Abstract
Nuclear Factor-κB (NF-κB)/Rel transcription factors form an integral part of innate immune defenses and are conserved throughout the animal kingdom. Studying the function, mechanism of activation and regulation of these factors is crucial for understanding host responses to microbial infections. The fruit fly Drosophila melanogaster has proved to be a valuable model system to study these evolutionarily conserved NF-κB mediated immune responses. Drosophila combats pathogens through humoral and cellular immune responses. These humoral responses are well characterized and are marked by the robust production of a battery of anti-microbial peptides. Two NF-κB signaling pathways, the Toll and the IMD pathways, are responsible for the induction of these antimicrobial peptides. Signal transduction in these pathways is strikingly similar to that in mammalian TLR pathways. In this chapter, we discuss in detail the molecular mechanisms of microbial recognition, signal transduction and NF-κB regulation, in both the Toll and the IMD pathways. Similarities and differences relative to their mammalian counterparts are discussed, and recent advances in our understanding of the intricate regulatory networks in these NF-κB signaling pathways are also highlighted.
1 Introduction
Nuclear Factor-kappaB/Rel proteins are an evolutionarily conserved class of transcription factors that play crucial roles in regulating many animal physiological processes such as, cell survival, proliferation, and most importantly, immune responses. In general, NF-κB/Rel proteins are found inactive in the cytoplasm associated with their inhibitory proteins or domains (IκBs), and various challenges trigger the degradation or cleavage of these IκBs thereby allowing the NF-κB transcription factor to translocate into the nucleus. NF-κB transcription factors play a central role in the induction of many cytokines and immune effector genes that initiate robust pro-inflammatory responses. Owing to the conserved structure and function of NF-κB/Rel proteins across the animal kingdom, various model systems have been extensively used to probe the molecular mechanisms of NF-κB activation and its role in inflammation, infection and disease.
Over the past 20 years, Drosophila melanogaster has been a favored model system for the study of NF-κB regulation and function in developmental biology and in immunity. The NF-κB factor Dorsal, and the Toll pathway which activates it, were first discovered in Drosophila because of their central role in establishing the dorsoventral pattern in the developing embryo (Nusslein-Volhard et al. 1987; Steward 1987). Moreover, the three Drosophila NF-κB factors, Dorsal, Dorsal-related immunity factor (DIF) and Relish, are the master regulators of the insect humoral immune response, which is characterized by the rapid and robust production of a battery of potent antimicrobial peptides (AMPs). These AMPs, along with other cellular defense strategies such as clotting, melanization, phagocytosis and encapsulation, effectively eliminate many microbes. The Drosophila NF-κB/Rel signaling modules display a striking degree of conservation with mammals, and this has made Drosophila a valuable experimental model for studying these critical NF-κB innate immune responses against both insect and human pathogens (Mansfield et al. 2003; Apidianakis et al. 2005; Scully and Bidochka 2006; Vonkavaara et al. 2008).
This review focuses on the role of NF-κB/Rel transcription factors in the immune defenses of Drosophila, highlighting both similarities and differences in the corresponding mammalian systems, and also, how studies in the Drosophila model system contribute to our understanding of these signaling networks in mammals.
2 Microbial Recognition and Humoral Responses
2.1 Anti-microbial Peptides
The Drosophila humoral immune system responds to microbial challenge by triggering the expression of anti-microbial peptide genes through NF-κB signaling pathways. In fact, NF-κB/Rel proteins control the transcription of almost one half of the immune responsive genes, including the collection of cationic AMPs (De Gregorio et al. 2001, 2002). As a result of infection-induced NF-κB activation, AMPs that are undetectable in the hemolymph (blood) of unchallenged flies are rapidly elevated to concentrations up to 100 μM (Hoffmann and Reichhart 2002). As best we know, the regulation of AMP production occurs at the level of gene expression. AMP genes are direct targets of NF-κB activation and their transcription is induced to very high levels rapidly after infection. Although the bulk of AMP production occurs in the insect fat body (similar to the mammalian liver), AMP genes are also expressed in circulating phagocytic hemocytes and local epithelial tissues, particularly, the gut and the trachea (Ferrandon et al. 1998; Tzou et al. 2000; Liehl et al. 2006). Two NF-κB signaling pathways control AMP gene expression—the Toll and the IMD pathways. These pathways are activated by microbial cell walls and/or other virulence determinants by circulating, cell surface, and/or cytosolic receptors. Each pathway responds to distinct microbial components and induces the expression of somewhat overlapping subsets of AMP and other immune responsive genes. For example, the antifungals Drosomycin and Metchnikowin are strongly induced by the systemic Toll pathway, while the IMD pathway induces antibacterial peptides such as Diptericin (Lemaitre et al. 1997; Tzou et al. 2002). On the other hand, some AMP genes such as Cecropin and Attacin are co-operatively regulated by both the pathways (Manfruelli et al. 1999). Details on these regulatory events are discussed in more detail below.
2.2 Peptidoglycan Recognition and Immune Activation
Similar to the mammalian innate immune responses, microbial cell walls are one of the most potent agonists of insect humoral immunity. Bacterial infections trigger either the Toll or the IMD pathway, depending on the structure of their peptidoglycan (PGN) cell wall. PGN is a polymeric glycopeptide that forms the cell wall of most bacteria. PGN contains long glycan chains usually composed of alternating residues of N-acetyl glucosamine (NAG) and N-acetylmuramic acid (NAM), with short stem-peptides of alternating l- and d-amino acids attached to the lactyl group of NAM. The third position in the stem peptide is highly variable with a meso-diaminopimelic acid (DAP) in most Gram-negatives and various Bacilli spp. and L-lysine (lys) in many other Gram-positive microbes (Schleifer and Kandler 1972). These stem peptides are often cross-linked to each other by short peptide bridges to create a rigid cell wall structure. The Toll signaling pathway is most robustly triggered by Lys-type peptidoglycan, while the IMD pathway is activated by DAP-type peptidoglycan (Leulier et al. 2003; Kaneko et al. 2004). The distinction is not completely black and white, and bacterial species with DAP-type PGN also activate Toll signaling, albeit more weakly (Leulier et al. 2003; Leone et al. 2008).
Activation of both Toll and IMD pathways by bacterial PGNs requires peptidoglycan recognition proteins (PGRPs). PGRP was initially identified in lepid-opterans as a protein capable of binding PGN (Yoshida et al. 1996; Kang et al. 1998; Ochiai and Ashida 1999). This was followed by the discovery of 13 PGRP genes (encoding 17 distinct proteins) in Drosophila, classified as short proteins (SA, SB1, SB2, SC1A, SC1B, SC2 and SD) and long proteins (LA, LB, LC, LD, LE and LF) (Werner et al. 2000). While the short proteins include a signal sequence and are secreted soluble proteins, the long PGRPs lack a signal sequence. Some long-form PGRPs encode a transmembrane domain and are cell surface receptors, e.g. PGRP-LC, while others are cytoplasmic, e.g. PGRP-LE. The PGRP domain is highly conserved from insects to mammals, whereas the other regions show very little similarity (Werner et al. 2000). The PGRP domain is structurally related to a class of PGN-digesting enzymes known as type 2 amidases (e.g. T7 lysozyme), which hydrolyzes the bond between the lactyl group in NAM and l-alanine in the stem-peptide of PGN (Mellroth et al. 2003). In fact, approximately one-half of the Drosophila PGRPs are type 2 amidases that degrade PGN and reduce its ability to stimulate immune responses (detailed in Sect. 6.1), while the other PGRPs lack key catalytic residue(s) and function as PGN receptors. In particular, PGRP-SA and PGRP-SD are two soluble PGN receptors that function in the Toll pathway (Michel et al. 2001; Bischoff et al. 2004), while PGRP-LC and PGRP-LE recognize DAP-type PGN and trigger the IMD pathway (Choe et al. 2002; Gottar et al. 2002; Takehana et al. 2004; Kaneko et al. 2006).
2.2.1 Activation of the Toll Pathway
In the Toll pathway, PGN recognition involves two circulating receptor proteins, PGRP-SA and PGRP-SD. Recognition of some Lys-type PGN (e.g. M. luteus) specifically requires PGRP-SA (Michel et al. 2001; Gottar et al. 2002), while other PGNs (e.g. S. aureus) may be detected by either PGRP-SA or PGRP-SD (Bischoff et al. 2004). PGRP-SA binds Lys-type PGN and the crystal structure of PGRP-SA complexed with a Lys-type muropeptide has been solved (Chang et al. 2004; Reiser et al. 2004). However, the precise biochemical mechanisms that underlie the differential recognition of various Lys-type PGNs by PGRP-SA and/or PGRP-SD are not completely understood. In particular, the exact function of PGRP-SD remains a mystery, as genetic studies link it to Lys-type PGN recognition, while structural studies indicate that it binds DAP-type PGN (Leone et al. 2008). In fact, PGRP-SD has been shown to be responsible for the weak Toll activation triggered by DAP-type PGN (Leone et al. 2008).
Lys-type PGN recognition also requires a third protein known as Gram-negative-binding protein (GNBP1). GNBP1 was originally identified in silkworms, as a protein that binds Gram-negative PGN/β-(1,3)-glucan (Yoshida et al. 1986). Conversely, later studies showed that GNBP1 is required for the recognition of Lys-type PGN and is dispensable for the activation of the IMD pathway by DAP-type PGN (Gobert et al. 2003; Pili-Floury et al. 2004). While GNBP1 and the above mentioned receptors, PGRP-SA and PGRP-SD, may form one large complex upon PGN recognition (Filipe et al. 2005; Wang et al. 2006a, b), the precise mechanistic function of GNBP1 is controversial. Some reports conclude that GNBP1 hydrolyzes Lys-type PGN into muropeptide dimers, for optimal recognition by PGRP-SA (Filipe et al. 2005; Wang et al. 2006a, b), while other groups report no such activity associated with GNBP1 (Park et al. 2007; Buchon et al. 2009). Instead, these groups conclude that GNBP1 may be critical for activating downstream signaling events (more details below). Thus, many Gram-positive bacteria are sensed by their Lys-type PGN cell wall by the proteins, PGRP-SA/PGRP-SD and GNBP1.
In addition to bacterial PGN, fungal cell walls are also potent activators of the Toll pathway (Gottar et al. 2006). A receptor known as GNBP3 recognizes β-(1,3)-glucans from the fungal cell wall, and robustly triggers the Toll pathway (Gottar et al. 2006). Moreover, GNBP3 mutant flies (hades) exhibit greatly reduced Drosomycin expression and reduced survival following Candida albicans infection (Gottar et al. 2006). Intriguingly, some fungi (e.g. Geotrichum candidum) also seem to activate the IMD pathway, through unknown mechanisms (Hedengren-Olcott et al. 2004). β-glucan recognition by GNBP3 and PGN recognition by PGRP-SA/SD/GNBP1 both feed into the Toll pathway via the same protease, Modular Serine Protease (Buchon et al. 2009).
The Modular Serine Protease (ModSP) was first discovered in biochemical analyses of the Toll pathway in the mealworm beetle, Tenebrio molitor (Kim et al. 2008). In this system, ModSP binds the PGRP-SA/GNBP1 complex following PGN recognition and triggers a serine protease cascade upon association with these receptors (Park et al. 2007; Kim et al. 2008). Likewise, in the Drosophila system, genetic experiments with a ModSP mutant demonstrate its central role in the Toll pathway activation by Lys-type PGN and β-glucans. Genetic experiments suggest that Drosophila ModSP also functions immediately downstream of both PGRP-SA/GNBP1 and GNBP3 (Buchon et al. 2009). Recruitment of ModSP to the PGN-receptor complex has been proposed to activate its proteolytic activity. Subsequently, ModSP initiates a cascade that proceeds through the sequential activation of CLIP domain containing serine proteases Grass and Spätzle processing enzyme (SPE) (Kambris et al. 2006; El Chamy et al. 2008). A third CLIP domain serine protease, Spirit, as well as some non-catalytic serine protease homologs (SPHs)—spheroide and sphinx1/2—may also function in this pathway. However, the role of these genes in the Toll pathway was discovered by RNAi-based approaches, and has not yet been confirmed by biochemistry or conventional genetics (Kambris et al. 2006). In any case, this proteolytic cascade culminates in the activation of SPE which directly cleaves pro-Spätzle and releases the active Toll ligand, Spätzle (Jang et al. 2006). Pro-Spätzle is a disulfide-linked dimer and upon processing the C-terminal 106 residue fragment binds Toll, induces receptor homodimerization, and activates intracellular signaling (Mizuguchi et al. 1998; Weber et al. 2003; Hu et al. 2004).
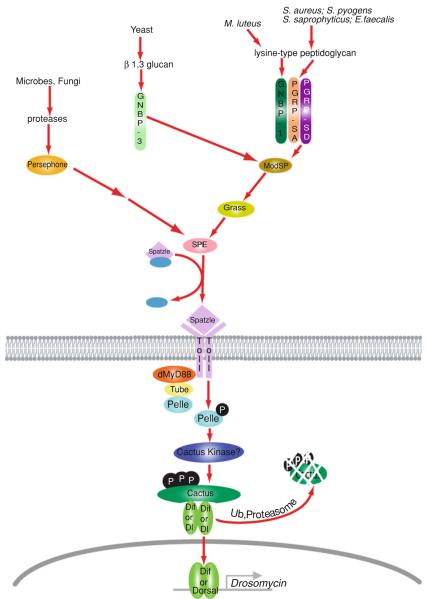
In addition to microbial cell walls, the Toll pathway can also be triggered by virulence factors, such as proteases released by some bacteria or fungi (Gottar et al. 2006; El Chamy et al. 2008). Entomopathogenic microbes often produce proteases to digest the insect cuticle (St Leger et al. 1992), and flies have evolved a mechanism to ‘guard’ against this signature of virulent infection. For example, both the PR1 protease of entomopathogenic fungi M. anisopliae and the subtilisin protease from B. subtilis can trigger the Toll pathway independent of the PGRP and GNBP-mediated cell wall-recognition pathways (Gottar et al. 2006; El Chamy et al. 2008). Instead, these virulent proteases appear to cleave and activate a host-encoded serine protease known as Persephone (Gottar et al. 2006). Persephone, in turn, converges on the same SPE utilized in the PGRP/GNBP pathways. However, this virulence protease-Persephone pathway is independent of ModSP and Grass (El Chamy et al. 2008; Buchon et al. 2009) (Refer to Fig. 1 for a model of how these microbial recognition pathways converge on Toll activation).
