Abstract
Birds use various vocalizations to communicate with one another, and some are acquired through learning. So far, three families of birds (songbirds, parrots, and hummingbirds) have been identified as having vocal learning ability. Previously, we found that cadherins, a large family of cell-adhesion molecules, show vocal control-area-related expression in a songbird, the Bengalese finch. To investigate the molecular basis of evolution in avian species, we conducted comparative analysis of cadherin expressions in the vocal and other neural systems among vocal learners (Bengalese finch and budgerigar) and a non-learner (quail and ring dove). The gene expression analysis revealed that cadherin expressions were more variable in vocal and auditory areas compared to vocally unrelated areas such as the visual areas among these species. Thus, it appears that such diverse cadherin expressions might have been related to generating species diversity in vocal behavior during the evolution of avian vocal learning.
Keywords: cadherin, evolution, gene expression, parrot, quail, ring dove, songbird, vocal learning
Introduction
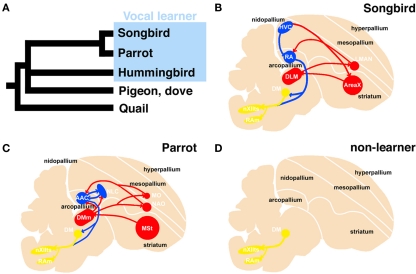
Vocal learning is the ability to acquire a new sound through imitation, and three families of birds (songbirds, parrots, and hummingbirds) have this ability (Jarvis, 2004). Because these birds are taxonomically distantly related, it has been suggested that they acquired this ability independently. In the brain of a vocal learner, a series of nuclei and a neural circuit exist called the “song system,” which is specialized for vocal learning and production (Figure 1; Nottebohm et al., 1976, 1982; Brauth et al., 1994; Striedter, 1994; Durand et al., 1997; Brainard and Doupe, 2000; Gahr, 2000; Jarvis and Mello, 2000; Jarvis et al., 2000; Jarvis, 2004; Bolhuis and Gahr, 2006; Bolhuis et al., 2010). In contrast, non-learners such as chickens and pigeons do not have such a system and only produce innate sounds (Bolhuis and Gahr, 2006). Because of these structure-related behavioral differences, the avian vocal system is a good model for studying brain evolution from a morphological and functional perspective (Matsunaga and Okanoya, 2009b).
Figure 1.
Phylogenetic tree of avian species and schematic representation of the vocal system in songbird, parrot, and non-learner. Phylogenetic relationship among vocal learners and non-learners (based on Hackett et al., 2008) (A). Sagittal view of a songbird (B), parrot (C), and non-learners such as quail and dove (D). The avian vocal system is composed of the telencephalic vocal learning pathway and the general vocal production pathway in the brainstem. The telencephalic vocal pathway is found only in vocal learning species. In the Bengalese finch, the telencephalic vocal pathway is composed of HVC-RA-(DM)-nXIIts posterior pathway and the HVC-AreaX-DLM-LMAN-RA anterior pathway (B). A similar but not the same vocal pathway is observed in the budgerigar: the NLC-AAC-(DM)-nXIIts posterior pathway and the AAC-DMm-NAO-AAC anterior pathway (C). In contrast, such neural circuits for vocal learning are not found in the quail brain (D). A similar neural circuit is found in the brainstems of these species to produce learned or innate vocalization. The red and blue lines indicate anterior and posterior pathways for vocal learning, respectively. The yellow line indicates the general vocalization pathway seen in the brainstem of avian species. AAC: central nucleus of the anterior arcopallium, DLM, dorsal lateral nucleus of the thalamus; DM, dorsal medial nucleus of the midbrain; DMm, magnocellular nucleus of the dorsal thalamus; LMAN, lateral magnocellular nucleus of the anterior nidopallium; NAO, oval nucleus of the anterior nidopallium; NLC, central nucleus of the lateral nidopallium; RA, robust nucleus of the arcopallium; nXIIts, tracheosyringeal hypoglossal nucleus.
We previously performed in situ hybridization screening in a songbird, the Bengalese finch, to explore the molecular basis of vocal system development and identified cadherins, neuropilin, and plexins as molecules whose expression is vocal-area related (Matsunaga et al., 2008). Among them, we particularly focused on cadherins, since cadherins show neural circuit-related expressions (each cadherins is expressed in some restricted population of neurons that are connected with each other; Suzuki et al., 1997; Takeichi, 2007). Actually, in the songbird brain, cadherins show vocal-system-related expressions (Matsunaga and Okanoya, 2008a). Cadherin-6B (cad6B) is broadly expressed in vocal control nuclei, whereas R-cadherin (Rcad) is expressed in the surrounding area. Cadherin-7 (Cad7) is transiently expressed in vocal control nuclei and its expression is downregulated during the phase from sensory to sensorimotor learning stage. Overexpression of cadherins affected vocal learning and production, suggesting the involvement in vocal development (Matsunaga and Okanoya, 2008c). Thus, it appears that cadherins are one of key regulators for vocal development.
Previously, we examined cadherin expressions in other vocal learner, budgerigar (Parrots) and non-learner quail (Galliformes), and published part of results in a short proceeding paper briefly (Matsunaga et al., 2008). Here, we performed more extensive comparative gene expression analysis of cadherins in vocal learner (Bengalese finch and budgerigar) and non-learner (quail and ring dove), and found that cadherin expressions are highly diverse in the vocal system. The auditory system is well connected to the vocal system and related to vocal learning. The visual system is unrelated to the vocal system and functionally conserved between vocal learners and non-learners. By analyzing gene expressions in these regions between vocal learners and non-learners, we can evaluate whether diverse cadherin expressions are related to vocal learning or not. Therefore, in this study, to examine whether such diverse cadherin expressions are vocal area-specific, we examined cadherin expressions not only in the vocal system, but also in other neural systems.
Materials and Methods
Animals
We used three 14-day postnatal (P14) and three P30 male Bengalese finches (Lonchura striata var. domestica), two P14, one P16, and three P30 male budgerigars (Melopsittacus undulatus), and three P14 and three P30 male quails (Coturnix japonica), two P24 and one P45 male ring doves (Streptopelia risoria), all of which were bred at our lab facilities. All birds were deeply anesthetized with an intramuscular injection of sodium pentobarbital (50 mg/kg) and then sacrificed. After decapitation, their brains were embedded in OCT compound (Tissue-Tek) and frozen on dry ice for cryosectioning. Frozen sections for in situ hybridization or thionine staining for neuroanatomical reference were cut serially in 20-μm thicknesses using a cryostat (Leica, Bannockburn, IL, USA). We made 16 slide sets from one brain to use in situ hybridization studies of various genes (the length between each neighboring sections on the same slide was 320 μm). To extract total RNA, brain tissues were dissected and placed in Qiazol Lysis reagent (Qiagen, Valencia, CA, USA), and the RNA was purified using an RNeasy Lipid Tissue Mini kit (Qiagen). The sex of the birds was determined by extracting genomic DNA from a portion of a digit with a DNeasy tissue kit (Qiagen) and performing a polymerase chain reaction (PCR) with primers that amplify the chromo-helicase-DNA binding gene (Ellegren, 1996), or verified by inspection of sex organs. Research protocols were approved by the animal care and use committee of RIKEN (#H18-2B002, #H20-2-231), and conformed to the National Institutes of Health (NIH, Bethesda) Guidelines.
Isolation and cloning of cdna
The budgerigar cDNA fragments for Rcad (Genbank accession no. AB329583), cad6B (AB329584), and cad7 (AB329582), ring dove cDNA fragments for Rcad (AB610760), cad6B (AB610761), and cad7 (AB610762) were isolated from the adult brain by the reverse transcription-PCR. The same primer sets used for Bengalese finch were used to isolate budgerigar and ring dove cDNAs, as previously described (Matsunaga and Okanoya, 2008a), except for ring dove cad6B. For ring dove cad6B, the primers were used as follows: 5′-CTCTTGTTGCCGTGATGAGA-3′ and 5′-GTTTATAGCCTGGGCACGAA-3′. Each cDNA fragment was inserted into the pGEM-T Easy Vector (Promega, Madison, WI, USA). We used the same plasmids for probe preparation that were used for Bengalese finch brain, as previously described (Matsunaga and Okanoya, 2008a). We used plasmids encoding chicken cadherin cDNAs for quail brain, kindly gifted by Dr. Masatoshi Takeichi (Inuzuka et al., 1991; Nakagawa and Takeichi, 1995). The plasmids were digested with enzymes to release the fragments, and probes were synthesized using SP6, T3, or T7 RNA polymerase (Roche, Indianapolis, IN, USA) with digoxigenin (DIG)-labeling mix (Roche).
