factors Elf1 and E74. Nucleic Acids Res. 1993 Nov 11;21(22):5184–5191. doi: 10.1093/nar/21.22.5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulukos K. E., Pognonec P., Begue A., Galibert F., Gesquière J. C., Stéhelin D., Ghysdael J. Identification in chickens of an evolutionarily conserved cellular ets-2 gene (c-ets-2) encoding nuclear proteins related to the products of the c-ets proto-oncogene. EMBO J. 1988 Mar;7(3):697–705. doi: 10.1002/j.1460-2075.1988.tb02865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepieux P., Leprince D., Flourens A., Albagli O., Ferreira E., Stéhelin D. The two functionally distinct amino termini of chicken c-ets-1 products arise from alternative promoter usage. Gene Expr. 1993;3(2):215–225. [PMC free article] [PubMed] [Google Scholar]
- Duterque-Coquillaud M., Niel C., Plaza S., Stehelin D. New human erg isoforms generated by alternative splicing are transcriptional activators. Oncogene. 1993 Jul;8(7):1865–1873. [PubMed] [Google Scholar]
- Galson D. L., Hensold J. O., Bishop T. R., Schalling M., D'Andrea A. D., Jones C., Auron P. E., Housman D. E. Mouse beta-globin DNA-binding protein B1 is identical to a proto-oncogene, the transcription factor Spi-1/PU.1, and is restricted in expression to hematopoietic cells and the testis. Mol Cell Biol. 1993 May;13(5):2929–2941. doi: 10.1128/mcb.13.5.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia A., LaMontagne K., Reavis D., Stober-Grässer U., Lipsick J. S. Determinants of sequence-specific DNA-binding by p48v-myb. Oncogene. 1991 Feb;6(2):265–273. [PubMed] [Google Scholar]
- Gegonne A., Punyammalee B., Rabault B., Bosselut R., Seneca S., Crabeel M., Ghysdael J. Analysis of the DNA binding and transcriptional activation properties of the Ets1 oncoprotein. New Biol. 1992 May;4(5):512–519. [PubMed] [Google Scholar]
- Ghysdael J., Gegonne A., Pognonec P., Dernis D., Leprince D., Stehelin D. Identification and preferential expression in thymic and bursal lymphocytes of a c-ets oncogene-encoded Mr 54,000 cytoplasmic protein. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1714–1718. doi: 10.1073/pnas.83.6.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golay J., Introna M., Graf T. A single point mutation in the v-ets oncogene affects both erythroid and myelomonocytic cell differentiation. Cell. 1988 Dec 23;55(6):1147–1158. doi: 10.1016/0092-8674(88)90259-0. [DOI] [PubMed] [Google Scholar]
- Graf T., McNagny K., Brady G., Frampton J. Chicken "erythroid" cells transformed by the Gag-Myb-Ets-encoding E26 leukemia virus are multipotent. Cell. 1992 Jul 24;70(2):201–213. doi: 10.1016/0092-8674(92)90096-u. [DOI] [PubMed] [Google Scholar]
- Gunther C. V., Nye J. A., Bryner R. S., Graves B. J. Sequence-specific DNA binding of the proto-oncoprotein ets-1 defines a transcriptional activator sequence within the long terminal repeat of the Moloney murine sarcoma virus. Genes Dev. 1990 Apr;4(4):667–679. doi: 10.1101/gad.4.4.667. [DOI] [PubMed] [Google Scholar]
- Hagman J., Grosschedl R. An inhibitory carboxyl-terminal domain in Ets-1 and Ets-2 mediates differential binding of ETS family factors to promoter sequences of the mb-1 gene. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):8889–8893. doi: 10.1073/pnas.89.19.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. C. A structural taxonomy of DNA-binding domains. Nature. 1991 Oct 24;353(6346):715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Janknecht R., Nordheim A. Elk-1 protein domains required for direct and SRF-assisted DNA-binding. Nucleic Acids Res. 1992 Jul 11;20(13):3317–3324. doi: 10.1093/nar/20.13.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janknecht R., Nordheim A. Gene regulation by Ets proteins. Biochim Biophys Acta. 1993 Dec 23;1155(3):346–356. doi: 10.1016/0304-419x(93)90014-4. [DOI] [PubMed] [Google Scholar]