Fig. 1.
Drosophila Toll pathway. Drosophila Toll pathway is a cytokine receptor pathway that responds indirectly to microbial infection. Distinct circulating receptor proteins recognize different microbial structures or activities and activate a protease cascade, which culminates in the processing of Spätzle. Lys-type PGN, from bacterial cell walls, is recognized by PGRP-SA/PGRP-SD and GNBP1, while GNBP3 detects β-(1, 3)-glucan from fungal cell walls. These upstream recognition events trigger a proteolytic cascade which proceeds through the serine proteases ModSP and Grass to activate Spätzle Processing Enzyme (SPE) which in turn cleaves pro-Spätzle to release the mature Toll ligand, Spätzle. In addition, virulence-associated proteases, released by pathogens, cleave and activate another pathway that requires the protease Persephone and converges on the activation of SPE. The intracellular Toll signaling pathway is very homologous to the mammalian MyD88-dependent signaling pathway. It signals through an upstream complex containing the adaptor proteins dMyD88, Tube and the kinase Pelle (an IRAK homolog). Pelle triggers the phosphorylation, K48-ubiquitination and proteasomal degradation of the IκB protein, Cactus, thereby releasing the Drosophila NF-κB/Rel factors DIF/Dorsal to translocate into the nucleus and transcribe target AMP genes, such as Drosomycin
2.2.2 Activation of the IMD Pathway
Detection of DAP-type PGN by two long form PGRPs, PGRP-LC and PGRP-LE triggers the IMD pathway (Choe et al. 2002; Gottar et al. 2002; Ramet et al. 2002; Takehana et al. 2002, 2004; Leulier et al. 2003; Kaneko et al. 2006). DAP-type PGN forms the cell wall of most Gram-negative bacteria as well as some Gram positive bacteria (e.g. Bacilli spp.) (Schleifer and Kandler 1972). Double mutants, lacking both PGRP-LC and LE, are unable to induce AMPs in response to Gram-negative bacteria and are highly susceptible to these infections (Takehana et al. 2004). PGRP-LC is a type 2 transmembrane receptor, with an extracellular PGRP domain that is critical for recognizing extracellular bacteria, while PGRP-LE lacks a transmembrane domain and functions as an intracellular receptor for DAP-type PGN (Kaneko et al. 2006; Yano et al. 2008). Interestingly, PGRP-LC encodes for three receptors via alternate splicing, known as PGRP-LCx, PGRP-LCy and PGRP-LCa (Werner et al. 2003). Each of these receptors has a unique extracellular domain fused to the identical intracellular signaling domain. Although the function of PGRP-LCy remains unclear, the other PGRP-LC splice forms have prominent roles in activating the IMD pathway. PGRP-LCx alone is sufficient to respond to polymeric PGN (as isolated from E. coli), whereas both PGRP-LCx and PGRP-LCa are required to recognize a monomeric PGN fragment known as TCT (Werner et al. 2003; Kaneko et al. 2004). In fact, TCT causes heterodimerization of PGRP-LCx and PGRP-LCa (Whalen and Steward 1993; Chang et al. 2005, 2006; Mellroth et al. 2005).
Additionally, small monomeric PGN fragments like TCT that enter cells are recognized by the cytosolic receptor PGRP-LE. TCT induces the formation of head-to-tail homo-oligomers of PGRP-LE (Lim et al. 2006). It is unclear how TCT gains access into the cytosol where it can be recognized by PGRP-LE. It has been hypothesized that a transmembrane transporter may aid in the entry of TCT into the cytoplasm, however, no such protein has been identified. PGRP-LE is also triggered by cytosolic bacteria with DAP-type PGN, like Listeria monocytogenes (Yano et al. 2008). While both PGRP-LC and PGRP-LE are potent activators of the IMD pathway, PGRP-LE can also trigger an autophagic response through a Relish (NF-κB)-independent pathway that is critical to protect the animal against intracellular pathogens like Listeria (Yano et al. 2008). Interestingly, a cleaved, PGRP-domain only form of PGRP-LE can be found extracellularly, where it aids the recognition of DAP-type PGN by the transmembrane receptor PGRP-LC (Kaneko et al. 2006). However, it is unclear how this cleaved form of PGRP-LE reaches the extracellular milieu. With either PGRP-LC or PGRP-LE, recognition of DAP-type PGN triggers the IMD signaling pathway, which in turn leads to the activation of the NF-κB precursor protein Relish (detailed later in the review).
As in Drosophila, mammalian innate immune signaling pathways respond to many immune stimuli, including microbial components, endogenous danger signals and pro-inflammatory cytokines, and converge on the activation of NF-κB. The receptors involved include Toll-like receptors (TLRs), NOD-like Receptors (NLRs), RIG-I-like Receptors (RLRs), and cytokine receptors (i.e. TNFR and IL-1R). These receptors are found in many different subcellular niches, including the cell surface, cytosol, or endosomal compartments. They recognize a variety of microbial signatures from bacterial, fungal, viral and parasitic pathogens and trigger overlapping signaling pathways that all lead to the activation of NF-κB and downstream responses.
The Drosophila NF-κB pathways display some striking similarities, as well as some notable differences, compared to these mammalian pathways. The paradigm for recognition of pathogens in the Drosophila Toll pathway is quite different from that exhibited by the mammalian TLRs. While mammalian TLRs are directly involved in detecting microbial derived products, such as LPS, lipopeptides, flagellin and nucleic acids, Drosophila Toll is a cytokine receptor that responds to microbial challenge indirectly. The key microbe detectors in the Drosophila Toll pathway are PGRP-SA, PGRP-SD and the GNBPs, which are not homologous to the TLRs. In case of the IMD pathway, PGRP-LC functions analogously to the TLRs as a cell surface receptor, and PGRP-LE is more similar to the cytosolic NOD receptor, although no molecular homology exists between TLRs or NLRs and the Drosophila PGRPs.
In the next section, we discuss in detail the more similar aspects of Drosophila NF-κB systems with that of mammals intracellular signal transduction and the mechanism of NF-κB activation in the Toll and the IMD pathways. These pathways show a remarkable degree of similarity to the MyD88-dependent and the MyD88-independent TLR pathways in mammals, respectively.
3 Intracellular Signaling in the Toll and IMD Pathways
Activation of the Toll or IMD pathways by any of the mechanisms outlined above leads to robust activation of Drosophila NF-κB/Rel family transcription factors. Recognition of DAP-type PGN through the IMD pathway triggers the activation of Relish. Activation of Toll, by Lys-type PGN, fungal cell walls or virulence-associated proteases, leads to the activation of either of the two p65-like factors—DIF or Dorsal. Each of these pathways uses distinct set of intracellular signaling components to drive NF-κB activation.
3.1 Toll Pathway
Activation Drosophila of Toll leads to the formation of a receptor proximal signaling complex containing the adaptor proteins MyD88, Tube and the kinase Pelle. Toll signals through its intracellular TIR domain, similar to the mammalian TLRs and IL-1R. Toll interacts with Drosophila MyD88 (homolog of mammalian MyD88) through a homotypic TIR interaction (Tauszig-Delamasure et al. 2002). MyD88 also has a death domain (DD) through which it assembles a heterotrimeric complex containing Tube and Pelle (homolog of the IRAK kinases in mammals) (Horng and Medzhitov 2001; Sun et al. 2002). Both Tube and Pelle also contain DDs. Tube associates with MyD88 through a homotypic DD interaction on one surface, while simultaneously recruiting Pelle, through another homotypic DD interaction, on the opposite surface (Schiffmann et al. 1999; Sun et al. 2002). Assembly of this signaling complex appears to be sufficient to activate the Toll pathway as over-expression of the MyD88 TIR domain triggers expression of the Toll pathway target gene Drosomycin (Tauszig-Delamasure et al. 2002). Recruitment of the kinase Pelle to this complex triggers its auto-phosphorylation and activation (Sun et al. 2004).
Activation of the kinase Pelle triggers the phosphorylation and degradation of Cactus. Cactus is the IκB-like protein which sequesters Dorsal and DIF in the cytoplasm. It is unclear whether Pelle directly phosphorylates Cactus or if other intermediate kinase(s) are involved. Although one study reported that Pelle phosphorylates Cactus in vitro (Grosshans et al. 1994), it remains unclear if Pelle phosphorylates the critical serine residues on Cactus which are known to be important for Toll signaling (see below for more detail) (Fernandez et al. 2001), or if Pelle-mediated phosphorylation of Cactus occurs in vivo. Experiments in which Pelle was forcibly anchored to the plasma membrane demonstrated that localization of this kinase to the membrane is sufficient to activate Toll signaling and this signaling requires kinase activity (Galindo et al. 1995). Thus, Pelle localization to the Toll receptor, via the MyD88 and Tube interaction network, is likely the critical event driving the activation of Pelle, the subsequent degradation of Cactus, and the nuclear translocation of DIF or Dorsal (Galindo et al. 1995; Reach et al. 1996; Edwards et al. 1997; Towb et al. 1998).
3.2 IMD Pathway
Intracellular signaling in the IMD pathway is also likely to involve a receptor–proximal multi-protein complex. In this pathway, PGRP-LC and PGRP-LE detect DAP-type PGN through their C-terminal PGN-binding PGRP domain and transduce signal through their extended N-terminal domains (Choe et al. 2005; Kaneko et al. 2006). Although PGRP-LC and LE are very similar in their PGRP domain, only a short stretch of about 20 amino acids is common to their N-terminal domains. In both receptors, deletion or mutation within this conserved domain abrogates signaling (Kaneko et al. 2006). This conserved domain has some weak similarity to the RHIM domain found in RIP1 and TRIF, signaling proteins that function in the mammalian TLR3 signaling pathway (Meylan et al. 2004). The molecular mechanism by which the RHIM-like domains in PGRP-LC and -LE regulate signaling is unclear. It may be involved in the dimerization of the entire N-terminal signaling domain, which has been reported to occur when PGRP-LC is over-expressed, and/or may interact with an unidentified factor (Choe et al. 2005). One factor that does bind both PGRP-LC and PGRP-LE is IMD, a death domain-containing protein (Georgel et al. 2001; Choe et al. 2005; Kaneko et al. 2006; Aggarwal et al. 2008). In the case of PGRP-LE, IMD binding requires the RHIM-like domain and this is a possible explanation for the function of the PGRP-LE RHIM-like motif. However, the association between IMD and PGRP-LC maps to another distinct region, which is not essential for signaling (Kaneko et al. 2006). Together, these data suggest that the RHIM-like domain of PGRP-LC has a function beyond IMD recruitment and that direct IMD recruitment to these receptors may not always be necessary for signaling. Regardless of the mechanism, IMD associates with these receptors and, in turn, is likely to recruit the Drosophila FADD homolog via a homotypic death domain interaction (Naitza et al. 2002). FADD, is then known to interact with the apical caspase DREDD (homolog of mammalian caspase-8) via a homotypic interaction between the death effector domain (DED) in these proteins (Hu and Yang 2000; Leulier et al. 2002). It will be interesting to learn whether recruitment of DREDD to the receptor–IMD–FADD complex is sufficient for its activation.
DREDD is critical for the initiation of downstream signaling events in the IMD pathway (Leulier et al. 2000; Stoven et al. 2003). In response to PGN stimulation, IMD is cleaved in a DREDD-dependent manner at a caspase cleavage site in the N-terminal region (LEKD30) (Paquette et al. 2010). Immediately following this cleavage site is a consensus IAP-binding motif (IBM, 31AAPV). Both the caspase site and the IBM are highly conserved in multiple species of Drosophila as well as the Anopheles gambiae mosquito (Paquette et al. 2010). DREDD-mediated cleavage of IMD exposes the IBM, allowing IMD to associate with the Drosophila inhibitor of apoptosis 2 protein (DIAP2). The BIR1 and BIR2 domains of DIAP2 are responsible for interacting with the IBM of cleaved IMD, similar to the IBM–BIR interactions observed in the regulation of caspase-mediated programmed cell death (Shi 2002). Further in vivo evidence demonstrating the important role for the IBM-mediated association is provided by the imd1 allele. This strong hypomorphic allele carries the relatively conserved substitution of Ala31 to Val, disrupting the IBM and markedly interfering with the association between cleaved-IMD and DIAP2 (Paquette et al. 2010).