In situ hybridization
Tissue sections were post-fixed for 10 min and then washed three times in PBS for 3 min. The slides were delipidated with acetone, acetylated, and washed in PBS with 1% Triton-X100 (Wako Pure Chemical, Osaka, Japan). The slides were incubated at room temperature with hybridization buffer containing 50% formamide (Wako), 5× SSC, 5× Denhardt's solution (Sigma, St. Louis, MO, USA), 250 yeast tRNA (Roche), and 500 μg/ml DNA (Roche). The sections were then hybridized at 72°C overnight in hybridization buffer with RNA probes. The sections were rinsed in 0.2× SSC for 2 h and then blocked for 2 h in a solution of 0.1 M Tris (pH 7.5) and 0.15 M NaCl with 10% sheep serum. The slides were incubated overnight with alkaline phosphatase (AP)-conjugated anti-DIG antibody (Roche). After washing, AP activity was detected by adding 337.5 mg/ml nitroblue tetrazolium chloride and 175 mg/ml 5-bromo-4-chloro-3-indolyl phosphate (Roche). All sections were visualized with an Eclipse E600 microscope (Nikon, Tokyo, Japan), and all images were scanned with a computer-based image processing system (Neurolucida; Microbrightfield, Inc., Williston, VT, USA). Images were processed with Photoshop CS2 software (Adobe Systems, Mountain View, CA, USA).
Results
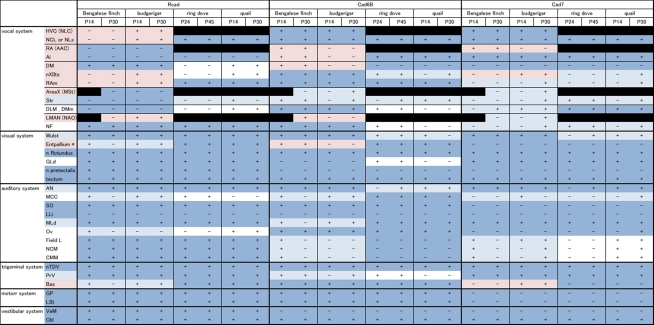
In this study, we analyzed Rcad, cad6B, and cad7 expression in two vocal learners (Bengalese finch and budgerigar) and non-learner (quail) at two different developmental stages (postnatal 2 and 4 weeks). Since gene expression patterns were similar in most regions between these different developmental stages, we only shows expression patterns of P30 brains in figures. Furthermore, we analyzed these cadherin expressions in the second non-learner, ring dove. All comparative gene expression results are summarized in Table 1 (we used the terminology of Reiner et al., 2004).
Table 1.
Summary of comparative gene expression among vocal learners (Bengalese finch and budgerigar) and non-learners (quail and ring dove).
Note that gene expression differed more among species in vocal-related areas than other areas. Dark blue filled region: highly similar expression pattern among species; light blue filled region: similar but developmentally different expression pattern; light red filled region: different expression between vocal learners; white region: different expression between non-learners.
Cadherin expression in the vocal system
We first examined cadherin expression in the vocal pathways among Bengalese finch, budgerigar, and quail.
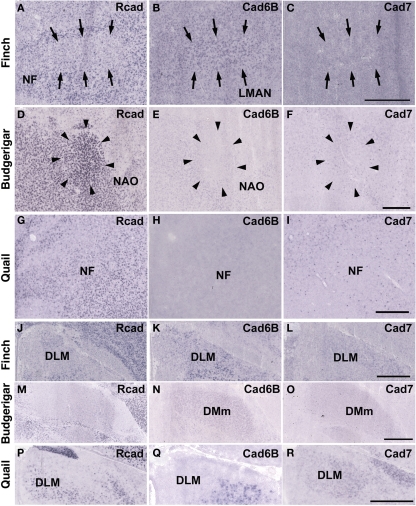
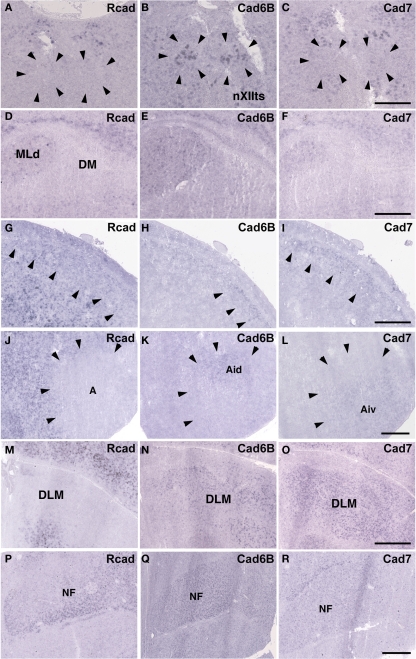
Cad6B was widely expressed in the vocal system of the Bengalese finch brain including in the HVC, robust nucleus of the arcopallium (RA), lateral magnocellular nucleus of the anterior nidopallium (LMAN), dorsal lateral nucleus of the thalamus (DLM), dorsal medial nucleus of the midbrain (DM), and nucleus retroambigularis (RAm) and it was weakly expressed in the tracheosyringeal hypoglossal nucleus (nXIIts; Figures 2B,K, 3B,E, and 4K), as previously described (Matsunaga and Okanoya, 2008a). In contrast, Rcad was not expressed in these vocal control nuclei (Figures 2A, 3A,D, and 4J), except for DM (Figure 2J). At P30, the LMAN had not been clearly formed yet, but cad6B expression and weak Rcad expression were already seen in the corresponding LMAN region (Figures 4A,B). In the juvenile stage, cad7 was also expressed in some vocal control nuclei such as the HVC and RA (Figures 3C,F), although their expression is downregulated during development (Matsunaga and Okanoya, 2008a).
Figure 2.
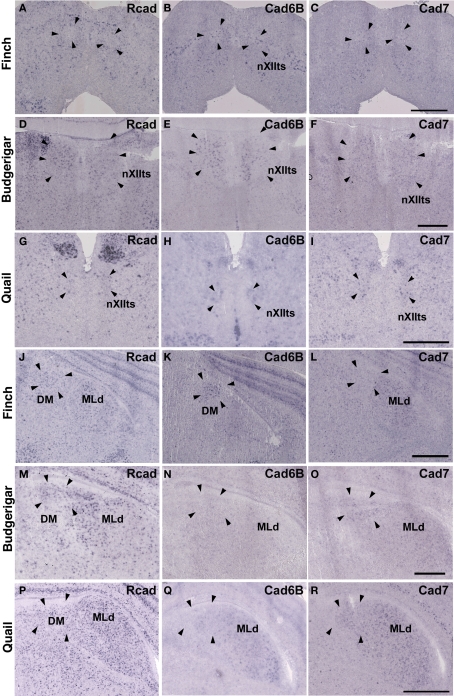
Expression of R-cadherin (Rcad), cadherin-6B (cad6B) and cadherin-7 (cad7) in the brainstem. (A–I) In situ hybridization for Rcad (A,D,G), cad6B (B,E,H), and cad7 (C,F,I) in the Bengalese finch (A–C), budgerigar (D–F), and quail (G–I) at the level of the nXIIts. (J–R) In situ hybridization for Rcad (J,M,P), cad6B (K,N,Q), and cad7 (L,O,R) in the Bengalese finch (J–L), budgerigar (M–O), and quail (P–R) at the level of the DM. Scale bars are 1 mm.
Figure 3.