- Janknecht R., Zinck R., Ernst W. H., Nordheim A. Functional dissection of the transcription factor Elk-1. Oncogene. 1994 Apr;9(4):1273–1278. [PubMed] [Google Scholar]
- Karim F. D., Urness L. D., Thummel C. S., Klemsz M. J., McKercher S. R., Celada A., Van Beveren C., Maki R. A., Gunther C. V., Nye J. A. The ETS-domain: a new DNA-binding motif that recognizes a purine-rich core DNA sequence. Genes Dev. 1990 Sep;4(9):1451–1453. doi: 10.1101/gad.4.9.1451. [DOI] [PubMed] [Google Scholar]
- Kraut N., Frampton J., McNagny K. M., Graf T. A functional Ets DNA-binding domain is required to maintain multipotency of hematopoietic progenitors transformed by Myb-Ets. Genes Dev. 1994 Jan;8(1):33–44. doi: 10.1101/gad.8.1.33. [DOI] [PubMed] [Google Scholar]
- Laget M. P., Callebaut I., de Launoit Y., Stehelin D., Mornon J. P. Predicted common structural features of DNA-binding domains from Ets, Myb and HMG transcription factors. Nucleic Acids Res. 1993 Dec 25;21(25):5987–5996. doi: 10.1093/nar/21.25.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudet V., Niel C., Duterque-Coquillaud M., Leprince D., Stehelin D. Evolution of the ets gene family. Biochem Biophys Res Commun. 1993 Jan 15;190(1):8–14. doi: 10.1006/bbrc.1993.1002. [DOI] [PubMed] [Google Scholar]
- Lautenberger J. A., Burdett L. A., Gunnell M. A., Qi S., Watson D. K., O'Brien S. J., Papas T. S. Genomic dispersal of the ets gene family during metazoan evolution. Oncogene. 1992 Sep;7(9):1713–1719. [PubMed] [Google Scholar]
- Lautenberger J. A., Papas T. S. Inversion of a chicken ets-1 proto-oncogene segment in avian leukemia virus E26. J Virol. 1993 Jan;67(1):610–612. doi: 10.1128/jvi.67.1.610-612.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince D., Crepieux P., Laudet V., Flourens A., Stehelin D. A new mechanism of oncogenic activation: E26 retroviral v-ets oncogene has inverted the C-terminal end of the transcription factor c-ets-1. Virology. 1993 Jun;194(2):855–857. doi: 10.1006/viro.1993.1330. [DOI] [PubMed] [Google Scholar]
- Leprince D., Crepieux P., Stehelin D. c-ets-1 DNA binding to the PEA3 motif is differentially inhibited by all the mutations found in v-ets. Oncogene. 1992 Jan;7(1):9–17. [PubMed] [Google Scholar]
- Leprince D., Duterque-Coquillaud M., Li R. P., Henry C., Flourens A., Debuire B., Stehelin D. Alternative splicing within the chicken c-ets-1 locus: implications for transduction within the E26 retrovirus of the c-ets proto-oncogene. J Virol. 1988 Sep;62(9):3233–3241. doi: 10.1128/jvi.62.9.3233-3241.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince D., Gegonne A., Coll J., de Taisne C., Schneeberger A., Lagrou C., Stehelin D. A putative second cell-derived oncogene of the avian leukaemia retrovirus E26. Nature. 1983 Nov 24;306(5941):395–397. doi: 10.1038/306395a0. [DOI] [PubMed] [Google Scholar]
- Lewin B. Oncogenic conversion by regulatory changes in transcription factors. Cell. 1991 Jan 25;64(2):303–312. doi: 10.1016/0092-8674(91)90640-k. [DOI] [PubMed] [Google Scholar]