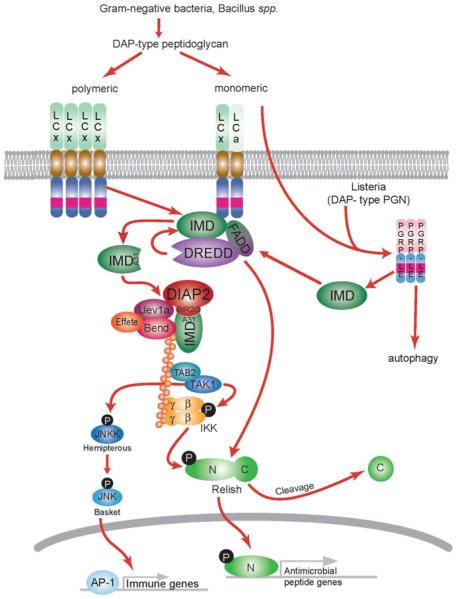
DIAP2, similar to other IAP proteins, also includes a C-terminal RING domain, which is also critical for the IMD signaling pathway (Huh et al. 2007). In fact, DIAP2 lacking a functional RING domain fails to support IMD signaling and the RING domain is essential for the robust ubiquitination of IMD that is observed following its cleavage and association with DIAP2. IMD is conjugated with K63-polyubiquitin chains, similar to what is observed with the mammalian RIP1 and TRAF6 proteins in TNFR and IL-1R signaling, respectively (Ea et al. 2006; Lamothe et al. 2007). IMD ubiquitination involves two distinct E2 ubiquitin-ligase enzymes, which appear to act in a partially redundant manner (Zhou et al. 2005; Paquette et al. 2010). Uev1a, Bendless (Ubc13) form a well-established K63-chain generating enzyme complex (Hofmann and Pickart 1999; Windheim et al. 2008). Effete (Ubc5) has recently been shown to generate K63-chains, but can also generate other types of polyubiquitin (including K48 and linear ubiquitin chains) depending on the context (Kirkpatrick et al. 2006; Jin et al. 2008; Bianchi and Meier 2009). IMD fails to be ubiquitinated only when both Bendless/Uev1a and Effete ubiquitin conjugation enzymes are knocked down (Paquette et al. 2010). It is not yet clear what types of polyubiquitin chains Effete generates in the IMD signaling pathway. Polyubiquitinated IMD has been proposed to act as a scaffold for recruiting the downstream kinases TAK1 and IKK, via ubiquitin binding domains found in their partners TAB 2 and IKKγ, respectively, similar to mammalian NF-κB signaling (Kanayama et al. 2004; Zhuang et al. 2006; Bianchi and Meier 2009; Iwai and Tokunaga 2009). Once recruited and activated, TAK1 activates IKK, which in turn phosphorylates the NF-κB precursor protein Relish (Vidal et al. 2001; Silverman et al. 2003; Stoven et al. 2003; Erturk-Hasdemir et al. 2009). Relish is also cleaved, likely by the caspase DREDD, resulting in the uncoupling of the C-terminal IκB-like domain from the N-terminal NF-κB module, thereby allowing the N-terminal Rel Homology Domain (RHD) to translocate into the nucleus, while the C-terminal domain remains in the cytoplasm (Stoven et al. 2003). Although Relish phosphorylation is dispensable for its cleavage, it appears to enhance the activity of Relish as a transcription factor in the nucleus (Erturk-Hasdemir et al. 2009). In a separate arm of the pathway, TAK1 also activates the JNK kinase, which in turn triggers the phosphorylation and nuclear translocation of the AP-1 transcription factor (refer to Fig. 2 for IMD pathway model).
Fig. 2.
Drosophila IMD pathway. Recognition of DAP-type PGN by the cell surface receptor PGRP-LC or the cytosolic receptor PGRP-LE triggers the IMD pathway. DAP-type PGN forms the cell wall of most Gram-negative bacteria and some Gram-positive bacteria, such as Bacillus spp. and Listeria. Extracellularly, detection of polymeric and monomeric PGN (TCT) occurs through homo or heterodimers of PGRP-LC splice forms, as depicted. PGRP-LE detects intracellular bacteria and stimulates the IMD pathway as well as an autophagic response. PGRP-LC and PGRP-LE signaling proceeds through IMD, FADD and the caspase DREDD, which are likely to form a complex on the activated receptor. DREDD-dependent cleavage of IMD exposes an IAP-binding motif in IMD, through which it binds DIAP2. This IMD-DIAP2 association leads to robust K63-ubiquitination of cleaved IMD by DIAP2, Uev1a, Bendless (Ubc13) and Effete (Ubc5). These K63-polyubiquitin chains are likely to serve as a scaffold for the recruitment and activation of the TAK1 kinase complex and the IKK complex. Once activated, TAK1 in turn activates both IKK and JNK kinase. Relish is phosphorylated by IKK and cleaved by DREDD to release the fully active N-terminal RHD for translocation into the nucleus and the transcriptional induction of AMP genes and other targets. The concurrent activation of JNKK leads to the phosphorylation and nuclear translocation of AP-1
Toll and IMD signaling pathways culminate in the activation of the Drosophila NF-κB transcription factors Dorsal, DIF and Relish. Since the structure and function of these NF-κB members are very similar to mammals, a quick glance at the NF-κB factors in general, will enable us to appreciate the similarities in the regulation of NF-κB proteins in these systems.
4 NF-κB Proteins
The NF-κB/Rel family of transcription factors are master regulators of host immune responses in mammals as well as in Drosophila. In mammals, this family includes the Rel proteins (RelA (p65), RelB, c-Rel) and NF-κB precursor proteins (NF-κB1 (p105) and NF-κB2 (p100)). In Drosophila, the NF-κB/Rel family includes two p65-like factors—dorsal and DIF, and one NF-κB precursor protein, Relish. NF-κB proteins are subdivided into four classes based on their structure and evolutionary divergence (Huguet et al. 1997; Liang et al. 2004). Class I includes c-Rel, RelA (p65) and RelB. The class I proteins contain a conserved 300 residue N-terminal RHD which includes a nuclear localization signal (NLS) and mediates their interaction with DNA, IκB inhibitory proteins, and other Rel proteins. IκB proteins contain ankyrin repeats through which they associate with the RHDs and mask their NLS, sequestering them in the cytosol of unstimulated cells. In addition, class I NF-κB factors contain a C-terminal transactivating domain. In the classical NF-κB pathway, pro-inflammatory stimuli (e.g.: LPS) trigger the activation of IKKβ, which in turn phosphorylates IκBα on serines 32 and 36 (Brown et al. 1995; Traenckner et al. 1995). Phosphorylated IκBα is then recognized by the β-TrCP-SCF ubiquitin ligase complex, K48-ubiquitinated, and degraded by the proteasome, thereby releasing NF-κB to translocate into the nucleus. Class I NF-κB factors form homo- or heterodimers, with p65-p50 heterodimers being the most common and well studied. While many of these dimers are transcriptionally active, some dimers, such as p50 or p52 homodimers, can be repressive (Ghosh et al. 1998). Drosophila Dorsal and DIF also belong to class I, and exhibit about 40% identity within the RHD of the mammalian Rel proteins in this group (Steward 1987; Meng et al. 1999; Minakhina and Steward 2006). The mechanism of their regulation is very similar to mammals, as discussed above and below.
In contrast to regulation of class I NF-κB members by IκB proteins, class II members are auto-regulated by their own inhibitory domains. For example, NF-κB1 and NF-κB2 (p105, p100) harbor inhibitory ankyrin repeats in their C-terminus, in addition to the N-terminal RHD domain. NF-κB1 and NF-κB2 are activated by phosphorylation, ubiquitination and partial proteasomal degradation of the ankyrin repeats. This processing results in the removal of the inhibitory C-terminal ankyrin repeats, to reveal the active subunits, p50 and p52, respectively (Perkins 2007). It should be noted that activation of p100 follows a non-canonical pathway that is independent of IKKβ and instead involves activation of IKKα. IKKα in turn phosphorylates p100 and marks it for controlled degradation (Senftleben et al. 2001). Proteasomal processing of p105 is thought to occur constitutively, by a co translational mechanism (Lin et al. 1998). Although Drosophila Relish is structurally similar to NF-κB1 and NF-κB2, phylogenetic analysis suggest it should be placed in a separate class, class III, because it may have evolved from an earlier ancestor of both the class I and class II factors (Huguet et al. 1997). Consistent with this molecular evolutionary analysis, Relish regulation is distinct from that of NF-κB1 and NF-κB2. It is not activated by partial proteasomal degradation, but instead is endoproteolytically cleaved by a caspase to uncouple the inhibitory C-terminal IκB-like ankyrin repeats from the N-terminal RHD. Both cleavage products appear to be relatively stable; the N-terminal RHD translocates to the nucleus to activate target genes while the C-terminal IκB-like domain remains in the cytosol (Stoven et al. 2003; Erturk-Hasdemir et al. 2009).
Class IV, is occupied by the related NF-AT family of transcription factors. Although NF-AT proteins are structurally and phylogenetically related to the NF-κB RHD domain, and function in the immune response, the mechanism of activation is distinct and their most well studied functions are in the adaptive immune response (Huguet et al. 1997). Therefore, they will not be discussed further here, but more information can be found in these reviews (Rao et al. 1997; Serfling et al. 2004).
4.1 Dorsal and DIF
Drosophila Dorsal was the first identified NF-κB protein in Drosophila, due to its role in early embryonic patterning. In particular, dorsal was identified as one of the 12 maternal effect genes that drive the expression of zygotic genes required for the normal development of the dorso-ventral pattern in the developing embryo. Loss of function mutation in 11 of these genes, results in dorsalized embryos, while loss of function of the 12th gene, cactus, results in a ventralized embryo (Anderson and Nusslein-Volhard 1984; Roth et al. 1989; Schupbach and Wieschaus 1989). Together, these genes identified the Toll pathway. During embryogenesis, developmental cues activate Toll signaling on the ventral side, leading to Cactus degradation and creating a gradient of nuclear Dorsal (Roth et al. 1989; Steward 1989). Dorsal, in turn, drives the transcription of key genes, required for creating the body axis, like twist and snail (Jiang et al. 1991; Pan et al. 1991; Thisse et al. 1991; Ip et al. 1992). Although dorsal and the Toll pathway were initially identified for their role during development, the identification of κB-binding motifs in the promoter/enhancer regions of several AMP genes suggested that humoral response may also be regulated by NF-κB factors (Engstrom et al. 1993; Georgel et al. 1993; Kappler et al. 1993).
Subsequently, two other Drosophila NF-κB factors, DIF and Relish, were discovered (Ip et al. 1993; Dushay et al. 1996). Rapidly thereafter, Relish was linked to Gram-negative response through the IMD signaling pathway (Hedengren et al. 1999), while Toll-mediated humoral responses were shown to be DIF and/or Dorsal dependent (Ip et al. 1993; Manfruelli et al. 1999; Meng et al. 1999; Rutschmann et al. 2000a, b). In larvae, DIF and Dorsal function in a redundant fashion to control AMP gene expression (humoral immunity) and promote blood cell survival (cellular immunity) (Ip et al. 1993; Manfruelli et al. 1999; Matova and Anderson 2006, 2010). However, in adults, humoral responses are primarily controlled by DIF (Rutschmann et al. 2000a, b).
Like p50/p65 in mammals, Dorsal and DIF are regulated by the IκB protein Cactus (Geisler et al. 1992). Cactus is the only identified IκB protein in Drosophila and has six ankyrin repeats. In unstimulated cells, Dorsal exists in a complex with Cactus in the cytoplasm mediated by an interaction between the RHD of Dorsal and the ankyrin repeats of Cactus (Govind et al. 1992; Kidd 1992; Isoda and Nusslein-Volhard 1994; Tatei and Levine 1995). Toll activation results in the phosphorylation, K48-ubiquitination and degradation of Cactus, thereby releasing Dorsal which leads to its translocation into the nucleus (Belvin et al. 1995; Bergmann et al. 1996; Govind et al. 1996; Reach et al. 1996; Wu and Anderson 1998; Fernandez et al. 2001; Sun et al. 2004). The kinase that phosphorylates Cactus remains unclear. As mentioned earlier, Pelle may directly phosphorylate Cactus or other unidentified kinases may function here. Although Drosophila IKKβ has been shown to phosphorylate Cactus in vitro (Kim et al. 2000), neither IKKβ (ird5) nor IKKγ (kenny) are required for Toll signaling in vivo (Rutschmann et al. 2000a, b; Silverman et al. 2000; Lu et al. 2001). Studies to identify the regulatory region in Cactus required for its signal-dependent degradation implicated serines at positions 74, 78 and 116 (Belvin et al. 1995; Bergmann et al. 1996; Reach et al. 1996; Fernandez et al. 2001). These residues are found in two motifs that are both similar to the phosphorylation sites at serine 32 and 36 of IκBα. However, regulation of Cactus degradation has been speculated to be much more complicated than IκBα, since mammalian IκBα has only three serines N-terminal of the ankyrin repeats while Cactus has 36 serines in this region. Moreover, these mechanisms should also allow for graded degradation of Cactus, as observed during dorsal–ventral patterning, while IκBα does not exhibit such a phenomenon (Fernandez et al. 2001). As mentioned above, β-TrCP, an F-box protein was originally found to be important for the K48-ubiquitination and degradation of phosphorylated IκBα in mammalian cells (Spencer et al. 1999; Winston et al. 1999). Similarly, Drosophila mutants deficient in Slimb (Drosophila homolog of β-TrCP), exhibit reduced expression of Dorsal target genes, twist and snail, suggesting that this is a shared mechanism of Cactus degradation in Drosophila and mammals (Spencer et al. 1999). One study indicated that Cactus degradation may not be sufficient to facilitate the robust nuclear translocation of Dorsal in embryos (Bergmann et al. 1996), and other studies suggest that Dorsal must also be phosphorylated for optimal nuclear localization (Whalen and Steward 1993; Gillespie and Wasserman 1994; Drier et al. 1999, 2000). Though both Dorsal and DIF are controlled by the Toll pathway, many of the above described specifics of signal-dependent activation of Dorsal have not been examined in detail for DIF. Once in the nucleus, DIF, and to a lesser degree Dorsal, activate a robust set of immune responsive genes (Lemaitre et al. 1996; De Gregorio et al. 2001; 2002; Irving et al. 2001).
4.2 Relish
Relish, a NF-κB precursor protein similar to p100 and p105, is activated by endoproteolytic cleavage at aspartate 545, within a caspase target site (LQHD) (Stoven et al. 2000; 2003; Cornwell and Kirkpatrick 2001). Relish is thought to be cleaved directly by the caspase DREDD for a number of reasons. DREDD directly interacts with Relish (Stoven et al. 2003), and Relish cleavage is completely abrogated in DREDD mutants (Stoven et al. 2003). Immunoprecipitated DREDD cleaves recombinant Relish in vitro (Erturk-Hasdemir et al. 2009). Moreover, RNAi knockdown of the other known Drosophila caspases have been reported to have no effect on Relish processing (Stoven et al. 2003; Erturk-Hasdemir et al. 2009). Following processing, the cleaved N-terminal portion of Relish which contains the DNA binding RHD domain translocates into the nucleus, while the C-terminal portion remains in the cytoplasm.