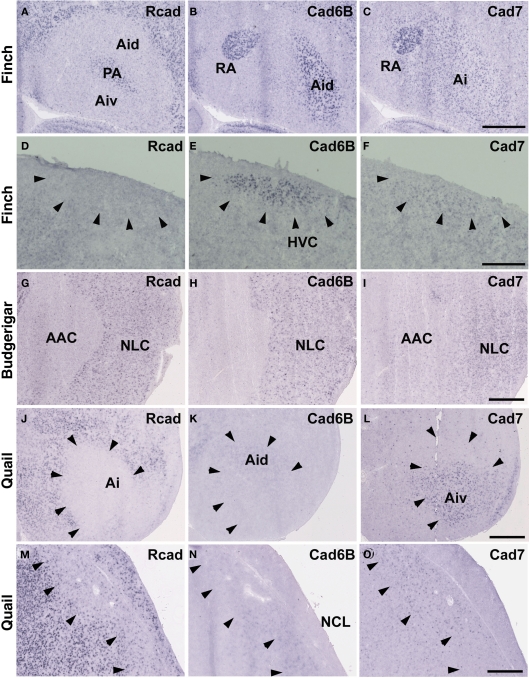
In situ hybridization for Rcad, cad6B, and cad7 at the telecephalic level. (A–F) Cadherin expression in P30 male Bengalese finch in the RA (A–C) and HVC (D–F). (G–I) Cadherin expression in P30 male budgerigar in the NLC and AAC. (J–O) Cadherin expression in P30 male quail in the intermediate arcopallium (Ai) and caudolateral nidopallium (NCL). Arrowheads indicate expression boundary between the strong Rcad expression domain and the strong cad6B and cad7 expression domain. Scale bars are 1 mm.
Figure 4.
In situ hybridization for Rcad, cad6B, and cad7 in the frontal nidopallium and dorsal thalamus. (A–C) Cadherin expression in the LMAN of the Bengalese finch. Although the LMAN is not clearly formed at P30, cad6B expression in the LMAN and Rcad expression in the surrounding region are beginning to show (A,B, arrows). (D–F) Cadherin expression in the NAO of the budgerigar, an area with functional correspondence to the LMAN. In contrast to the LMAN of Bengalese finch, Rcad was expressed, but Cad6B was not expressed in the NAO (D,E). (G–I) Cadherin expression in the frontal nidopallium of quail at the level corresponding to the LMAN and the NAO. (J–L) Cadherin expression in the DLM of Bengalese finch. (M–O) Cadherin expression in the DMm of budgerigar. (P–R) Cadherin expression in the dorsolateral region of the thalamus of quail. Scale bars are 1 mm.
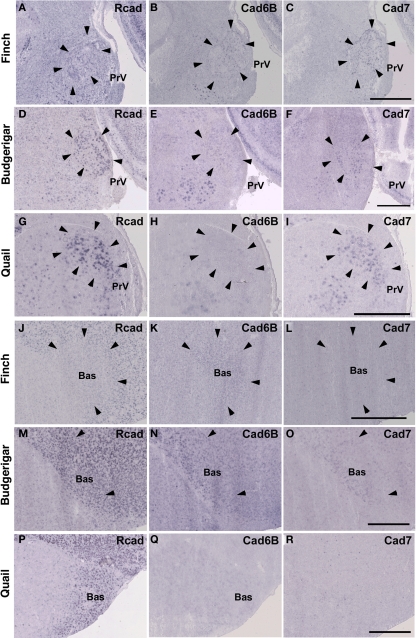
Similar to the Bengalese finch, cad6B and cad7 were expressed in the vocal control nuclei of budgerigar; however, the expression pattern differed. Cad6B was expressed in the nXIIts, RAm, magnocellular nucleus of the dorsomedial thalamus (DMm), and central nucleus of the lateral nidopallium (NLC; Figures 2E, 3H, and 4N) as in Bengalese finch, but cad6B expression was not observed in the DM, central nucleus of the anterior arcopallium (AAC) and oval nucleus of the anterior nidopallium (NAO; Figures 2N, 3H, and 4E). As in the Bengalese finch, Cad7 was expressed in the nXIIts, DMm, and NLC, and not expressed in DM (Figures 2C,F,L,O, 3E,H, and 4L,O). However, cad7 was not expressed in the AAC (Figure 3I). Sparse cad7 expression was seen in the NAO (Figures 4C,F). Additionally, some of the vocal control nuclei did not express Rcad in the budgerigar brain (Figures 3G and 4M), but, in contrast to the Bengalese finch, Rcad was expressed in some vocal control nuclei such as the nXIIts, RAm, DM, NLC, and NAO (Figures 2D,M, 3G, and 4D).
Although no telencephalic vocal control nuclei occur in quail, they have vocal nuclei in the brainstem to produce innate vocalization. Rcad was expressed in the RAm (data not shown) and DM, but not expressed in the nXIIts of the quail brainstem (Figures 2G,P). As cad6B, cad7 expression was observed in the nXIIts (Figures 2H,I), but no cad7 expression was seen in the DM (Figures 2Q, R). Although no vocal nuclei occur in the thalamus of quail brain, a corresponding area exists. Rcad was expressed, but cad6B was not expressed, in the dorsolateral area of the thalamus (Figures 4P,Q). This expression pattern was different to that in the Bengalese finch and budgerigar (Rcad was not expressed but cad6B was expressed in both species), though cad7 expression was similar (Figures 4L,O,R).
Cadherin expression in the region surrounding the vocal system
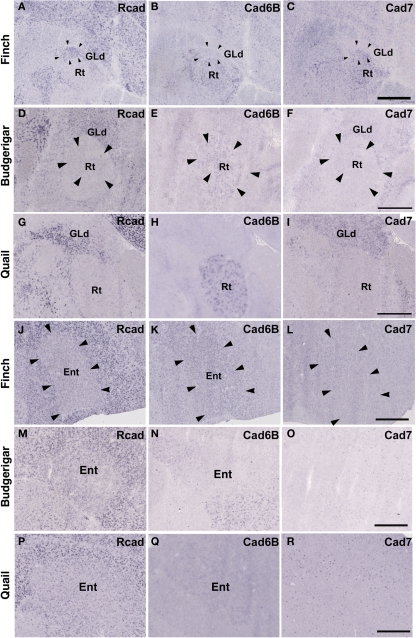
Consistent with the idea that avian vocal learning system might be evolved from neighboring motor learning system (Feenders et al., 2008), cadherin expression patterns in the vocal control nuclei were similar to those in surrounding regions of Bengalese finch, although the expression level and density were different (Matsunaga and Okanoya, 2008a; Table 1). The RA was located in the dorsomedial region of the arcopallium (Ai). Cad6B was expressed in the dorsal region of the intermediate Ai, whereas cad7 was expressed in the dorsal and ventral regions of the Ai (Figures 3B,C). Cad6B and cad7 were weakly expressed in the nidopallium around the HVC, although cad6 expression is upregulated and cad7 expression is downregulated during development (Matsunaga and Okanoya, 2008a; Figures 3E,F).
The expression patterns in the vocal control nuclei and their surrounding regions were basically similar in budgerigar brain, as in the Bengalese finch. For example, no clear expression border was found between the NLC vocal control nucleus and the supracentral nucleus of the lateral neostriatum (NLs; data not shown), the surrounding area of NLC (Plummer and Striedter, 2002). However, in contrast to Bengalese finch, the expression pattern in the NAO and AAC was quite different in the budgerigar brain. Cad7 expression was detected in the medial region of the Ai, but its expression was absent in the AAC (Figure 3I). Cad6B was expressed in the frontal nidopallium, but its expression was lacking in the NAO (Figure 4E).
Although quail have no telencephalic vocal control nuclei, it seems that other brain regions share similar features between vocal learners and non-learners. In the quail brain, no clear Rcad expression was seen in the arcopallium, and both cad6B and cad7 were expressed in the Ai. However, the expression domains were mostly separated (Figures 3J–L); Cad6B was expressed in the dorsal region, whereas cad7 was expressed in the ventral region. Whereas Rcad was weakly expressed in the caudolateral nidopallium (NCL) of the quail brain, cad6B and cad7 were expressed in the NCL, although their expression domains were generally separated, with some overlap; Cad6B was expressed in the ventral region, whereas Cad7 was expressed in the dorsal region of the NCL (Figures 3M–O). In addition, no clear cad6B expression was observed in the frontal region of the nidpallium (NF; Figure 4H), though Rcad expression was similar to vocal learners (Figure 4G). Only sparse Cad7 expression was seen in this region (Figure 4I).