- Lim F., Kraut N., Framptom J., Graf T. DNA binding by c-Ets-1, but not v-Ets, is repressed by an intramolecular mechanism. EMBO J. 1992 Feb;11(2):643–652. doi: 10.1002/j.1460-2075.1992.tb05096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Sodeoka M., Lane W. S., Verdine G. L. Evidence for a non-alpha-helical DNA-binding motif in the Rel homology region. Proc Natl Acad Sci U S A. 1994 Feb 1;91(3):908–912. doi: 10.1073/pnas.91.3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod K., Leprince D., Stehelin D. The ets gene family. Trends Biochem Sci. 1992 Jul;17(7):251–256. doi: 10.1016/0968-0004(92)90404-w. [DOI] [PubMed] [Google Scholar]
- Mavrothalassitis G., Fisher R. J., Smyth F., Watson D. K., Papas T. S. Structural inferences of the ETS1 DNA-binding domain determined by mutational analysis. Oncogene. 1994 Feb;9(2):425–435. [PubMed] [Google Scholar]
- Metz T., Graf T. Fusion of the nuclear oncoproteins v-Myb and v-Ets is required for the leukemogenicity of E26 virus. Cell. 1991 Jul 12;66(1):95–105. doi: 10.1016/0092-8674(91)90142-l. [DOI] [PubMed] [Google Scholar]
- Metz T., Graf T. v-myb and v-ets transform chicken erythroid cells and cooperate both in trans and in cis to induce distinct differentiation phenotypes. Genes Dev. 1991 Mar;5(3):369–380. doi: 10.1101/gad.5.3.369. [DOI] [PubMed] [Google Scholar]
- Nunn M. F., Seeburg P. H., Moscovici C., Duesberg P. H. Tripartite structure of the avian erythroblastosis virus E26 transforming gene. Nature. 1983 Nov 24;306(5941):391–395. doi: 10.1038/306391a0. [DOI] [PubMed] [Google Scholar]
- Nye J. A., Petersen J. M., Gunther C. V., Jonsen M. D., Graves B. J. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992 Jun;6(6):975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- Shin M. K., Koshland M. E. Ets-related protein PU.1 regulates expression of the immunoglobulin J-chain gene through a novel Ets-binding element. Genes Dev. 1993 Oct;7(10):2006–2015. doi: 10.1101/gad.7.10.2006. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Urness L. D., Thummel C. S. Molecular interactions within the ecdysone regulatory hierarchy: DNA binding properties of the Drosophila ecdysone-inducible E74A protein. Cell. 1990 Oct 5;63(1):47–61. doi: 10.1016/0092-8674(90)90287-o. [DOI] [PubMed] [Google Scholar]
- Wang C. Y., Petryniak B., Ho I. C., Thompson C. B., Leiden J. M. Evolutionarily conserved Ets family members display distinct DNA binding specificities. J Exp Med. 1992 May 1;175(5):1391–1399. doi: 10.1084/jem.175.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Hahn S. L., Giovane A. The Ets family of transcription factors. Eur J Biochem. 1993 Jan 15;211(1-2):7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Flores P., Begue A., Leprince D., Stehelin D. The c-ets proto-oncogenes encode transcription factors that cooperate with c-Fos and c-Jun for transcriptional activation. Nature. 1990 Jul 12;346(6280):191–193. doi: 10.1038/346191a0. [DOI] [PubMed] [Google Scholar]
- Wasylyk C., Gutman A., Nicholson R., Wasylyk B. The c-Ets oncoprotein activates the stromelysin promoter through the same elements as several non-nuclear oncoproteins. EMBO J. 1991 May;10(5):1127–1134. doi: 10.1002/j.1460-2075.1991.tb08053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk C., Wasylyk B. Oncogenic conversion alters the transcriptional properties of ets. Cell Growth Differ. 1992 Sep;3(9):617–625. [PubMed] [Google Scholar]
- Wasylyk C., Wasylyk B. Oncogenic conversion of Ets affects redox regulation in-vivo and in-vitro. Nucleic Acids Res. 1993 Feb 11;21(3):523–529. doi: 10.1093/nar/21.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke M., Muñoz A., Sap J., Vennström B., Beug H. v-erbA oncogene activation entails the loss of hormone-dependent regulator activity of c-erbA. Cell. 1990 Jun 15;61(6):1035–1049. doi: 10.1016/0092-8674(90)90068-p. [DOI] [PubMed] [Google Scholar]