In addition to cleavage, Relish is also regulated by signal-induced phosphorylation. In particular, the Relish N-terminus is phosphorylated at two adjacent serines, 528 and 529, by the Drosophila IKK complex (Erturk-Hasdemir et al. 2009). This phosphorylation of Relish does not play a role in its cleavage, as evident from the normal cleavage observed in the non-phosphorylatable SS528/529AA Relish mutant. Instead, phosphorylation of Relish facilitates the recruitment of RNA polymerase II to the transcription start site of the AMP gene Diptericin (Erturk-Hasdemir et al. 2009).
Although IKK induced phosphorylation of Relish is not required for its cleavage, the IKK complex has a critical, non-catalytic role in Relish cleavage. Relish cleavage is abolished in ird5 (IKKβ) or kenny (IKKγ) null mutants, but is rescued by expression of a kinase-dead IKKβ (Stoven et al. 2003; Erturk-Hasdemir et al. 2009). The mechanism by which the IKK complex functions non-catalytically to support Relish cleavage is unclear. One possible explanation may lie in the connection between IKKβ and the very C-terminal 107 amino acids of Relish. This domain is C-terminal to the IκB-like ankyrin repeats (Fig. 3), and is required for both phosphorylation and cleavage of Relish (Stoven et al. 2003). However, mutation of all serines and threonines in this region did not interfere with Relish phosphorylation and cleavage, indicating that this region is not a target for IKK catalytic activity (Erturk-Hasdemir et al. 2009). On the other hand, this C-terminal 107 amino acid region is responsible for the interaction between Relish and IKKβ, suggesting that a lack of association between these two proteins prevents both signaling and cleavage.
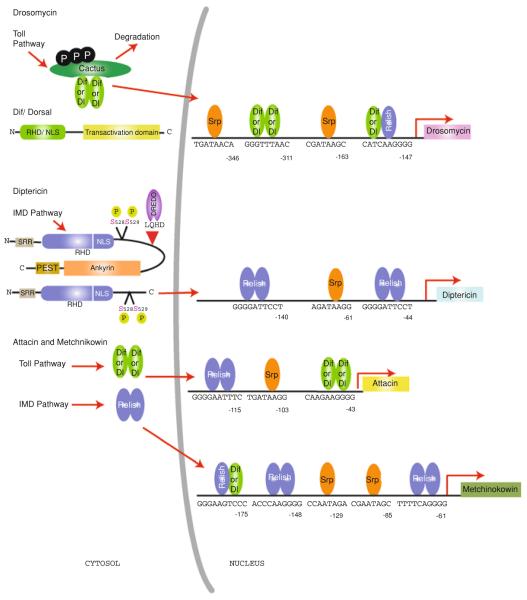
Fig. 3.
κB site specificity of Drosophila NF-κB proteins: NF-κB factors, upon activation by the Toll and IMD pathways, translocate into the nucleus and bind to specific κB sites in the promoter/enhancer region of immune responsive AMP genes. DIF and Dorsal are activated by Toll signaling through degradation of the IκB inhibitor protein Cactus, while Relish activation is triggered by IMD signaling through DREDD-dependent endoproteolytic cleavage to remove the C-terminal IκB-like domain and the IKK-mediated phosphorylation of serines 528 and 529. In this figure, selected AMP genes Drosomycin,Diptericin, AttacinA and Metchnikowin with their proximal DIF, Dorsal or Relish binding κB sites and Serpent (Srp) binding GATA sites are represented (Senger et al. 2004; Busse et al. 2007; Tanji et al. 2007). Expression of AttacinA requires both DIF and Relish, and hence contains separate binding sites for each of them, while Diptericin requires only Relish and has two Relish specific sites. Drosomycin and Metchnikowin each contain a distinct DIF/Relish site, which is responsive to both DIF and Relish. It is not clear if this site binds a heterodimer (as suggested by this figure) or if it functions by binding either homodimer. In addition, the Metchnikowin enhancer includes separate Relish-specific sites, while Drosomycin has another DIF/Dorsal-specific site only. Refer to Sect. 4.3 in the text for details on the consensus κB and GATA sites and the complex regulation involved in the expression of AMPs
In addition to the caspase target site, the IKK phosphorylation sites and the IKK interaction site, other regions in Relish also modulate its activity. Relish contains a PEST domain near its C-terminus (Fig. 3), similar to mammalian p105. Although the p105 PEST domain contains IKK phosphorylation sites and is important for its proteasomal degradation, the Relish PEST domain is not involved in signal-induced cleavage or activation. On the other hand, PEST deleted Relish seems to exhibit enhanced nuclear localization and target gene expression (Stoven et al. 2003). A similar phenotype is also observed with deletion of a serine rich stretch in the Relish N-terminus (Fig. 3). Therefore, these two domains have been proposed to negatively regulate the nuclear translocation of Relish (Stoven et al. 2003). However, the specific mechanism involved in this regulation is not clear.
Cleavage of Relish results in a 68 kDa N-terminal fragment which includes the RHD domain and a 49 kDa C-terminal fragment containing the ankyrin repeat domain. Over expression of the N-terminal 69 kDa portion is sufficient to induce the expression of some AMP genes, but does not recapitulate the response seen with fully activated Relish, consistent with the notion that Relish must be phosphorylated for its full activity. However, the expression of the C-terminal ankyrin repeat domain separately, doesn’t hinder the activation of Relish (Wiklund et al. 2009), unlike the inhibitory effect exerted on NF-κB activation when IκBα is overexpressed in mammalian cells.
4.3 Transcriptional Regulation of NF-κB Targets
Once inside the nucleus, Dorsal, DIF and Relish bind to their corresponding κB sites in the promoter/enhancer region of AMP and other target genes. Mammalian NF-κB/Rel factors bind a similar κB consensus motif [GGG(G/A)NN(T/C)(T/C)CC] (Baeuerle 1991). However, the exact composition of Rel protein dimers and associated co-activators bound to particular elements depends on the specific sequence of the κB site, as well as on other nearby transcription factors (Gross et al. 1996; Leung et al. 2004). One elegant study in the Drosophila immune response combined expression profiling of wild type, relish, or spz, or relish, spz animals and bioinformatic analysis of promoter sequences to identify Relish- and/or DIF-responsive κB sites (Busse et al. 2007). In particular, DIF prefers a sequence with three Gs followed by 4–5 AT-rich nucleotides [GGGAAA(A/T/G)(C/T)CC] while Relish prefers 4 Gs followed by a shorter AT-rich stretch [GGGGATT(T/C)(T/C)(T/C)]. Some sites were also found to bind both Relish and DIF, e.g. perfect palindromes [GGGAATTCCC] (Busse et al. 2007). The nature of the κB-sites found upstream of individual AMP genes (and other immune responsive genes) determines whether each gene will respond to Toll or IMD signaling (or both). For example, Tanji et al. 2007 reported that Drosomycin has two κB sites, one of which preferentially responds to Toll/DIF while the other also responds to IMD/Relish signaling. Optimal Drosomycin induction involves activation of both pathways and signaling through both κB sites. Similar complexity is found at other AMP genes, too; Attacin-A has two separate κB sites, one for DIF and one for Relish binding, and mutation of the DIF site renders it completely IMD pathway dependent (Busse et al. 2007). The enhancer region of Metchnikowin contains one κB site, which allows the binding of both DIF and Relish, in addition to two other Relish-specific sites (Levashina et al. 1998; Senger et al. 2004; Busse et al. 2007) (Fig. 3).
While DIF and Dorsal were believed to predominantly form homodimers, studies by Han and lp (1999) indicated that DIF, Dorsal and Relish may form heterodimers and cooperatively regulate the expression of certain AMPs (Gross et al. 1996; Han and Ip 1999). For example, expression of Drosomycin, a Toll-dependent gene is greatly enhanced by the co-expression of Relish and DIF. Moreover, Relish was found to interact with both DIF and Dorsal under overexpressed conditions (Han and Ip 1999). This idea is further supported by the recent evidence that synergistic activation of both Toll and IMD pathways is important for Drosomycin expression (Tanji et al. 2007). Consistent with this idea, a novel Dif/Relish heterodimer binding sequence, GGGA(A/T)TC(C/A)C, distinct from that of Dif or Relish homodimers was identified by Senger et al. 2004. Interestingly, the −147 κB site GGGGAACTAC in the enhancer region of Drosomycin closely resembles this DIF/Relish site (Senger et al. 2004). As in mammals, permutation of different homo/heterodimers of Drosophila Rel proteins may expand the regulatory potential of these proteins and further experimental data are needed to clarify these possibilities.
Optimal expression of AMP genes relies not only on the Rel-binding sites, but also depends on other regulatory regions in their promoter/enhancers (Uvell and Engstrom 2003; Senger et al. 2004). GATA sites (consensus: (no G) GATAA (no A) (no T)) have been found in close proximity to Rel binding sites and oriented in the same direction as the Rel sites in almost one-third of the immune responsive genes in Drosophila (Senger et al. 2004). Inversion of this orientation upstream of Metchnikowin or CecropinA1 genes reduces their expression (Kadalayil et al. 1997; Senger et al. 2004). The authors propose that this peculiar orientation might provide a platform for the appropriate interaction between the GATA-binding transcription factor Serpent and the Rel proteins. Moreover, it may also allow the cooperative assembly of other factors required for transcription (Petersen et al. 1999; Senger et al. 2004).
Another distinct regulatory region found in many AMP genes is the ‘R1 motif’. The R1 element was best studied in the CecropinA gene where it is found in addition to κB and GATA sites. κB and R1 sites independently regulate CecA expression through different transcription factors. Similar to κB, R1 binding activity is induced by immune challenge and R1 sites are essential for the proper expression of CecA, although, it is not clear how these R1 factors are regulated in response to immune challenge. R1 motif is also found in the upstream regions of other AMPs such as Defensin, Drosomycin and Metchnikowin (Uvell and Engstrom 2003). Many AMP genes also harbor binding sites for the JNK pathway transcription factor AP-1. Their functions remain controversial, but most likely they act as repressors of the NF-κB-mediated Drosophila humoral response, as discussed below (Kim et al. 2005; 2007).
5 Co-activators of Drosophila Rel Proteins
Rel transcription factors in Drosophila function in concert with co-activator proteins to drive the transcription of their target genes. DIF interacts with the co-activator dTRAP80, to induce transcription of target genes such as Drosomycin. dTRAP80 is a component of the multi-protein transcriptional Mediator co-activator complex (Park et al. 2003). In mammals, the co-activator protein called CREB-binding protein (CBP) assists NF-κB in the expression of certain immune effectors like IL-6 and IFNβ (Merika et al. 1998; Vanden Berghe et al. 1999; Qin et al. 2005). Similarly, maternally expressed Drosophila CBP (dCBP) is essential for the expression of the dorsal target gene, twist, during larval development. Dorsal interacts directly with dCBP through its RHD domain (Akimaru et al. 1997). The role of dCBP in the immune response has not been thoroughly examined. Another regulator of Dorsal and DIF dependent transcription is Deformed epidermal autoregulatory factor 1(DEAF1) (Reed et al. 2008; Kuttenkeuler et al. 2010). Many AMP genes such as Drosomycin and Metchnikowin have Deaf1-binding sites (“TTCGGNT”) in their upstream regions and mutating these sites reduces their signal-dependent expression. Moreover, its strict nuclear localization and epistatic analysis have indicated that it might function together with DIF and Dorsal to drive AMP gene expression. Although it does not seem to influence Relish dependent transcription, Deaf1 regulation is not completely clear (Kuttenkeuler et al. 2010). Another possible co-activator family, involved in the immune response is the “POU” domain proteins Pdm1, Pdm2 and Dfr, which were identified as potential regulators of DIF-mediated CecropinA expression (Junell et al. 2007).
Akirins are a conserved class of proteins that modulate NF-κB activation in both Drosophila and mammalian innate immune pathways. Akirin has been proposed to be a co-activator of Relish, since knockdown of Akirin decreases Diptericin expression and epistatic analysis has placed it at the level of or downstream of Relish in the IMD pathway. Its strict nuclear localization further adds support to this hypothesis (Goto et al. 2008). Akirin does not contain any known DNA-binding domains and a direct interaction between Relish and Akirin was not detected. Mammals express two nuclear Akirin proteins. Although, the function of Akirin1 remains unclear, with no obvious phenotypes, the Akirin2 knockout is embryonic lethal. Fibroblasts from these knockout embryos showed defects in the production of an NF-κB-dependent cytokine, IL-6 in response to TNF, IL-1 and LPS, suggesting a function similar to Drosophila Akirin (Goto et al. 2008).
Another possible co-activator, known as Helicase89B, is critical for Rel protein-dependent transcription of both Toll and IMD target anti-microbial genes in Drosophila larvae. Curiously, Helicase89B does not appear to be critical for immune responses in the adult fly (Yagi and Ip 2005). Helicase89B is a SNF-like ATPase, most similar to yeast Mot1p. Like Mot1p, Hel89B was also reported to interact with TATA-binding protein (TBP) and thus, may function as an adaptor protein that links the Rel protein with the basal transcription complex. However, no direct interaction was observed between Hel89B and Rel proteins so its exact function requires further clarification (Yagi and Ip 2005).
6 Negative Regulators of the Toll and IMD Pathways
6.1 Catalytic PGRPs
While non-catalytic PGRPs such as PGRP-SA/SD, LC/LE function as PGN-binding receptors and activate the Toll and the IMD pathways, the catalytically active PGRPs negatively regulate the IMD pathway by degrading microbial PGN (Bischoff et al. 2006; Zaidman-Remy et al. 2006). As mentioned earlier, these PGRPs are type 2 amidases that cleave the amide bond separating the L-Ala in the stem peptide from the glycan backbone of PGN, greatly diminishing its immunostimulatory activity. In Drosophila, PGRP-LB, PGRP-SB1, PGRP-SC1a/b are known PGN processing amidases. Also, PGRP-SB2, and PGRP-SC2 are predicted to have amidase activity, based on the presence of a complete catalytic site (Werner et al. 2000; Kim et al. 2003; Mellroth et al. 2003; Bischoff et al. 2006; Mellroth and Steiner 2006; Zaidman-Remy et al. 2006). In addition, genetic studies using RNAi to knockdown either PGRP-LB or PGRP-SC1/2 in vivo firmly established that these amidases are important to down-modulate the IMD pathway, both systemically and in the gut (Bischoff et al. 2006; Zaidman-Remy et al. 2006).