Cadherin expression in the visual system
In avian species, the visual information is transmitted from the retina to the telencephalon via two distinct pathways. One is the tectofugal pathway via the tectum, nucleus rotundus (Rt) and entopallium (Ent; Benowitz and Karten, 1976), and the other is the thalamofugal pathway via dorsal lateral geniculate nucleus (GLd) and visual wulst (Shimizu and Karten, 1993). Since these pathways are seen in various avian species (Mey and Thanos, 2000; Heyers et al., 2007), it appears that the visual system is evolutionally conserved among all avian species. Hence, it was suspected that gene expression in visual nuclei would be tightly constrained. To examine this possibility, we next analyzed cadherin expression in these visual areas.
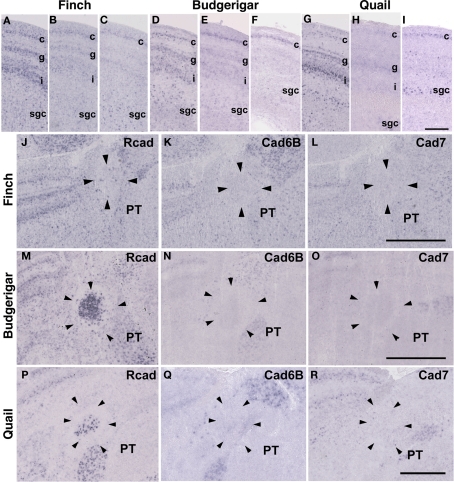
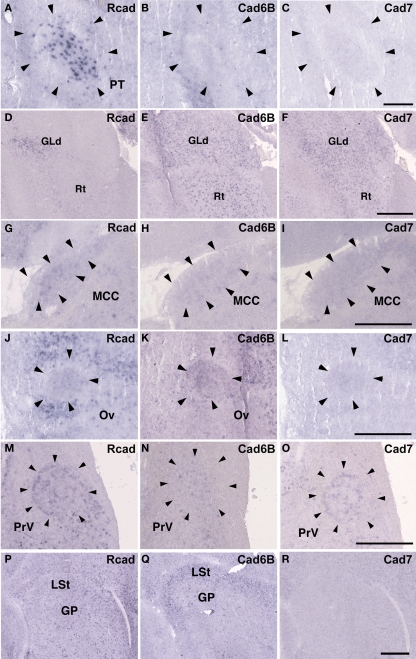
The Rcad, cad6B, and cad7 expression patterns were basically similar among the three species. Rcad was expressed in the tectum, nucleus pretectalis (PT), GLd, Ent, and visual wulst and was sparsely expressed in the Rt (Figures 5A,D,G,J,M,P and 6A,D,G,J,M,P). Cad7 was expressed in the tectum, GLd, and visual wulst and sparsely expressed in the Rt, whereas cad7 was not expressed in the PT or Et (Figures 5C,F,I,L,O,R and 6C,F,I,L,O,R). Cad6B expression patterns were also basically similar, although the expression pattern in the Et was exceptionally different (Figures 5B,E,H,K,N,Q and 6B,E,H,K,N,Q).
Figure 5.
Cadherin expression in the visual system at the level of the tectum and pretectum. (A–I) In situ hybridization for Rcad (A,D,G), cad6B (B,E,H), and cad7 (C,F,I) in the Bengalese finch (A–C), budgerigar (D–F), and quail (G–I) in the caudolateral tectum. Cadherins are strongly expressed in layer c, g and i of the stratum griseum et fibrosum superficiale (sfgs) and the stratum griseum centrale (sgc). (J–R) In situ hybridization for Rcad (J,M,P), Cad6B (K,N,Q), and Cad7 (L,O,R) of Bengalese finch (J–L), budgerigar (M–O), and quail (P–R) in the nucleus pretectalis (PT). Scale bars are 250 μm (I) and 500 μm (L,O,R).
Figure 6.
Cadherin expression in the visual system at the level of the thalamus and telencephlon. (A–I) In situ hybridization for Rcad (A,D,G), cad6B (B,E,H), and cad7 (C,F,I) in the Bengalese finch (A–C), budgerigar (D–F), and quail (G–I) in the nucleus rotondus. (J–R) In situ hybridization for Rcad (J,M,P), Cad6B (K,N,Q), and Cad7 (L,O,R) in the Bengalese finch (J–L), budgerigar (M–O), and quail (P–R) in the Entopallium. Scale bars are 1 mm.
Cadherin expressions in the auditory system
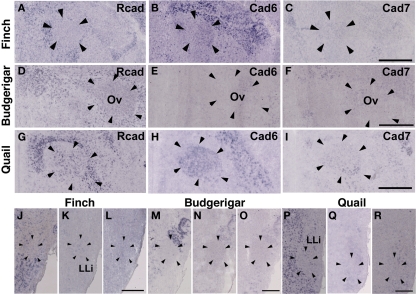
In the avian species, the auditory information is transmitted from the inner ear to telencephalic auditory areas Field L via a series of auditory nuclei such as the cochlear nucleus (CN), superior olivary nucleus (SO), the intermediate lateral lemniscal nucleus (LLi), lateral mesencephalic nucleus (MLd), and nucleus ovoidalis (Ov). These ascending auditory pathway is seen both in vocal learners and non-learners (Correia et al., 1982; Mello et al., 1998). However, in contrast to the visual system, the auditory system is related to the vocal control system, particularly in the telencephalon (Bauer et al., 2008). Therefore, it is possible that cadherin expression in the auditory system would differ to some extent among species. As expected, cadherin expression in the auditory system was different in some parts of the brain.
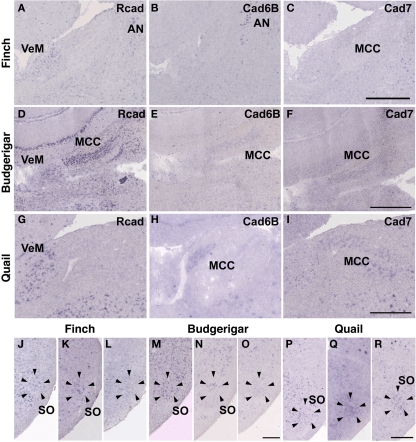
In the CN, though the cadherin expression pattern in the magnocellular sub-nucleus (MCC) varied among the three species (Figures 7A–I), Rcad, cad6B, and cad7-expressed cells were distributed in the angular sub-nucleus (AN) of the three species (data not shown). The cadherin expression pattern was similar in the SO and LLi (Figures 7J–R and 8J–R). However, in the MLd and Ov, the cadherin expression patterns differed among species (Figures 2J–R and 8A–I). In the telencephalic auditory area, Field L, the caudal medial nidopallium (NCM), and the caudomedial mesopallium (CMM), cadherin expression patterns were diverse (Table 1).
Figure 7.
Cadherin expression in the cochlear nucleus and superior olivary nucleus (SO) of the auditory system. (A–I) In situ hybridization for Rcad (A,D,G), cad6B (B,E,H), and cad7 (C,F,I) in the Bengalese finch (A–C), budgerigar (D–F), and quail (G–I) in the cochlear magnocellular nucleus (MCC). (J–R) In situ hybridization for Rcad (J,M,P), cad6B (K,N,Q), and cad7 (L,O,R) in the Bengalese finch (J–L), budgerigar (M–O), and quail (P–R) in the SO. Scale bars are 1 mm (C,F,I) and 500 μm (O,R).
Figure 8.
Cadherin expression in the nucleus ovoidalis and intermediate nucleus of the lateral lemniscus in the auditory system. (A–I) In situ hybridization for Rcad (A,D,G), cad6B (B,E,H), and cad7 (C,F,I) in the Bengalese finch (A–C), budgerigar (D–F), and quail (G–I) in the nucleus ovoidalis (Ov). (J–R) In situ hybridization for Rcad (J,M,P), cad6B (K,N,Q), and cad7 (L,O,R) in the Bengalese finch (J–L), budgerigar (M–O), and quail (P–R) in the intermediate lateral lemniscal nucleus (LLi). Scale bars are 1 mm (C,F,I) and 500 μm (L,O,R).