Distinct from the type 2 amidase activity discussed above, PGRP-SA was shown to possess an L,D-carboxypeptidase activity, which enables it to specifically cleave DAP-type PGN. It was proposed that this activity functions as an ‘editor’ allowing PGRP-SA to signal Toll activation following binding of lysine-type PGN while binding to DAP-type PGN would instead, lead to the digestion of the PGN without signaling (Chang et al. 2004). However, this model is yet to be tested.
The four human PGRPs (also known as PGLYRPs) also function in host defense. PGLYRP-2 is a catalytic amidase and likely functions by down-modulating PGN-induced innate immune response in mammals, much like the catalytic Drosophila PGRPs (Royet and Dziarski 2007). On the other hand, the three non-catalytic mouse PGRPs, PGLYRP1, 3, and 4, do not appear to function as PGN receptors, as observed in the Drosophila system. Instead, these PGN-binding proteins appear to be directly bactericidal and, for example, contribute to microbial killing by DNA NETS released by activated neutrophils (Cho et al. 2005; Royet and Dziarski 2007).
6.2 Negative Regulators of Toll Signaling
Necrotic (nec) is a member of the SERine Protease INhibitor (serpin) family. nec mutants display constitutive activation of Toll and spontaneous Drosomycin expression and melanization (Levashina et al. 1999; Ligoxygakis et al. 2002). In fact, the previously mentioned protease-recognition system featuring Persephone (psh) was identified in a screen for suppressors of the necrotic (nec) phenotype (Ligoxygakis et al. 2002). Toll activation leads to the cleavage of Nec, removing its N-terminal polyglutamine extension. In vitro, cleaved Nec is still an active serpin, in some cases displaying increased activity. Thus, this feed back mechanism may regulate the protease pathways that control Spätzle processing (Pelte et al. 2006).
The Drosophila genome encodes for 29 serpins, including nec, some of which are also involved in regulating Toll activation and other aspects of the immune response. For example, Serpin-27A and Serpin-28D are major regulators of melanization (Ligoxygakis et al. 2002; Scherfer et al. 2008) while Serpin5 was recently linked to controlling Toll (Ahmad et al. 2009). Serpin-27A also controls the Spätzle processing protease Easter in the dorsal–ventral patterning pathway (Hashimoto et al. 2003; Ligoxygakis et al. 2003). Serpin 77Ba regulates the protease cascade that controls melanization, but seems to function specifically in the trachea. Interestingly, the up-regulation of tracheal melanization observed in the Spn77Ba mutant indirectly activates the Toll pathway and Drosomycin expression, through the Persephone serine protease cascade (Tang et al. 2008).
Cactus and Wnt inhibitor of Dorsal (WntD) are both feedback inhibitors of the intracellular Toll signaling pathway (Nicolas et al. 1998; Ganguly et al. 2005; Gordon et al. 2005). As described previously, Cactus sequesters Dorsal in the cytoplasm of unstimulated cells and prevents its nuclear localization. Cactus is also an early target of DIF/Dorsal transcriptional activation. Newly synthesized Cactus can then sequester DIF or Dorsal in the cytoplasm, down-modulating the response. Similarly, WntD also inhibits Dorsal activation, although the mechanism is not clearly understood. wntD mutants display elevated levels of nuclear Dorsal even in unstimulated conditions. Loss of WntD alters the transcriptional profile of embryonic genes, AMPs and other immune genes in response to infection (Ganguly et al. 2005; Gordon et al. 2005, 2008). For example, Listeria infection causes elevated expression of certain immune genes in wntD mutants, among which edin (elevated during infection) and eiger, in particular, have been shown to be necessary to fight the infection efficiently. At the same time, over-expression of these same proteins can also reduce the survival of flies (Dionne and Schneider 2008; Gordon et al. 2008), indicating that WntD acts a checkpoint to ensure controlled activation of immune responses against pathogens. It should be noted that Drosophilaeiger is a TNF family member. eiger has been shown to play a protective role particularly against extracellular pathogens, by activating cellular defenses such as phagocytosis. As mentioned above, when expressed at higher levels, eiger can also exacerbate the pathology of disease, probably through its ability to promote programmed cell death, similar to TNF (Schneider et al. 2007; Narasimamurthy 2009). However, eiger does not seem to be a component of the conventional Toll and IMD pathways, and a link to Drosophila Rel proteins, if any, is yet to be identified.
6.3 Negative Regulators of IMD Signaling
Rudra/PIMS/Pirk (hereafter Rudra) is a negative feedback regulator of IMD signaling that was identified as a PGRP-LC interacting protein in a yeast two-hybrid screen. Rudra interacts with both the PGN receptors of the IMD pathway, PGRP-LC and PGRP-LE, at least in part, through the conserved RHIM domain that is critical for signaling by both receptors (Aggarwal et al. 2008). Rudra is induced by PGN stimulation and associates with both the receptors as well as with the IMD protein, and disrupts the receptor proximal signaling complex (Aggarwal et al. 2008; Kleino et al. 2008). In addition, Rudra may also traffic PGRP-LC to the lysosome, for degradation (Lhocine et al. 2008). Rudra not only prevents the hyper-activation of the IMD pathway in response to pathogens, but also maintains intestinal homeostasis by preventing the induction of AMPs by commensal bacteria (Lhocine et al. 2008).
Caudal, a homeobox encoding transcription factor, is another negative regulator of AMP expression, e.g. Diptericin and Cecropin, that functions in the gut to prevent IMD activation by commensal bacteria (Ryu et al. 2008). Gut-specific knock-down of caudal perturbs the balance of gut commensal populations, resulting in the dominance of certain microbes that are only minor members of the gut microbiota in wildtype animals. The dominance of these microbes causes an apoptotic pathology in the Drosophila midgut epithelium. This phenomenon is seen in conventionally reared flies, but not in germ free flies which are devoid of gut commensal flora (Ryu et al. 2008). Caudal also regulates constitutive, NF-κB-independent expression of Drosomycin and Cecropin in certain local epithelial tissues such as salivary glands, through Caudal binding sites in their promoter regions (Ryu et al. 2004).
Caspar is another negative regulator of the IMD pathway. Caspar is homologous to human Fas associated factor (FAF1) protein, which inhibits NF-κB activation by various inflammatory signals such as TNF, LPS, IL-1β. FAF1 is thought to function by interacting with p65/RelA and retaining p65 in cytoplasm (Park et al. 2004a, b). Caspar-deficient flies produce Diptericin in the absence of immune challenge and are more resistant to bacterial infections (Kim et al. 2006). Epistasis experiments with over-expressed Caspar and IMD signaling components suggested that Caspar functions by inhibiting DREDD (Kim et al. 2006). Further, Caspar contains a DED interaction domain, similar to FAF1, and this may mediate its interaction with the DED domain of Drosophila DREDD. Caspar also contains ubiquitin-associated (UAS) domains and ubiquitin-like domains, but whether any of these domains regulate IMD signaling awaits experimental data.
Several reports suggest that IMD pathway may also be regulated by the ubiquitin–proteasome pathway. RNAi knockdown of Defense repressor protein (DNR1), both in vitro and in vivo results in constitutive Diptericin expression even in non-immune challenged conditions (Foley and O’Farrell 2004; Guntermann et al. 2009). Although its exact role in the IMD pathway is not clear, DNR1 contains a conserved RING domain, characteristic of ubiquitin E3 ligases, suggesting that DNR1 may mediate ubiquitination of some IMD pathway component (Foley and O’Farrell 2004). In fact, a recent report showed that DNR1 RING domain could target DREDD for proteasomal degradation (Guntermann et al. 2009). Another finding that suggests a role for ubiquitin-/proteasome-mediated degradation in the down-modulation of the IMD pathway is that flies deficient in SCF ubiquitin-ligase components Slimb and dcullin1 exhibit spontaneous Relish dependent Diptericin expression. This indicates that K48-ubiquitination and proteasomal degradation of certain signaling components may be involved in tuning down the signal (Khush et al. 2002). How, or if, DNR1 and the SCF function together is unknown.
Removal of K63-ubiquitin linkages associated with IMD is another mechanism by which the IMD pathway is regulated. As described above, following PGN stimulation, IMD is K63 ubiquitinated in a DREDD- and DIAP2-dependent manner. dUSP36, a de-ubiquitinase (DUB), targets this ubiquitination event to down-regulate Diptericin expression in response to PGN. USP36 removes K63-polyubiquitin chains from IMD and then appears to increase the addition of K48-ubiquitin chains and degradation of IMD (Thevenon et al. 2009). The E3 ligase for K48-ubiquitination of IMD is not yet identified, but DNR1 or Slimb are both candidates. This is reminiscent of RIP1 regulation by A20 in mammalian cells. A20 exhibits both similar K63 de-ubiquitinating and a separate K48 ubiquitin ligase activity, and is often referred to as an ‘ubiquitin editor’ (Wertz et al. 2004). In mammals, in addition to A20, several other DUBs such as CYLD, DUBA, and Caezanne down-regulate NF-κB activation by removing K63 ubiquitin linkage in key signaling proteins such as RIP1, TRAF6 and IKKγ (Evans et al. 2001; Kayagaki et al. 2007; Enesa et al. 2008; Friedman et al. 2008). In this context, it would be interesting to know whether IKKγ also gets ubiquitinated in the IMD pathway. Interestingly, Drosophila also expresses a homolog of CYLD, with deubiquitinase activity, and among its other functions, Drosophila CYLD has also been implicated in anti-bacterial responses (Tsichritzis et al. 2007; Xue et al. 2007). These studies show a strong conservation of negative regulatory mechanisms in NF-κB pathways from flies to mammals.
The two branches of the IMD signaling cascade, leading to JNK and Relish activation, have been shown to down-regulate each other (Park et al. 2004a, b). Certain genes induced by Relish can target TAK1 for degradation and thus, reduce the activation of the JNK pathway (Park et al. 2004a, b). A specific example is Plenty of SH3s (POSH), which is up-regulated by Relish. POSH, in turn, regulates the duration of JNK and Relish activation, downstream of TAK1 in the IMD pathway. POSH is a RING finger containing E3 ligase. On one hand, it is important for the timely initiation of JNK and Relish dependent transcription, while on the other hand, it triggers the proteasomal degradation of TAK1 by the E3 ligase activity of its RING domain (Tsuda et al. 2005). Thus, POSH seems to play two contradictory roles in IMD signaling by regulating the amount of activated TAK1. JNK signaling also inhibits Relish-mediated transcription. In particular, the Drosophila AP-1 and STAT proteins (Jra and STAT92E), along with a HMG protein Dsp1 form a repressome complex on the Attacin-A promoter replacing Relish, thereby inhibiting Relish-dependent transcription. This repressome was reported to function by recruiting a histone deacetylase dHDAC1 and altering the chromatin structure of the Relish target promoter region (Kim et al. 2005, 2007). Although many Relish target genes contain AP-1 sites, this regulatory antagonism, between AP-1/STAT and Relish, has not yet been examined in detail at other AMP gene promoters. A similar negative regulatory mechanism has been observed with the CREB family transcription factor ATF3 in mammals. ATF3, induced by TLR4 signaling, inhibits LPS induced NF-κB transcription of certain cytokines such as IL-6 and IL-12 in a negative feedback loop. Similar to the Drosophila AP-1/STAT proteins, ATF3 also functions by associating with HDAC1 and altering the chromatin acetylation status (Gilchrist et al. 2006).
7 Beyond AMPs
7.1 Anti-viral Immunity
Drosophila mounts potent responses not only against bacterial and fungal pathogens, but also against viruses. However, it appears that these anti-viral effects may not be mediated by the AMPs that are induced by the Toll and the IMD pathways. Infection of flies with Cricket Paralysis virus (CrPV) and Drosophila C virus (DCV) does not induce AMP expression, and forced expression of single AMPs during the course of infection also does not protect the host (Sabatier et al. 2003; Dostert et al. 2005; Zambon et al. 2005; Tsai et al. 2008). However, mutants lacking various components of the IMD pathway are less efficient in clearing CrPV, indicating that other immune related functions of the IMD pathway, such as apoptosis may be playing a role (Costa et al. 2009). Similarly, in the case of Drosophila X virus, Toll pathway mutants were more sensitive (Zambon et al. 2005). The JAK-STAT pathway, a separate immune response pathway not involving NF-κB factors, is critical to control other viral infections, e.g. DCV infection (Dostert et al. 2005; Hedges and Johnson 2008). The RNAi pathway is also triggered by the genome and replication intermediates of viruses and has been shown to have important antiviral effects in insects (Li et al. 2002; Galiana-Arnoux et al. 2006; Wang et al. 2006a, b; Zambon et al. 2006).
7.2 Adaptive Immunity in Drosophila
Some studies have also indicated that Drosophila immune responses may not be limited to innate and non-specific mechanisms. Pham et al. (2007), demonstrated that flies, primed with a sublethal dose of the pathogen S. pnuemoniae are protected life-long, from subsequent infections by lethal doses of the same microbe, but not by other microbes (Pham et al. 2007). This indicates that Drosophila immunity also allows for pathogen-specific responses and the development of immunological ‘memory’. However, the mechanisms underlying this specificity and adaptation have not yet been characterized. Another finding that supports the existence of specific immune responses in flies (and mosquitoes) is the multiple splice isoforms of a gene known as Down’s syndrome cell adhesion molecule (Dscam). Dscam is an immunoglobulin superfamily protein which can generate up to 18,000 different isoforms in hemocytes (blood cells). Interestingly, these isoforms have been proposed to mediate pathogen-specific recognition and the expression of various isoforms is modulated by the presence of different infectious microbes (Watson et al. 2005; Dong et al. 2006).
8 Concluding Remarks
Innate immune NF-κB signaling in Drosophila shares a great deal of similarity with mammalian systems (Silverman and Maniatis 2001; Girardin et al. 2002). In the broadest sense, the signal relay culminating in NF-κB activation follows a similar pattern in both insects and mammals. Microbial recognition by innate immune receptors is followed by the formation of receptor proximal signaling complexes, generated primarily by homotypic interactions between conserved domains in adapter proteins. These receptor complexes induce regulatory ubiquitination events and activation of kinases, which trigger the degradation or processing of IκB like inhibitory proteins/domains leading to activation and nuclear translocation of NF-κB/Rel transcription factors. In fact, the signaling events in the Toll and IMD pathways share a striking amount of molecular similarity with the mammalian MyD88-dependent and MyD88-independent TLR pathways, respectively. Hence, studying these pathways in detail will likely provide further insights on the molecular mechanisms involved in the related mammalian signaling pathways.
However, on detailed inspection, some notable differences are evident. While the mammalian TLRs directly recognize their corresponding microbial ligands, Drosophila Toll is a cytokine receptor, with an endogenous ligand, Spätzle, that is processed from its precursor in response to infection. Another notable difference is that the NF-κB precursor Relish is activated by endoproteolytic caspase-dependent cleavage, which has not been observed with mammalian NF-κB precursors, p100 and p105. These differences, in combination with the overall similarities, make the Drosophila innate immune response, a fascinating model system for the study of innate immune NF-κB signaling.
Contributor Information
Sandhya Ganesan, Division of Infectious Diseases, Department of Medicine, University of Massachusetts Medical School, Worcester MA, 01605, USA.
Kamna Aggarwal, Division of Infectious Diseases, Department of Medicine, University of Massachusetts Medical School, Worcester MA, 01605, USA.
Nicholas Paquette, Program of Developmental Immunology, Department of Pediatrics, Massachusetts General Hospital/Harvard Medical School, Boston MA, 02114, USA.
Neal Silverman, Division of Infectious Diseases, Department of Medicine, University of Massachusetts Medical School, Worcester MA, 01605, USA.
References
- Aggarwal K, et al. Rudra interrupts receptor signaling complexes to negatively regulate the IMD pathway. PLoS Pathog. 2008;4(8):e1000120. doi: 10.1371/journal.ppat.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad ST, et al. Genetic screen identifies serpin5 as a regulator of the toll pathway and CHMP2B toxicity associated with frontotemporal dementia. Proc Natl Acad Sci USA. 2009;106(29):12168–12173. doi: 10.1073/pnas.0903134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimaru H, et al. Drosophila CBP is required for dorsal-dependent twist gene expression. Nat Genet. 1997;17(2):211–214. doi: 10.1038/ng1097-211. [DOI] [PubMed] [Google Scholar]
- Anderson KV, Nusslein-Volhard C. Information for the dorsal–ventral pattern of the Drosophila embryo is stored as maternal mRNA. Nature. 1984;311(5983):223–227. doi: 10.1038/311223a0. [DOI] [PubMed] [Google Scholar]
- Apidianakis Y, et al. Profiling early infection responses: Pseudomonas aeruginosa eludes host defenses by suppressing antimicrobial peptide gene expression. Proc Natl Acad Sci USA. 2005;102(7):2573–2578. doi: 10.1073/pnas.0409588102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle PA. The inducible transcription activator NF-kappa B: regulation by distinct protein subunits. Biochim Biophys Acta. 1991;1072(1):63–80. doi: 10.1016/0304-419x(91)90007-8. [DOI] [PubMed] [Google Scholar]
- Belvin MP, et al. Cactus protein degradation mediates Drosophila dorsal–ventral signaling. Genes Dev. 1995;9(7):783–793. doi: 10.1101/gad.9.7.783. [DOI] [PubMed] [Google Scholar]
- Bergmann A, et al. A gradient of cytoplasmic cactus degradation establishes the nuclear localization gradient of the dorsal morphogen in Drosophila. Mech Dev. 1996;60(1):109–123. doi: 10.1016/s0925-4773(96)00607-7. [DOI] [PubMed] [Google Scholar]
- Bianchi K, Meier P. A tangled web of ubiquitin chains: breaking news in TNF-R1 signaling. Mol Cell. 2009;36(5):736–742. doi: 10.1016/j.molcel.2009.11.029. [DOI] [PubMed] [Google Scholar]
- Bischoff V, et al. Function of the drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat Immunol. 2004;5(11):1175–1180. doi: 10.1038/ni1123. [DOI] [PubMed] [Google Scholar]
- Bischoff V, et al. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006;2(2):e14. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K, et al. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267(5203):1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- Buchon N, et al. A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc Natl Acad Sci USA. 2009;106(30):12442–12447. doi: 10.1073/pnas.0901924106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse MS, et al. A kappaB sequence code for pathway-specific innate immune responses. Embo J. 2007;26(16):3826–3835. doi: 10.1038/sj.emboj.7601798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CI, et al. A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual l, d-carboxypeptidase activity. PLoS Biol. 2004;2(9):E277. doi: 10.1371/journal.pbio.0020277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CI, et al. Structure of the ectodomain of Drosophila peptidoglycan-recognition protein LCa suggests a molecular mechanism for pattern recognition. Proc Natl Acad Sci USA. 2005;102(29):10279–10284. doi: 10.1073/pnas.0504547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CI, et al. Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science. 2006;311(5768):1761–1764. doi: 10.1126/science.1123056. [DOI] [PubMed] [Google Scholar]
- Cho JH, et al. Human peptidoglycan recognition protein S is an effector of neutrophil-mediated innate immunity. Blood. 2005;106(7):2551–2558. doi: 10.1182/blood-2005-02-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KM, et al. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296(5566):359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- Choe KM, et al. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci USA. 2005;102(4):1122–1126. doi: 10.1073/pnas.0404952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornwell WD, Kirkpatrick RB. Cactus-independent nuclear translocation of Drosophila RELISH. J Cell Biochem. 2001;82(1):22–37. doi: 10.1002/jcb.1144. [DOI] [PubMed] [Google Scholar]
- Costa A, et al. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS One. 2009;4(10):e7436. doi: 10.1371/journal.pone.0007436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E, et al. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci USA. 2001;98(22):12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E, et al. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. Embo J. 2002;21(11):2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne MS, Schneider DS. Models of infectious diseases in the fruit fly Drosophila melanogaster. Dis Model Mech. 2008;1(1):43–49. doi: 10.1242/dmm.000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, et al. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biol. 2006;4(7):e229. doi: 10.1371/journal.pbio.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert C, et al. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat Immunol. 2005;6(9):946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- Drier EA, et al. Nuclear import of the Drosophila Rel protein Dorsal is regulated by phosphorylation. Genes Dev. 1999;13(5):556–568. doi: 10.1101/gad.13.5.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drier EA, et al. Cactus-independent regulation of dorsal nuclear import by the ventral signal. Curr Biol. 2000;10(1):23–26. doi: 10.1016/s0960-9822(99)00267-5. [DOI] [PubMed] [Google Scholar]
- Dushay MS, et al. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc Natl Acad Sci USA. 1996;93(19):10343–10347. doi: 10.1073/pnas.93.19.10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ea CK, et al. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22(2):245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Edwards DN, et al. An activity-dependent network of interactions links the Rel protein Dorsal with its cytoplasmic regulators. Development. 1997;124(19):3855–3864. doi: 10.1242/dev.124.19.3855. [DOI] [PubMed] [Google Scholar]
- El Chamy L, et al. Sensing of ‘danger signals’ and pathogen-associated molecular patterns defines binary signaling pathways ‘upstream’ of Toll. Nat Immunol. 2008;9(10):1165–1170. doi: 10.1038/ni.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enesa K, et al. NF-kappaB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro-inflammatory signaling. J Biol Chem. 2008;283(11):7036–7045. doi: 10.1074/jbc.M708690200. [DOI] [PubMed] [Google Scholar]
- Engstrom Y, et al. kappa B-like motifs regulate the induction of immune genes in Drosophila. J Mol Biol. 1993;232(2):327–333. doi: 10.1006/jmbi.1993.1392. [DOI] [PubMed] [Google Scholar]
- Erturk-Hasdemir D, et al. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci USA. 2009;106(24):9779–9784. doi: 10.1073/pnas.0812022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PC, et al. Isolation and characterization of two novel A20-like proteins. Biochem J. 2001;357(Pt 3):617–623. doi: 10.1042/0264-6021:3570617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez NQ, et al. Separable and redundant regulatory determinants in Cactus mediate its dorsal group dependent degradation. Development. 2001;128(15):2963–2974. doi: 10.1242/dev.128.15.2963. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, et al. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. Embo J. 1998;17(5):1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipe SR, et al. Requirements of peptidoglycan structure that allow detection by the Drosophila Toll pathway. EMBO Rep. 2005;6(4):327–333. doi: 10.1038/sj.embor.7400371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E, O’Farrell PH. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2004;2(8):E203. doi: 10.1371/journal.pbio.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman CS, et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9(9):930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiana-Arnoux D, et al. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat Immunol. 2006;7(6):590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- Galindo RL, et al. Interaction of the pelle kinase with the membrane-associated protein tube is required for transduction of the dorsoventral signal in Drosophila embryos. Development. 1995;121(7):2209–2218. doi: 10.1242/dev.121.7.2209. [DOI] [PubMed] [Google Scholar]
- Ganguly A, et al. Drosophila WntD is a target and an inhibitor of the Dorsal/Twist/Snail network in the gastrulating embryo. Development. 2005;132(15):3419–3429. doi: 10.1242/dev.01903. [DOI] [PubMed] [Google Scholar]
- Geisler R, et al. Cactus, a gene involved in dorsoventral pattern formation of Drosophila, is related to the I kappa B gene family of vertebrates. Cell. 1992;71(4):613–621. doi: 10.1016/0092-8674(92)90595-4. [DOI] [PubMed] [Google Scholar]
- Georgel P, et al. Insect immunity: the diptericin promoter contains multiple functional regulatory sequences homologous to mammalian acute-phase response elements. Biochem Biophys Res Commun. 1993;197(2):508–517. doi: 10.1006/bbrc.1993.2508. [DOI] [PubMed] [Google Scholar]
- Georgel P, et al. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell. 2001;1(4):503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- Ghosh S, et al. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Gilchrist M, et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441(7090):173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- Gillespie SK, Wasserman SA. Dorsal, a Drosophila Rel-like protein, is phosphorylated upon activation of the transmembrane protein Toll. Mol Cell Biol. 1994;14(6):3559–3568. doi: 10.1128/mcb.14.6.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin SE, et al. Intracellular vs extracellular recognition of pathogens—common concepts in mammals and flies. Trends Microbiol. 2002;10(4):193–199. doi: 10.1016/s0966-842x(02)02334-x. [DOI] [PubMed] [Google Scholar]
- Gobert V, et al. Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science. 2003;302(5653):2126–2130. doi: 10.1126/science.1085432. [DOI] [PubMed] [Google Scholar]
- Gordon MD, et al. WntD is a feedback inhibitor of Dorsal/NF-kappaB in Drosophila development and immunity. Nature. 2005;437(7059):746–749. doi: 10.1038/nature04073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MD, et al. Pathogenesis of listeria-infected Drosophila wntD mutants is associated with elevated levels of the novel immunity gene edin. PLoS Pathog. 2008;4(7):e1000111. doi: 10.1371/journal.ppat.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto A, et al. Akirins are highly conserved nuclear proteins required for NF-kappaB-dependent gene expression in Drosophila and mice. Nat Immunol. 2008;9(1):97–104. doi: 10.1038/ni1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottar M, et al. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416(6881):640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- Gottar M, et al. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell. 2006;127(7):1425–1437. doi: 10.1016/j.cell.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind S, et al. In vivo self-association of the Drosophila rel-protein dorsal. Proc Natl Acad Sci USA. 1992;89(17):7861–7865. doi: 10.1073/pnas.89.17.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind S, et al. Regulated nuclear import of the Drosophila rel protein dorsal: structure-function analysis. Mol Cell Biol. 1996;16(3):1103–1114. doi: 10.1128/mcb.16.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross I, et al. Drosophila immunity: a comparative analysis of the Rel proteins dorsal and Dif in the induction of the genes encoding diptericin and cecropin. Nucleic Acids Res. 1996;24(7):1238–1245. doi: 10.1093/nar/24.7.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans J, et al. Activation of the kinase Pelle by Tube in the dorsoventral signal transduction pathway of Drosophila embryo. Nature. 1994;372(6506):563–566. doi: 10.1038/372563a0. [DOI] [PubMed] [Google Scholar]
- Guntermann S, et al. Dnr1-dependent regulation of the Drosophila immune deficiency signaling pathway. Dev Comp Immunol. 2009;33(1):127–134. doi: 10.1016/j.dci.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Han ZS, Ip YT. Interaction and specificity of Rel-related proteins in regulating Drosophila immunity gene expression. J Biol Chem. 1999;274(30):21355–21361. doi: 10.1074/jbc.274.30.21355. [DOI] [PubMed] [Google Scholar]
- Hashimoto C, et al. Spatial regulation of developmental signaling by a serpin. Dev Cell. 2003;5(6):945–950. doi: 10.1016/s1534-5807(03)00338-1. [DOI] [PubMed] [Google Scholar]