Cadherin expression in the trigeminal system
Among trigeminal nuclei, we analyzed cadherin expression in the principal sensory nucleus of the pons (PrV), the nucleus et tractus descendens nervi trigemini (nTDV), and the basorostral pallial nucleus (Bas). PrV neurons project to the Bas to regulate tongue and beak movement (Wild et al., 1984, 1997). The Bas receives trigeminal input from the PrV and auditory input from the LLi (Farabaugh and Wild, 1997). Auditory input from the Bas is connected with vocal control nuclei via the NF in the budgerigar (Farabaugh and Wild, 1997), but is not connected in songbirds (Wild and Farabaugh, 1996). Therefore, the trigeminal system seems to differ among the three species, and in the case of the budgerigar, this system is connected to the vocal control system.
As expected, cadherin expression in the PrV and Bas were different in some extent (Figures 9A–R), whereas Rcad, cad6B, and cad7 expression patterns were similar among the three species in the nTDv (data not shown). At P30, although cad6B expression in the Bas was similar in all species, Rcad was expressed in the budgerigar and quail and cad7 was only expressed in the budgerigar (Figures 9J–R).
Figure 9.
Cadherin expression in the trigeminal pathway. (A–I) In situ hybridization for Rcad (A,D,G), cad6B (B,E,H), and cad7 (C,F,I) in the Bengalese finch (A–C), budgerigar (D–F), and quail (G–I) in the principal sensory nucleus of the pons (PrV). (J–R) In situ hybridization for Rcad (J,M,P), Cad6B (K,N,Q), and Cad7 (L,O,R) in the Bengalese finch (J–L), budgerigar (M–O), and quail (P–R) in the nucleus basorostralis (Bas). Scale bars are 1 mm.
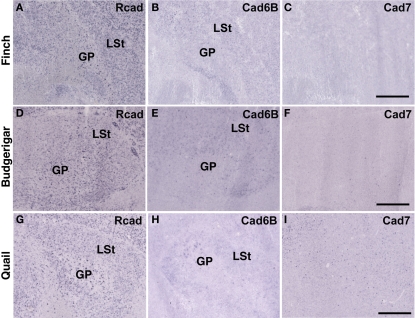
Cadherin expressions in other system
In telencephalic motor nuclei such as GP and LSt, cadherin expressions were similar among three species (Figures 10). In vestibular system, nucleus vestibularis medialis (VeM) and lateral nucleus of cerebellum (Cbl), cereberum nucleus, cadherin expressions were also similar (data not shown).
Figure 10.
Cadherin expression in the motor system of the basal ganglia. (A–I) In situ hybridization for Rcad (A,D,G), cad6B (B,E,H), and cad7 (C,F,I) in the Bengalese finch (A–C), budgerigar (D–F), and quail (G–I). Scale bars are 1 mm.
Developmental difference in cadherin expressions
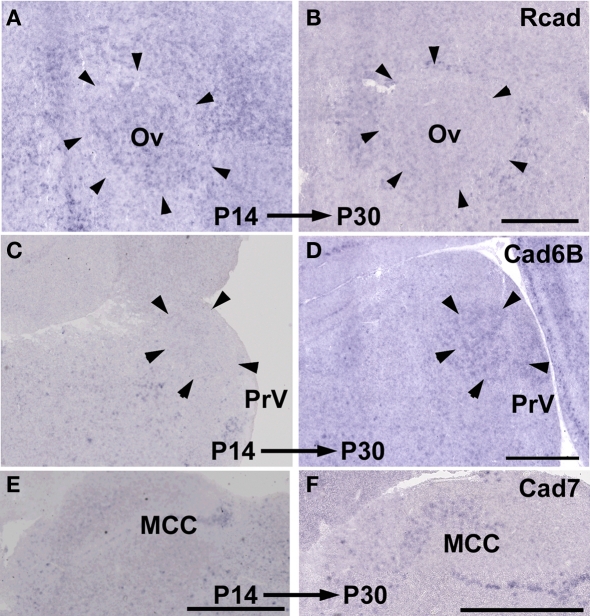
By comparing gene expressions among three species, it seemed that cadherin expressions were diverse particularly in the vocal and auditory system. However, it was suspected that these differences did not reflect phylogenic differences among three species but differences in developmental progress among three species (e.g., budgerigars develop more slowly than Bengalese finches). To examine the possibility, we compared gene expressions between two different developmental stages P14 and P30.
As expected, some cadherin expressions were upregulated or downregulated in some brain areas of the same species between P14 and P30 (Figure 11; summarized in Table 1). By comparing between these different stages, cadherin expressions were particularly variable in the auditory areas during the development (Table 1). Difference in these cadherin expressions among three species may be due to differences in developmental progress rather than phylogenic differences (summarized in Table 2).
Figure 11.
Developmental difference in cadherin expressions between P14 and P30. Rcad expression in Ov of Bengalese finch (A,B). Cad6B expression in PrV of budgerigar (C,D). Cad7 expression in MCC of quail (E,F). Scale bars are 1 mm.
Table 2.
Summary of species difference in cadherin expressing areas.
| Species difference in cadherin expressing areas | ||
|---|---|---|
| Vocal learners | Non-learners | |
| Telencephalic vocal areas | 5/12 (42%) | − |
| 0/12 (0%) | − | |
| Surrounding areas | 2/12 (17%) | 1/12 (8%) |
| 1/12 (8%) | 4/12 (33%) | |
| Vocal areas in the brainstem | 4/12 (33%) | 4/12 (33%) |
| 2/12 (17%) | 2/12 (17%) | |
| Visual areas | 1/18 (6%) | 0/18 (0%) |
| 1/18 (6%) | 2/18 (11%) | |
| Auditory areas | 0/27 (0%) | 5/27 (19%) |
| 11/27 (41%) | 2/27 (7%) | |
| Other areas | 1/21 (5%) | 1/21 (5%) |
| 2/21 (10%) | 0/21 (0%) | |
The value shown in bold indicates the percentage of species difference in cadherin expressions that may be due to phylogenic differences, and the value shown in light letters indicates the percentage of species difference that may be due to developmental stage differences among species. Note that diverse cadherin expressions are prominent in vocal and auditory areas.
Cadherin expression in another non-learner ring dove
By comparing among two vocal learners (Bengalese finch and budgerigar) and an non-learner quail, we found diverse cadherin expressions in the vocal system. However, if cadherin expressions differ between two different non-learners, the differences might have been caused by evolutional changes unrelated to vocal learning. To exclude the possibility, we also examined cadherin expressions in another non-learner, ring dove.
Some cadherin expressions were different between quail and ring dove [Rcad in nXIIts (Figures 2G and 12A), DM (Figures 2P and 12D), and DLM (Figures 4P and 12M); cad6B in NF (Figures 4H and 12Q), DLM (Figures 4Q and 12N), GLd (Figures 6H and 13E), and PrV (Figures 9H and 13N); cad7 in the second auditory areas]. However, most cadherin expression patters were similar between two species (Figures 2H,I,Q,R, 3J–O, 4G,I,R, 5P–R, 6G,I, 7G–I, 8G–I, 9G,I, 10G–I, 12B,C,E–L,O,P,R and 13A–D,F–M,O–R).
Figure 12.
Cadherin expressions in vocal areas of the brainstem and corresponding areas of ring dove to telencephalic vocal areas. In situ hybridization for Rcad (A,D,G,J,M,P), cad6B (B,E,H,K,N,Q), and cad7 (C,F,I,L,O,R). Cadherin expressions in nXIIts (A–C), DM (D–F), NCL (G–I), Ai (J–L), DLM (M–O), and NF (P–R). Scale bars are 1 mm.
Figure 13.
Cadherin expressions in non-vocal areas of ring dove. In situ hybridization for Rcad (A,D,G,J,M,P), cad6B (B,E,H,K,N,Q), and cad7 (C,F,I,L,O,R). Cadherin expressions in PT (A–C), Rt (D–F), MCC (G–I), Ov (J–L), PrV (M–O), and GP (P–R). Scale bars are 1 mm (F,I,L,O,R), and 500 μm (C).