- Hedengren M, et al. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell. 1999;4(5):827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- Hedengren-Olcott M, et al. Differential activation of the NF-kappaB-like factors Relish and Dif in Drosophila melanogaster by fungi and Gram-positive bacteria. J Biol Chem. 2004;279(20):21121–21127. doi: 10.1074/jbc.M313856200. [DOI] [PubMed] [Google Scholar]
- Hedges LM, Johnson KN. Induction of host defence responses by Drosophila C virus. J Gen Virol. 2008;89(Pt 6):1497–1501. doi: 10.1099/vir.0.83684-0. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA, Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nat Immunol. 2002;3(2):121–126. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96(5):645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- Horng T, Medzhitov R. Drosophila MyD88 is an adapter in the Toll signaling pathway. Proc Natl Acad Sci USA. 2001;98(22):12654–12658. doi: 10.1073/pnas.231471798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Yang X. dFADD, a novel death domain-containing adapter protein for the Drosophila caspase DREDD. J Biol Chem. 2000;275(40):30761–30764. doi: 10.1074/jbc.C000341200. [DOI] [PubMed] [Google Scholar]
- Hu X, et al. Multimerization and interaction of Toll and Spatzle in Drosophila. Proc Natl Acad Sci USA. 2004;101(25):9369–9374. doi: 10.1073/pnas.0307062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguet C, et al. Rel/NF-kappa B transcription factors and I kappa B inhibitors: evolution from a unique common ancestor. Oncogene. 1997;15(24):2965–2974. doi: 10.1038/sj.onc.1201471. [DOI] [PubMed] [Google Scholar]
- Huh JR, et al. The Drosophila inhibitor of apoptosis (IAP) DIAP2 is dispensable for cell survival, required for the innate immune response to gram-negative bacterial infection, and can be negatively regulated by the reaper/hid/grim family of IAP-binding apoptosis inducers. J Biol Chem. 2007;282(3):2056–68. doi: 10.1074/jbc.M608051200. [DOI] [PubMed] [Google Scholar]
- Ip YT, et al. Dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev. 1992;6(8):1518–1530. doi: 10.1101/gad.6.8.1518. [DOI] [PubMed] [Google Scholar]
- Ip YT, et al. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell. 1993;75(4):753–763. doi: 10.1016/0092-8674(93)90495-c. [DOI] [PubMed] [Google Scholar]
- Irving P, et al. A genome-wide analysis of immune responses in Drosophila. Proc Natl Acad Sci USA. 2001;98(26):15119–15124. doi: 10.1073/pnas.261573998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoda K, Nusslein-Volhard C. Disulfide cross-linking in crude embryonic lysates reveals three complexes of the Drosophila morphogen dorsal and its inhibitor cactus. Proc Natl Acad Sci USA. 1994;91(12):5350–5354. doi: 10.1073/pnas.91.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-kappaB activation. EMBO Rep. 2009;10(7):706–713. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IH, et al. A Spatzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev Cell. 2006;10(1):45–55. doi: 10.1016/j.devcel.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Jiang J, et al. The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes Dev. 1991;5(10):1881–1891. doi: 10.1101/gad.5.10.1881. [DOI] [PubMed] [Google Scholar]
- Jin L, et al. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133(4):653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junell A, et al. Isolation of regulators of Drosophila immune defense genes by a double interaction screen in yeast. Insect Biochem Mol Biol. 2007;37(3):202–212. doi: 10.1016/j.ibmb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Kadalayil L, et al. Adjacent GATA and kappa B-like motifs regulate the expression of a Drosophila immune gene. Nucleic Acids Res. 1997;25(6):1233–1239. doi: 10.1093/nar/25.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambris Z, et al. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr Biol. 2006;16(8):808–813. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- Kanayama A, et al. TAB 2 and TAB 3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15(4):535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Kaneko T, et al. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20(5):637–649. doi: 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- Kaneko T, et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol. 2006;7(7):715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- Kang D, et al. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc Natl Acad Sci USA. 1998;95(17):10078–10082. doi: 10.1073/pnas.95.17.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappler C, et al. Insect immunity. Two 17 bp repeats nesting a kappa B-related sequence confer inducibility to the diptericin gene and bind a polypeptide in bacteria-challenged Drosophila. Embo J. 1993;12(4):1561–1568. doi: 10.1002/j.1460-2075.1993.tb05800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, et al. DUBA: a deubiquitinase that regulates type I interferon production. Science. 2007;318(5856):1628–1632. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- Khush RS, et al. A ubiquitin-proteasome pathway represses the rosophila immune deficiency signaling cascade. Curr Biol. 2002;12(20):1728–1737. doi: 10.1016/s0960-9822(02)01214-9. [DOI] [PubMed] [Google Scholar]
- Kidd S. Characterization of the Drosophila cactus locus and analysis of interactions between cactus and dorsal proteins. Cell. 1992;71(4):623–635. doi: 10.1016/0092-8674(92)90596-5. [DOI] [PubMed] [Google Scholar]
- Kim YS, et al. Lipopolysaccharide-activated kinase, an essential component for the induction of the antimicrobial peptide genes in Drosophila melanogaster cells. J Biol Chem. 2000;275(3):2071–2079. doi: 10.1074/jbc.275.3.2071. [DOI] [PubMed] [Google Scholar]
- Kim MS, et al. Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat Immunol. 2003;4(8):787–793. doi: 10.1038/ni952. [DOI] [PubMed] [Google Scholar]
- Kim T, et al. Downregulation of lipopolysaccharide response in Drosophila by negative crosstalk between the AP1 and NF-kappaB signaling modules. Nat Immunol. 2005;6(2):211–218. doi: 10.1038/ni1159. [DOI] [PubMed] [Google Scholar]
- Kim M, et al. Caspar, a suppressor of antibacterial immunity in Drosophila. Proc Natl Acad Sci USA. 2006;103(44):16358–16363. doi: 10.1073/pnas.0603238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim LK, et al. Down-regulation of NF-kappaB target genes by the AP-1 and STAT complex during the innate immune response in Drosophila. PLoS Biol. 2007;5(9):e238. doi: 10.1371/journal.pbio.0050238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, et al. A three-step proteolytic cascade mediates the activation of the peptidoglycan-induced toll pathway in an insect. J Biol Chem. 2008;283(12):7599–7607. doi: 10.1074/jbc.M710216200. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8(7):700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Kleino A, et al. Pirk is a negative regulator of the Drosophila Imd pathway. J Immunol. 2008;180(8):5413–5422. doi: 10.4049/jimmunol.180.8.5413. [DOI] [PubMed] [Google Scholar]
- Kuttenkeuler NP, et al. A large-scale RNAi screen identifies Deaf1 as a regulator of innate immune responses in Drosophila. J Innate Immun. 2010;2(2):181–194. doi: 10.1159/000248649. [DOI] [PubMed] [Google Scholar]
- Lamothe B, et al. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. J Biol Chem. 2007;282(6):4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, et al. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86(6):973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, et al. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci USA. 1997;94(26):14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone P, et al. Crystal structure of Drosophila PGRP-SD suggests binding to DAP-type but not lysine-type peptidoglycan. Mol Immunol. 2008;45(9):2521–30. doi: 10.1016/j.molimm.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Leulier F, et al. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 2000;1(4):353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F, et al. Inducible expression of double-stranded RNA reveals a role for dFADD in the regulation of the antibacterial response in Drosophila adults. Curr Biol. 2002;12(12):996–1000. doi: 10.1016/s0960-9822(02)00873-4. [DOI] [PubMed] [Google Scholar]
- Leulier F, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. 2003;4(5):478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- Leung TH, et al. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell. 2004;118(4):453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Levashina EA, et al. Two distinct pathways can control expression of the gene encoding the Drosophila antimicrobial peptide metchnikowin. J Mol Biol. 1998;278(3):515–527. doi: 10.1006/jmbi.1998.1705. [DOI] [PubMed] [Google Scholar]
- Levashina EA, et al. Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science. 1999;285(5435):1917–1919. doi: 10.1126/science.285.5435.1917. [DOI] [PubMed] [Google Scholar]
- Lhocine N, et al. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe. 2008;4(2):147–158. doi: 10.1016/j.chom.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Li H, et al. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296(5571):1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- Liang Y, et al. NF-kappaB and its regulation on the immune system. Cell Mol Immunol. 2004;1(5):343–350. [PubMed] [Google Scholar]
- Liehl P, et al. Prevalence of local immune response against oral infection in a Drosophila/Pseudomonas infection model. PLoS Pathog. 2006;2(6):e56. doi: 10.1371/journal.ppat.0020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligoxygakis P, et al. A serpin mutant links Toll activation to melanization in the host defence of Drosophila. Embo J. 2002;21(23):6330–6337. doi: 10.1093/emboj/cdf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligoxygakis P, et al. A serpin regulates dorsal-ventral axis formation in the Drosophila embryo. Curr Biol. 2003;13(23):2097–2102. doi: 10.1016/j.cub.2003.10.062. [DOI] [PubMed] [Google Scholar]
- Lim JH, et al. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J Biol Chem. 2006;281(12):286–295. doi: 10.1074/jbc.M513030200. [DOI] [PubMed] [Google Scholar]
- Lin L, et al. Cotranslational biogenesis of NF-kappaB p50 by the 26S proteasome. Cell. 1998;92(6):819–828. doi: 10.1016/s0092-8674(00)81409-9. [DOI] [PubMed] [Google Scholar]
- Lu Y, et al. The antibacterial arm of the Drosophila innate immune response requires an IkappaB kinase. Genes Dev. 2001;15(1):104–110. doi: 10.1101/gad.856901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfruelli P, et al. A mosaic analysis in Drosophila fat body cells of the control of antimicrobial peptide genes by the Rel proteins Dorsal and DIF. Embo J. 1999;18(12):3380–3391. doi: 10.1093/emboj/18.12.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield BE, et al. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell Microbiol. 2003;5(12):901–911. doi: 10.1046/j.1462-5822.2003.00329.x. [DOI] [PubMed] [Google Scholar]
- Matova N, Anderson KV. Rel/NF-kappaB double mutants reveal that cellular immunity is central to Drosophila host defense. Proc Natl Acad Sci USA. 2006;103(44):16424–16429. doi: 10.1073/pnas.0605721103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matova N, Anderson KV. Drosophila Rel proteins are central regulators of a robust, multi-organ immune network. J Cell Sci. 2010;123(Pt 4):627–633. doi: 10.1242/jcs.060731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellroth P, Steiner H. PGRP-SB1: an N-acetylmuramoyl l-alanine amidase with antibacterial activity. Biochem Biophys Res Commun. 2006;350(4):994–999. doi: 10.1016/j.bbrc.2006.09.139. [DOI] [PubMed] [Google Scholar]
- Mellroth P, et al. A scavenger function for a Drosophila peptidoglycan recognition protein. J Biol Chem. 2003;278(9):7059–7064. doi: 10.1074/jbc.M208900200. [DOI] [PubMed] [Google Scholar]
- Mellroth P, et al. Ligand-induced dimerization of Drosophila peptidoglycan recognition proteins in vitro. Proc Natl Acad Sci USA. 2005;102(18):6455–6460. doi: 10.1073/pnas.0407559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, et al. Toll receptor-mediated Drosophila immune response requires Dif, an NF-kappaB factor. Genes Dev. 1999;13(7):792–797. doi: 10.1101/gad.13.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika M, et al. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1(2):277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- Meylan E, et al. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat Immunol. 2004;5(5):503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- Michel T, et al. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414(6865):756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- Minakhina S, Steward R. Nuclear factor-kappa B pathways in Drosophila. Oncogene. 2006;25(51):6749–6757. doi: 10.1038/sj.onc.1209940. [DOI] [PubMed] [Google Scholar]
- Mizuguchi K, et al. Getting knotted: a model for the structure and activation of Spatzle. Trends Biochem Sci. 1998;23(7):239–242. doi: 10.1016/s0968-0004(98)01216-x. [DOI] [PubMed] [Google Scholar]
- Naitza S, et al. The Drosophila immune defense against gram-negative infection requires the death protein dFADD. Immunity. 2002;17(5):575–581. doi: 10.1016/s1074-7613(02)00454-5. [DOI] [PubMed] [Google Scholar]
- Narasimamurthy R. Structure-function analysis of Eiger, the Drosophila TNF homolog. Cell Res. 2009;19:392–394. doi: 10.1038/cr.2009.16. [DOI] [PubMed] [Google Scholar]
- Nicolas E, et al. In vivo regulation of the IkappaB homologue cactus during the immune response of Drosophila. J Biol Chem. 1998;273(17):10463–10469. doi: 10.1074/jbc.273.17.10463. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, et al. Determination of anteroposterior polarity in Drosophila. Science. 1987;238(4834):1675–1681. doi: 10.1126/science.3686007. [DOI] [PubMed] [Google Scholar]
- Ochiai M, Ashida M. A pattern recognition protein for peptidoglycan. Cloning the cDNA and the gene of the silkworm, Bombyx mori. J Biol Chem. 1999;274(17):11854–11858. doi: 10.1074/jbc.274.17.11854. [DOI] [PubMed] [Google Scholar]
- Pan DJ, et al. Functional analysis of the Drosophila twist promoter reveals a dorsal-binding ventral activator region. Genes Dev. 1991;5(10):1892–1901. doi: 10.1101/gad.5.10.1892. [DOI] [PubMed] [Google Scholar]
- Paquette N, et al. Caspase-mediated cleavage, IAP binding, and ubiquitination: linking three mechanisms crucial for Drosophila NF-kappaB signaling. Mol Cell. 2010;37(2):172–182. doi: 10.1016/j.molcel.2009.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, et al. Signal-induced transcriptional activation by Dif requires the dTRAP80 mediator module. Mol Cell Biol. 2003;23(4):1358–1367. doi: 10.1128/MCB.23.4.1358-1367.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, et al. Targeting of TAK1 by the NF-kappa B protein Relish regulates the JNK-mediated immune response in Drosophila. Genes Dev. 2004;18(5):584–594. doi: 10.1101/gad.1168104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MY, et al. Fas-associated factor-1 inhibits nuclear factor-kappaB (NF-kappaB) activity by interfering with nuclear translocation of the RelA (p65) subunit of NF-kappaB. J Biol Chem. 2004;279(4):2544–2549. doi: 10.1074/jbc.M304565200. [DOI] [PubMed] [Google Scholar]
- Park JW, et al. Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc Natl Acad Sci USA. 2007;104(16):6602–6607. doi: 10.1073/pnas.0610924104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelte N, et al. Immune challenge induces N-terminal cleavage of the Drosophila serpin Necrotic. Insect Biochem Mol Biol. 2006;36(1):37–46. doi: 10.1016/j.ibmb.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8(1):49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- Petersen UM, et al. Serpent regulates Drosophila immunity genes in the larval fat body through an essential GATA motif. Embo J. 1999;18(14):4013–4022. doi: 10.1093/emboj/18.14.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham LN, et al. A specific primed immune response in Drosophila is dependent on phagocytes. PLoS Pathog. 2007;3(3):e26. doi: 10.1371/journal.ppat.0030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pili-Floury S, et al. In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J Biol Chem. 2004;279(13):12848–12853. doi: 10.1074/jbc.M313324200. [DOI] [PubMed] [Google Scholar]
- Qin BY, et al. Crystal structure of IRF-3 in complex with CBP. Structure. 2005;13(9):1269–1277. doi: 10.1016/j.str.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Ramet M, et al. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416(6881):644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- Rao A, et al. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- Reach M, et al. A gradient of cactus protein degradation establishes dorsoventral polarity in the Drosophila embryo. Dev Biol. 1996;180(1):353–364. doi: 10.1006/dbio.1996.0308. [DOI] [PubMed] [Google Scholar]
- Reed DE, et al. DEAF-1 regulates immunity gene expression in Drosophila. Proc Natl Acad Sci USA. 2008;105(24):8351–8356. doi: 10.1073/pnas.0802921105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser JB, et al. Crystal structure of the Drosophila peptidoglycan recognition protein (PGRP)-SA at 1.56 A resolution. J Mol Biol. 2004;340(4):909–917. doi: 10.1016/j.jmb.2004.04.077. [DOI] [PubMed] [Google Scholar]
- Roth S, et al. A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell. 1989;59(6):1189–1202. doi: 10.1016/0092-8674(89)90774-5. [DOI] [PubMed] [Google Scholar]
- Royet J, Dziarski R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol. 2007;5(4):264–277. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- Rutschmann S, et al. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity. 2000;12(5):569–580. doi: 10.1016/s1074-7613(00)80208-3. [DOI] [PubMed] [Google Scholar]
- Rutschmann S, et al. Role of Drosophila IKK gamma in a toll-independent antibacterial immune response. Nat Immunol. 2000;1(4):342–347. doi: 10.1038/79801. [DOI] [PubMed] [Google Scholar]
- Ryu JH, et al. The homeobox gene Caudal regulates constitutive local expression of antimicrobial peptide genes in Drosophila epithelia. Mol Cell Biol. 2004;24(1):172–185. doi: 10.1128/MCB.24.1.172-185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JH, et al. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319(5864):777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- Sabatier L, et al. Pherokine-2 and -3. Eur J Biochem. 2003;270(16):3398–3407. doi: 10.1046/j.1432-1033.2003.03725.x. [DOI] [PubMed] [Google Scholar]
- Scherfer C, et al. Drosophila Serpin-28D regulates hemolymph phenoloxidase activity and adult pigmentation. Dev Biol. 2008;323(2):189–196. doi: 10.1016/j.ydbio.2008.08.030. [DOI] [PubMed] [Google Scholar]
- Schiffmann DA, et al. Formation and biochemical characterization of tube/pelle death domain complexes: critical regulators of postreceptor signaling by the Drosophila toll receptor. Biochemistry. 1999;38(36):11722–11733. doi: 10.1021/bi9904252. [DOI] [PubMed] [Google Scholar]
- Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36(4):407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DS, et al. Drosophila eiger mutants are sensitive to extracellular pathogens. PLoS Pathog. 2007;3(3):e41. doi: 10.1371/journal.ppat.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics. 1989;121(1):101–117. doi: 10.1093/genetics/121.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully LR, Bidochka MJ. Developing insect models for the study of current and emerging human pathogens. FEMS Microbiol Lett. 2006;263(1):1–9. doi: 10.1111/j.1574-6968.2006.00388.x. [DOI] [PubMed] [Google Scholar]
- Senftleben U, et al. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 2001;293(5534):1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- Senger K, et al. Immunity regulatory DNAs share common organizational features in Drosophila. Mol Cell. 2004;13(1):19–32. doi: 10.1016/s1097-2765(03)00500-8. [DOI] [PubMed] [Google Scholar]
- Serfling E, et al. NFAT and NF-kappaB factors-the distant relatives. Int J Biochem Cell Biol. 2004;36(7):1166–1170. doi: 10.1016/j.biocel.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Shi Y. A conserved tetrapeptide motif: potentiating apoptosis through IAP-binding. Cell Death Differ. 2002;9(2):93–95. doi: 10.1038/sj.cdd.4400957. [DOI] [PubMed] [Google Scholar]
- Silverman N, Maniatis T. NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001;15(18):2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- Silverman N, et al. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14(19):2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman N, et al. Immune activation of NF-kappaB and JNK requires Drosophila TAK1. J Biol Chem. 2003;278(49):48928–48934. doi: 10.1074/jbc.M304802200. [DOI] [PubMed] [Google Scholar]
- Spencer E, et al. Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes Dev. 1999;13(3):284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Leger RJ, et al. Molecular cloning and regulatory analysis of the cuticle-degrading-protease structural gene from the entomopathogenic fungus Metarhizium anisopliae. Eur J Biochem. 1992;204(3):991–1001. doi: 10.1111/j.1432-1033.1992.tb16721.x. [DOI] [PubMed] [Google Scholar]
- Steward R. Dorsal, an embryonic polarity gene in Drosophila, is homologous to the vertebrate proto-oncogene, c-rel. Science. 1987;238(4827):692–694. doi: 10.1126/science.3118464. [DOI] [PubMed] [Google Scholar]
- Steward R. Relocalization of the dorsal protein from the cytoplasm to the nucleus correlates with its function. Cell. 1989;59(6):1179–1188. doi: 10.1016/0092-8674(89)90773-3. [DOI] [PubMed] [Google Scholar]
- Stoven S, et al. Activation of the Drosophila NF-kappaB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 2000;1(4):347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoven S, et al. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc Natl Acad Sci USA. 2003;100(10):5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, et al. A heterotrimeric death domain complex in Toll signaling. Proc Natl Acad Sci USA. 2002;99(20):12871–12876. doi: 10.1073/pnas.202396399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, et al. Regulated assembly of the Toll signaling complex drives Drosophila dorsoventral patterning. Embo J. 2004;23(1):100–110. doi: 10.1038/sj.emboj.7600033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana A, et al. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophe-noloxidase cascade in Drosophila larvae. Proc Natl Acad Sci USA. 2002;99(21):13705–13710. doi: 10.1073/pnas.212301199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana A, et al. Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. Embo J. 2004;23(23):4690–4700. doi: 10.1038/sj.emboj.7600466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, et al. A serpin that regulates immune melanization in the respiratory system of Drosophila. Dev Cell. 2008;15(4):617–626. doi: 10.1016/j.devcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji T, et al. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol Cell Biol. 2007;27(12):4578–4588. doi: 10.1128/MCB.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatei K, Levine M. Specificity of Rel-inhibitor interactions in Drosophila embryos. Mol Cell Biol. 1995;15(7):3627–3634. doi: 10.1128/mcb.15.7.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauszig-Delamasure S, et al. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat Immunol. 2002;3(1):91–97. doi: 10.1038/ni747. [DOI] [PubMed] [Google Scholar]
- Thevenon D, et al. The Drosophila ubiquitin-specific protease dUSP36/Scny targets IMD to prevent constitutive immune signaling. Cell Host Microbe. 2009;6(4):309–320. doi: 10.1016/j.chom.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Thisse C, et al. Sequence-specific transactivation of the Drosophila twist gene by the dorsal gene product. Cell. 1991;65(7):1191–1201. doi: 10.1016/0092-8674(91)90014-p. [DOI] [PubMed] [Google Scholar]
- Towb P, et al. Recruitment of Tube and Pelle to signaling sites at the surface of the Drosophila embryo. Development. 1998;125(13):2443–2450. doi: 10.1242/dev.125.13.2443. [DOI] [PubMed] [Google Scholar]
- Traenckner EB, et al. Phosphorylation of human I kappa B-alpha on serines 32 and 36 controls I kappa B-alpha proteolysis and NF-kappa B activation in response to diverse stimuli. Embo J. 1995;14(12):2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CW, et al. Drosophila melanogaster mounts a unique immune response to the Rhabdovirus sigma virus. Appl Environ Microbiol. 2008;74(10):3251–3256. doi: 10.1128/AEM.02248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsichritzis T, et al. A Drosophila ortholog of the human cylindromatosis tumor suppressor gene regulates triglyceride content and antibacterial defense. Development. 2007;134(14):2605–2614. doi: 10.1242/dev.02859. [DOI] [PubMed] [Google Scholar]
- Tsuda M, et al. The RING-finger scaffold protein Plenty of SH3 s targets TAK1 to control immunity signalling in Drosophila. EMBO Rep. 2005;6(11):1082–1087. doi: 10.1038/sj.embor.7400537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzou P, et al. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity. 2000;13(5):737–748. doi: 10.1016/s1074-7613(00)00072-8. [DOI] [PubMed] [Google Scholar]
- Tzou P, et al. Constitutive expression of a single antimicrobial peptide can restore wild-type resistance to infection in immunodeficient Drosophila mutants. Proc Natl Acad Sci USA. 2002;99(4):2152–2157. doi: 10.1073/pnas.042411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uvell H, Engstrom Y. Functional characterization of a novel promoter element required for an innate immune response in Drosophila. Mol Cell Biol. 2003;23(22):8272–8281. doi: 10.1128/MCB.23.22.8272-8281.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Berghe W, et al. The nuclear factor-kappaB engages CBP/p300 and histone acetyltransferase activity for transcriptional activation of the interleukin-6 gene promoter. J Biol Chem. 1999;274(45):32091–32098. doi: 10.1074/jbc.274.45.32091. [DOI] [PubMed] [Google Scholar]
- Vidal S, et al. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-kappaB-dependent innate immune responses. Genes Dev. 2001;15(15):1900–1912. doi: 10.1101/gad.203301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonkavaara M, et al. Drosophila melanogaster as a model for elucidating the pathogenicity of Francisella tularensis. Cell Microbiol. 2008;10(6):1327–1338. doi: 10.1111/j.1462-5822.2008.01129.x. [DOI] [PubMed] [Google Scholar]
- Wang L, et al. Sensing of Gram-positive bacteria in Drosophila: GNBP1 is needed to process and present peptidoglycan to PGRP-SA. Embo J. 2006;25(20):5005–5014. doi: 10.1038/sj.emboj.7601363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312(5772):452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson FL, et al. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309(5742):1874–1878. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- Weber AN, et al. Binding of the Drosophila cytokine Spatzle to Toll is direct and establishes signaling. Nat Immunol. 2003;4(8):794–800. doi: 10.1038/ni955. [DOI] [PubMed] [Google Scholar]
- Werner T, et al. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97(25):13772–13777. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, et al. Functional diversity of the Drosophila PGRP-LC gene cluster in the response to lipopolysaccharide and peptidoglycan. J Biol Chem. 2003;278(29):26319–26322. doi: 10.1074/jbc.C300184200. [DOI] [PubMed] [Google Scholar]
- Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430(7000):694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Whalen AM, Steward R. Dissociation of the dorsal-cactus complex and phosphorylation of the dorsal protein correlate with the nuclear localization of dorsal. J Cell Biol. 1993;123(3):523–534. doi: 10.1083/jcb.123.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiklund ML, et al. The N-terminal half of the Drosophila Rel/NF-kappaB factor Relish, REL-68, constitutively activates transcription of specific Relish target genes. Dev Comp Immunol. 2009;33(5):690–696. doi: 10.1016/j.dci.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Windheim M, et al. Two different classes of E2 ubiquitin-conjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology. Biochem J. 2008;409(3):723–729. doi: 10.1042/BJ20071338. [DOI] [PubMed] [Google Scholar]
- Winston JT, et al. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999;13(3):270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LP, Anderson KV. Regulated nuclear import of Rel proteins in the Drosophila immune response. Nature. 1998;392(6671):93–97. doi: 10.1038/32195. [DOI] [PubMed] [Google Scholar]
- Xue L, et al. Tumor suppressor CYLD regulates JNK-induced cell death in Drosophila. Dev Cell. 2007;13(3):446–454. doi: 10.1016/j.devcel.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Yagi Y, Ip YT. Helicase89B is a Mot1p/BTAF1 homologue that mediates an antimicrobial response in Drosophila. EMBO Rep. 2005;6(11):1088–1094. doi: 10.1038/sj.embor.7400542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, et al. Autophagic control of listeria through intracellular innate immune recognition in Drosophila. Nat Immunol. 2008;9(8):908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, et al. Beta-1,3-glucan receptor and peptidoglycan receptor are present as separate entities within insect prophenoloxidase activating system. Biochem Biophys Res Commun. 1986;141(3):1177–1184. doi: 10.1016/s0006-291x(86)80168-1. [DOI] [PubMed] [Google Scholar]
- Yoshida H, et al. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J Biol Chem. 1996;271(23):3854–3860. doi: 10.1074/jbc.271.23.13854. [DOI] [PubMed] [Google Scholar]
- Zaidman-Remy A, et al. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24(4):463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Zambon RA, et al. The Toll pathway is important for an antiviral response in Drosophila. Proc Natl Acad Sci USA. 2005;102(20):7257–7262. doi: 10.1073/pnas.0409181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon RA, et al. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol. 2006;8(5):880–889. doi: 10.1111/j.1462-5822.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- Zhou R, et al. The role of ubiquitination in Drosophila innate immunity. J Biol Chem. 2005;280(40):34048–34055. doi: 10.1074/jbc.M506655200. [DOI] [PubMed] [Google Scholar]
- Zhuang ZH, et al. Drosophila TAB 2 is required for the immune activation of JNK and NF-kappaB. Cell Signal. 2006;18(7):964–970. doi: 10.1016/j.cellsig.2005.08.020. [DOI] [PubMed] [Google Scholar]