Discussion
Highly diverse cadherin expressions in the vocal control system
We performed a comparative gene expression analysis among two vocal learners, the Bengalese finch (oscine songbirds) and budgerigar (parrots), and non-vocal learners, the quail (Galliformes) and ring dove (Columbiformes). Both vocal learners had a similar but not the same series of telencephalic nuclei and neural circuits specialized for vocal learning and production, whereas non-learners had no such structure in the telencephalon (Bolhuis and Gahr, 2006). Consistent with diverse vocal system among avian species, cadherin expressions were highly variable, not only in the vocal telencephalic nuclei, but also in the vocal nuclei at the level of the brainstem. In contrast, the visual system may be more tightly conserved and less variable than the vocal system during evolution. Actually, cadherin expression in the visual system was almost similar among all species.
Diverse cadherin expressions in the vocal system were also seen between two non-learners quail and dove. The ratio of different cadherin expressions in brainstem vocal areas between these species were almost similar to the ratio of different cadherin expressions between two vocal learners (see Table 2), suggesting that diverse cadherin expressions are not vocal learner-specific and change of cadherin expressions may not have been related to the acquisition of vocal learning ability itself, rather, related to diversity in vocal behaviors among avian species.
Some cadherin expression patterns change during development. For example, cadherin expression was not the same between chick embryos (Redies et al., 2001) and postnatal quail in the present study. Cad7 downregulation and Cad6B upregulation are observed in the RA nucleus during the transition from the sensory to sensorimotor learning stage (Matsunaga and Okanoya, 2008a). In this study, to verify the possibility that some gene expression differences were caused by developmental differences among species, we used juvenile birds at two developmental stages to perform the comparative gene expression analysis. We found cadherin expressions were changed in some areas between two different developmental stages of the same species (light blue region, Table 1). However, in the vocal system, cadherin expressions differed in many areas among different species, even their expressions were similar between two different developmental stages of the same species (dark blue region, Table 1), suggesting that these gene expression differences were caused by species difference rather than developmental differences. Thus, it seems to be sure that there is a clear tendency that cadherin expressions are highly diverse particularly in the vocal system (see Table 1).
Variable and conserved cadherin expressions in other system
Since the vocal system is closely related to auditory system (Margoliash et al., 1994; Fortune and Margoliash, 1995; Mello et al., 1998; Bauer et al., 2008) and vocal learning is auditory-dependent (Konishi, 1965), changes of gene expression in auditory system might have been related to species differences in vocal learning and behavior. Actually, our results showed that cadherin expressions were diverse in auditory nuclei such as MLd, Ov, telencephalic auditory area. In avian species, there are two auditory systems: primary auditory pathway via thalamus and secondary auditory pathway without thalamic relay via Bas. Songbirds only use the former, while parrots use both auditory pathways (Wild and Farabaugh, 1996; Farabaugh and Wild, 1997). Thus, not only the vocal system but also the auditory system are different between songbirds and parrots. In accordance with this, some cadherin expressions were different in the Bas between Bengalese finch and budgerigar. However, comparing between different stages, cadherin expressions were diverse between the early and late developmental stages in Bengalese finch and budgerigar, and it seems that these differences were mainly due to ontogenic differences rather than phylogenic differences (see Table 2). In contrast, comparing between two non-learners quail and dove, expression patterns were similar between two different developmental stages, though their expressions differed between these species. Some phylogenic variations in cadherin expressions may also exist in the non-learner's auditory system.
In contrast to vocal and auditory system, cadherin expressions were less variable in the visual, trigeminal, motor, and vestibular system (see Table 2). Compared to vocal and auditory system, similar anatomical structures are seen in these systems among avian species. Since these systems are mutually seen and directly involved in survival for avian species, their anatomical structure and gene expressions may have been tightly constrained during the evolution. In contrast, the vocal and auditory-vocal systems may have more acceptable range for diverse anatomical structure and gene expressions, resulting in variety of species-specific vocal behaviors.
Diverse cadherin expression and evolution in the vocal learning system
Songbirds, parrots, and hummingbirds have neural circuits for vocal learning. Because these birds are taxonomically distantly related, it has been suggested that these families acquired vocal learning ability independently (Jarvis, 2000, 2004). Anatomical and molecular analyses have suggested the possibility that each vocal system evolved from general primordial structures (Margoliash et al., 1994; Mello et al., 1998; Farries, 2001; Feenders et al., 2008; Matsunaga et al., 2008; Matsunaga and Okanoya, 2009b). Actually, some expressed genes are similar in the vocal systems of songbirds and parrots. The androgen receptor, a transcription factor, is widely expressed in vocal control nuclei of the budgerigar as well as in songbirds (Matsunaga and Okanoya, 2008b). The axon guidance molecules nueropilin-1 and plexin-A4 show similar expression patterns between the Bengalese finch and budgerigar (Matsunaga and Okanoya, 2009a). In contrast, cadherin expression in the vocal system is highly diverse between songbirds and parrots. Cadherins, originally isolated as cell-adhesion molecules, are involved not only in synapse formation, but also in synapse function. For example, cad8 mutant mice show reductions in miniature excitatory postsynaptic currents in temperature-sensitive neurons (Suzuki et al., 2007). Cad11 mutant mice show enhanced long-term potentiation in CA1 neurons (Manabe et al., 2000). Cadehrin overexpression by lentiviral vectors affects vocal learning and production (Matsunaga and Okanoya, 2008c). In vitro analysis using rat hippocampal culture neurons revealed that cad6B increased but cad7 decreased the number of spines and frequency of miniature excitatory post synaptic current (mEPSC; our unpublished data). Therefore, the diversity of cadherin expression may modulate neuronal activity and plasticity to generate diversification in the processes of vocal learning and production. Glutamate receptors also showed diverse expression in the vocal system among three vocal learners (Wada et al., 2004). It appears that transcription factors induce the expression of various genes involved in synapse formation of vocal control nuclei, and subsequently, that the differential expression of synaptic genes such as neurotransmitter receptors and cadherins modulate neuronal activity and plasticity in vocal control nuclei to create species-specific vocal behaviors and evolution.
Many vocal control-area-related genes have been recently identified in the zebra finch and Bengalese finch using comprehensive cDNA chip gene expression analysis (Wade et al., 2004; Lombardino et al., 2005; Wada et al., 2006; Li et al., 2007; Lovell et al., 2008; Replogle et al., 2008; Kato and Okanoya, 2010), and the genomic sequence of the zebra finch has been recently published (Warrant et al., 2010). Using this new technology and knowledge, similarities and differences among vocal learners and non-learners will become clear, and the molecular mechanisms of avian vocal system evolution will be elucidated.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Drs. Masatoshi Takeichi for reagent, and the Support Unit for Bio-material Analysis, RIKEN BSI Research Resources Center, for DNA sequence analysis. Eiji Matsunaga was supported by RIKEN Special Postdoctoral researcher program, Grant-in-aid for young scientist (B) 21700365 from Japan Society of promotion of Science, and Takeda Science Foundation.
Abbreviations
A, arcopallium; AAC, central nucleus of the anterior arcopallium; Ai, intermediate arcopallium; Aid, dorsal region of the intermediate arcopallium; Aiv, ventral region of the intermediate arcopallium; AN, nucleus angularis; Bas, nucleus basorostralis; Cbl, lateral nucleus of the cerebellum; CMM, caudomedial mesopallium; CN, cochlear nucleus; DLM, dorsal lateral nucleus of the thalamus; DM, dorsal medial nucleus of the midbrain; DMm, magnocellular nucleus of the dorsal thalamus; HVC, high vocal center; GLd, dorsal lateral geniculate nucleus; GP, globus pallidus; LLi, intermediate lateral lemniscal nucleus; LMAN, lateral magnocellular nucleus of the anterior nidopallium; LSt, Lateral striatum; MCC, cochlear magnocellular nucleus; MLd, lateral mesencephalic nucleus; NAO, oval nucleus of the anterior nidopallium; NCL, caudolateral nidopallium; NCM, caudal medial nidopallium; NF, frontal nidopallium; NLC, central nucleus of the lateral nidopallium; nXIIts, tracheosyringeal hypoglossal nucleus; Ov, nucleus ovoidalis; PrV, principal sensory nucleus of the pons; PT, nucleus pretectalis; RA, robust nucleus of the arcopallium; RAm, nucleus retroambigularis; Rt, nucleus rotundus; SGC, stratum griseum central; SO, superior olivary nucleus; Str, striatum; tec, tectum; nTDV, nucleus et tractus descendens nervi trigemini; VeM, nucleus vestibularis medialis.
References
- Bauer E. E., Coleman M. J., Roberts T. F., Roy A., Prather J. F., Mooney R. (2008). A synaptic basis for auditory-vocal integration in the songbird. J. Neurosci. 28, 1509–1522 10.1523/JNEUROSCI.3838-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benowitz L. I., Karten H. J. (1976). Organization of the tectofugal pathway in the pigeon: a retrograde transport study, J. Comp. Neurol. 167, 503–520 [DOI] [PubMed] [Google Scholar]
- Bolhuis J. J., Gahr M. (2006). Neural mechanisms of birdsong memory. Nat. Rev. Neurosci. 7, 347–357 [DOI] [PubMed] [Google Scholar]
- Bolhuis J. J., Okanoya K., Scharff C. (2010). Twitter evolution: converging mechanisms in birdsong and human speech. Nat. Rev. Neurosci. 11, 747–759 [DOI] [PubMed] [Google Scholar]
- Brainard M. S., Doupe A. J. (2000). Interruption of a basal ganglia-forebrain circuit prevents plasticity of learned vocalizations. Nature 404, 762–766 10.1038/35008083 [DOI] [PubMed] [Google Scholar]
- Brauth S. E., Heaton J. T., Durand S. E., Liang W., Hall W. S. (1994). Functional anatomy of forebrain auditory pathways in the budgerigar (Melopsittacus undulatus). Brain Behav. Evol. 44, 210–233 [DOI] [PubMed] [Google Scholar]
- Correia M. J., Eden A. R., Westlund K. N., Coulter J. D. (1982). Organization of ascending auditory pathways in the pigeon (Columba livia) as determined by autoradiographic methods. Brain Res. 234, 205–212 10.1016/0006-8993(82)90862-9 [DOI] [PubMed] [Google Scholar]
- Durand S. E., Heaton J. T., Amateau S. K., Brauth S. E. (1997). Vocal control pathways through the anterior forebrain of a parrot (Melopsittacus undulatus). J. Comp. Neurol. 13, 179–206 [DOI] [PubMed] [Google Scholar]
- Ellegren H. (1996). First gene on the avian W chromosome (CHD) provides a tag for universal sexing of non-ratite birds. Proc. R. Soc. Lond. B. 263, 1635–1641 [DOI] [PubMed] [Google Scholar]
- Farabaugh S. M., Wild J. M. (1997). Reciprocal connections between primary and secondary auditory pathways in the telencephalon of the budgerigar (Melopsittacus undulatus). Brain Res. 747, 18–25 10.1016/S0006-8993(96)01143-2 [DOI] [PubMed] [Google Scholar]
- Farries M. A. (2001). The oscine song system considered in the context of the avian brain: lessons learned from comparative neurobiology. Brain Behav. Evol. 58, 80–100 [DOI] [PubMed] [Google Scholar]
- Feenders G., Liedvogel M., Rivas M., Zapka M., Horita H., Hara E., Wada K., Mouritsen H., Jarvis E. D. (2008). Molecular mapping of movement-associated areas in the avian brain: a motor theory for vocal learning origin. PLoS ONE 3, e1768. 10.1371/journal.pone.0001768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune E. S., Margoliash D. (1995). Parallel pathways and convergence onto HVc and adjacent neostriatum of adult zebra finches (Taeniopygia guttata). J. Comp. Neurol. 360, 413–441 [DOI] [PubMed] [Google Scholar]
- Gahr M. (2000). Neural song control system of hummingbirds: comparison to swifts, vocal learning (Songbirds) and nonlearning (Suboscines) passerines, and vocal learning (Budgerigars) and nonlearning (Dove, owl, gull, quail, chicken) nonpasserines. J. Comp. Neurol. 426,182–196 [PubMed] [Google Scholar]
- Hackett S. J., Kimball R. T., Reddy S., Bowie R. C., Braun E. L., Braun M. J., Chojnowski J. L., Cox W. A., Han K. L., Harshman J., Huddleston C. J., Marks B. D., Miglia K. J., Moore W. S., Sheldon F. H., Steadman D. W., Witt C. C., Yuri T. (2008). A phylogenomic study of birds reveals their evolutionary history. Science 320, 1763–1768 10.1126/science.1157704 [DOI] [PubMed] [Google Scholar]
- Heyers D., Manns M., Luksch H., Güntürkün O., Mouritsen H. (2007). A visual pathway links brain structures active during magnetic compass orientation in migratory birds. PLoS ONE 2, e937. 10.1371/journal.pone.0000937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H., Miyatani S., Takeichi M. (1991). R-cadherin: a novel Ca2+-dependent cell-cell adhesion molecule expressed in the retina. Neuron 7, 1–20 10.1016/0896-6273(91)90075-B [DOI] [PubMed] [Google Scholar]
- Jarvis E. D. (2004). Learned birdsong and the neurobiology of human language. Ann. N.Y. Acad. Sci. 1016, 749–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis E. D., Mello C.V. (2000). Molecular mapping of brain areas involved in parrot vocal communication. J. Comp. Neurol. 419, 1–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis E. D., Ribeiro S., da Silva M. L., Ventura D., Vielliard J., Mello C. V. (2000). Behaviourally driven gene expression reveals song nuclei in hummingbird brain. Nature 406, 628–632 10.1038/35020570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Okanoya K. (2010). Molecular characterization of the song control nucleus HVC in Bengalese finch brain. Brain Res. 1360, 56–76 10.1016/j.brainres.2010.09.014 [DOI] [PubMed] [Google Scholar]
- Konishi M. (1965). The role of auditory feedback in the control of vocalization in the white-crowned sparrow. Z. Tierpsychol. 22, 770–783 [PubMed] [Google Scholar]
- Li X., Wang X. J., Tannenhauser J., Podell S., Mukherjee P., Hertel M., Biane J., Masuda S., Nottebohm F., Gaasterland T. (2007). Genomic resources for songbird research and their use in characterizing gene expression during brain development. Proc. Natl. Acad. Sci. U.S.A. 104, 6834–6839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardino A. J., Li X. C., Hertel M., Nottebohm F. (2005). Replaceable neurons and neurodegenerative disease share depressed UCHL1 levels. Proc. Natl. Acad. Sci. U.S.A. 102, 8036–8041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell P. V., Clayton D. F., Replogle K. L., Mello C. V. (2008). Birdsong ‘‘Transcriptomics’’: neurochemical specializations of the Oscine song system. PLoS ONE 3, e3440. 10.1371/journal.pone.0003440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe T., Togashi H., Uchida N., Suzuki S. C., Hayakawa Y., Yamamoto M., Yoda H., Miyakawa T., Takeichi M., Chisaka O. (2000). Loss of cadherin-11 adhesion receptor enhances plastic changes in hippocampal synapses and modifies behavioral responses. Mol. Cell Neurosci. 15, 534–546 10.1006/mcne.2000.0849 [DOI] [PubMed] [Google Scholar]
- Margoliash D., Fortune E. S., Sutter M. L., Yu A. C., Wren-Hardin B. D., Dave A. (1994). Distributed representation in the song system of oscines: evolutionary implications and functional consequences. Brain Behav. Evol. 44, 247–264 [DOI] [PubMed] [Google Scholar]
- Matsunaga E., Kato M., Okanoya K. (2008). Comparative analysis of gene expressions among avian brains: a molecular approach to the evolution of vocal learning. Brain Res. Bull. 75, 474–479 [DOI] [PubMed] [Google Scholar]
- Matsunaga E., Okanoya K. (2008a). Expression analysis of cadherins in the songbird brain: relationship to vocal system evolution. J. Comp. Neurol. 508, 329–342 10.1002/cne.21676 [DOI] [PubMed] [Google Scholar]
- Matsunaga E., Okanoya K. (2008b). Vocal area-related expression of the androgen receptor in the budgerigar (Melopsittacus undulatus) brain. Brain Res. 1208, 87–94 10.1016/j.brainres.2008.02.076 [DOI] [PubMed] [Google Scholar]
- Matsunaga E., Okanoya K. (2008c). Cadherins and vocal learning in avian species. Soc. Neurosci. Abstr. 38, 80, 87. [Google Scholar]
- Matsunaga E., Okanoya K. (2009a). Vocal control area-related expression of neuropilin-1, plexin-A4, and the ligand semaphorin-3A has implications for the evolution of the avian vocal system. Dev. Growth Differ. 51, 45–54 10.1111/j.1440-169X.2008.01080.x [DOI] [PubMed] [Google Scholar]
- Matsunaga E., Okanoya K. (2009b). Evolution and diversity in avian vocal system: an Evo-Devo model from the morphological and behavioral perspectives. Dev. Growth Differ. 51, 355–367 10.1111/j.1440-169X.2009.01091.x [DOI] [PubMed] [Google Scholar]
- Mello C. V., Vates G. E., Okuhata S., Nottebohm F. (1998). Descending auditory pathways in the adult male zebra finch (Taeniopygia guttata). J. Comp. Neurol. 395, 137–160 [PubMed] [Google Scholar]
- Mey J., Thanos S. (2000). Development of the visual system of the chick. I. Cell differentiation and histogenesis. Brain Res. Brain Res. Rev. 32, 343–379 [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Takeichi M. (1995). Neural crest cell-cell adhesion controlled by sequential and subpopulation-specific expression of novel cadherins. Development 121, 1321–1332 [DOI] [PubMed] [Google Scholar]
- Nottebohm F., Kelley D. B., Paton J. A. (1982). Connections of vocal control nuclei in the canary telencephalon. J. Comp. Neurol. 207, 344–357 [DOI] [PubMed] [Google Scholar]
- Nottebohm F., Stokes T. M., Leonard C. M. (1976). Central control of song in the canary, Serinus canarius. J. Comp. Neurol. 165, 457–486 [DOI] [PubMed] [Google Scholar]
- Plummer T. K., Striedter G. F. (2002). Brain lesions that impair vocal imitation in adult budgerigars. J. Neurobiol. 53, 413–428 10.1002/neu.10120 [DOI] [PubMed] [Google Scholar]
- Replogle K., Arnold A. P., Ball G. F., Band M., Bensch S., Brenowitz E. A., Dong S., Drnevich J., Ferris M., George J. M., Gong G., Hasselquist D., Hernandez A. G., Kim R., Lewin H. A., Liu L., Lovell P. V., Mello C. V., Naurin S., Rodriguez-Zas S., Thimmapuram J., Wade J., Clayton D. F. (2008). The songbird neurogenomics (SoNG) initiative: community-based tools and strategies for study of brain gene function and evolution. BMC Genomics 9, 131. 10.1186/1471-2164-9-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redies C., Medina L., Puelles L. (2001). Cadherin expression by embryonic divisions and derived gray matter structures in the telencephalon of the chicken. J. Comp. Neurol. 438, 253–285 [PubMed] [Google Scholar]
- Reiner A., Perkel D. J., Bruce L. L., Butler A. B., Csillag A., Kuenzel W., Medina L., Paxinos G., Shimizu T., Striedter G., Wild M., Ball G. F., Durand S., Gunturkun O., Lee D. W., Mello C. V., Powers A., White S. A., Hough G., Kubikova L., Smulders T. V., Wada K., Dugas-Ford J., Husband S., Yamamoto K., Yu J., Siang C., Jarvis E. D. and Avian Brain Nomenclature Forum. (2004). Revised nomenclature for avian telencephalon and some related brainstem nuclei. J. Comp. Neurol. 473, 377–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Karten H. J. (1993). The Avian Visual System and the Evolution of the Neocortex. Vision, Brain, and Behavior in Birds. Cambridge: MIT Press [Google Scholar]
- Striedter G. F. (1994). The vocal control pathways in budgerigars differ from those in songbirds. J. Comp. Neurol. 343, 35–56 [DOI] [PubMed] [Google Scholar]
- Suzuki S. C., Furue H., Koga K., Jiang N., Nohmi M., Shimazaki Y., Katoh-Fukui Y., Yokoyama M., Yoshimura M., Takeichi M. (2007). Cadherin-8 is required for the first relay synapses to receive functional inputs from primary sensory afferents for cold sensation. J. Neurosci. 27, 3466–3476 10.1523/JNEUROSCI.0243-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S. C., Inoue T., Kimura Y., Tanaka T., Takeichi M. (1997). Neuronal circuits are subdivided by differential expression of type-II classic cadherins in postnatal mouse brains. Mol. Cell Neurosci. 9, 433–447 10.1006/mcne.1997.0626 [DOI] [PubMed] [Google Scholar]
- Takeichi M. (2007). The cadherin superfamily in neuronal connections and interactions. Nat. Rev. Neurosci. 8, 11–20 [DOI] [PubMed] [Google Scholar]
- Wada K., Howard J. T., McConnell P., Whitney O., Lints T., Rivas M. V., Horita H., Patterson M. A., White S. A., Scharff C., Haesler S., Zhao S., Sakaguchi H., Hagiwara M., Shiraki T., Hirozane-Kishikawa T., Skene P., Hayashizaki Y., Carninci P., Jarvis E. D. (2006). A molecular neuroethological approach for identifying and characterizing a cascade of behaviorally regulated genes. Proc. Natl. Acad. Sci. U.S.A. 103, 15212–15217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K., Sakaguchi H., Jarvis E. D., Hagiwara M. (2004). Differential expression of glutamate receptors in avian neural pathways for learned vocalization. J. Comp. Neurol. 476, 44–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade J., Peabody C., Coussens P., Tempelman R. J., Clayton D. F., Liu L., Arnold A. P., Agate R. (2004). A cDNA microarray from the telencephalon of juvenile male and female zebra finches. J. Neurosci. Methods 138, 199–206 [DOI] [PubMed] [Google Scholar]
- Warrant W. C., Clayton D. F., Ellegren H., Arnold A. P., Hillier L. W., Künstner A., Searle S., White S., Vilella A. J., Fairley S., Heger A., Kong L., Ponting C. P., Jarvis E. D., Mello C. V., Minx P., Lovell P., Velho T. A., Ferris M., Balakrishnan C. N., Sinha S., Blatti C., London S. E., Li Y., Lin Y. C., George J., Sweedler J., Southey B., Gunaratne P., Watson M., Nam K., Backström N., Smeds L., Nabholz B., Itoh Y., Whitney O., Pfenning A. R., Howard J., Völker M., Skinner B. M., Griffin D. K., Ye L., McLaren W.M., Flicek P., Quesada V., Velasco G., Lopez-Otin C., Puente X. S., Olender T., Lancet D., Smit A. F., Hubley R., Konkel M. K., Walker J.A., Batzer M.A., Gu W., Pollock D. D., Chen L., Cheng Z., Eichler E. E., Stapley J., Slate J., Ekblom R., Birkhead T., Burke T., Burt D., Scharff C., Adam I., Richard H., Sultan M., Soldatov A., Lehrach H., Edwards S. V., Yang S. P., Li X., Graves T., Fulton L., Nelson J., Chinwalla A., Hou S., Mardis E. R., Wilson R. K. (2010). The genome of a songbird. Nature 464, 757–762 10.1038/nature08819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild J. M., Arends J. J., Zeigler H. P. (1984). A trigeminal sensorimotor circuit for pecking, grasping and feeding in the pigeon (Columba livia). Brain Res. 300, 146–151 10.1016/0006-8993(84)91349-0 [DOI] [PubMed] [Google Scholar]
- Wild J. M., Farabaugh S. M. (1996). Organization of afferent and efferent projections of the nucleus basalis prosencephali in a passerine, Taeniopygia guttata. J. Comp. Neurol. 365, 306–328 [DOI] [PubMed] [Google Scholar]
- Wild J. M., Reinke H., Farabaugh S. M. (1997). A non-thalamic pathway contributes to a whole body map in the brain of the budgerigar. Brain Res. 755, 137–141 10.1016/S0006-8993(97)00026-7 [DOI] [PubMed] [Google Scholar]