Abstract
We report the design and construction of a nanometer-sized tetrahedron from a single strand of DNA that is 286 nucleotides long. The formation of the tetrahedron was verified by restriction enzyme digestion, Ferguson analysis, and atomic force microscopy (AFM) imaging. We further demonstrate that synthesis of the tetrahedron can be easily scaled up through in vivo replication using standard molecular cloning techniques. We found that the in vivo replication efficiency of the tetrahedron is significantly higher in comparison to in vitro replication using rolling-circle amplification (RCA). Our results suggest that it is now possible to design and replicate increasingly complex, single-stranded DNA nanostructures in vivo.
Introduction
With highly specific Watson–Crick base pairing and a well-characterized double-helical structure, DNA has been utilized as a programmable building material to construct designer nanoscale architectures for a broad range of applications, such as organizing nanoparticles and proteins and confining the motions of DNA-based nanomotors.1–10 To date, a large variety of one- and two-dimensional (1D and 2D) DNA nanostructures have been successfully designed and assembled.11–25 Recently, a series of three-dimensional (3D) polyhedral DNA nanoarchitectures26–35 were generated through either one-step or hierarchical assembly approaches, further enriching the vast library of artificial DNA constructions. Nevertheless, these DNA polyhedrons were constructed from multiple oligonucleotides with deliberately designed sequences. In one case, Shih et al.29 synthesized an octahedron by folding a 1.7-kb single-stranded DNA (ssDNA) with the help of five short DNA strands, suggesting the possibility of folding a ssDNA molecule into a well-defined 3D nanostructure. However, the minimum number of DNA strands required to build a complete 3D polyhedron remained to be determined. In addition, recent progress in replicating artificial DNA nanostructures revealed that ssDNA molecules with complicated secondary structures can be amplified efficiently and with high fidelity by biological methods,36 making the replication of a single-stranded 3D polyhedron an appealing objective to pursue. Here we present the facile preparation and in vivo replication of a DNA tetrahedron folded from one ssDNA molecule that is 286 nucleotides (nt) long. This study demonstrates a reliable method that can be used for the design and replication of other types of single-stranded, 3D DNA nanostructures of considerable complexity.
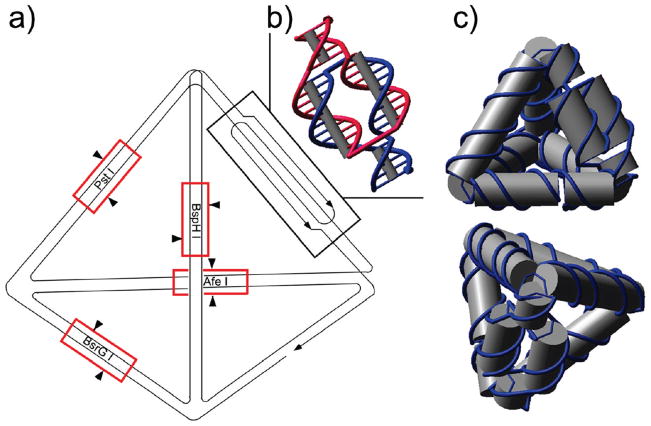
The folding pathway of the single-stranded tetrahedron is illustrated in Figure 1a. Among its six edges, five are composed of 21-base-pair (bp) double helices, while the remaining edge contains a “twin double-helical” motif (Figure 1b) to accommodate the required reverse polarity of complementary DNA strands. Four cleavable sites, specific to the restriction enzymes PstI, BsrGI, AfeI, and BspHI, were designed in the middle of four edges of the DNA tetrahedron (Figure 1a) for restriction digestion characterization of the assembly product. An unpaired thymine base was incorporated at each vertex to allow adequate flexibility for folding. When annealed, the DNA strand self-assembled into the desired tetrahedron (Figure 1c) through designated intramolecular base pairing.
Figure 1.
Design of the ssDNA tetrahedron. (a) Folding pathway of the single-stranded tetrahedron. Five edges are composed of 21-bp double helices, while the remaining edge contains a “twin double helical” component. In the middle of four edges of the DNA tetrahedron, four restriction enzyme sites (PstI, BsrGI, AfeI, and BspHI) are designed. The restriction digestion sites of the corresponding enzymes are indicated by red boxes and black arrowheads. (b) Structural design of the twin double-helical component of the remaining edge. (c) Front and top views of the 3D molecular model of the tetrahedron.
Materials and Methods
Materials
Detailed information about the materials used in this study can be found in the Supporting Information.
Structural Design and Assembly
The tetrahedron structure was modeled by use of Nanoengineer-1 (www.nanorex.com) and the DNA sequence was generated by Uniquimer (Figure S1 in Supporting Information).37 Due to the extremely low yield of the synthesis of DNA oligonucleotides longer than 200 bases, the 286-nt ssDNA was divided into three segments (Table 1); they were first synthesized separately and subsequently ligated to yield the complete strand. Equal molar amounts of component strands 1, 2, and 3 were mixed at 0.5 μM in 1× TAE/Mg2+ buffer [Tris–acetic acid 40 mM, pH 8.0, magnesium acetate 12.5 mM, and ethylene-diaminetetraacetic acid (EDTA) 1 mM] and annealed in a water bath from 95 °C to room temperature for approximately 48 h. Ten units of T4 DNA ligase in 1× T4 DNA ligase buffer was added to 100 pmol of annealed sample and left at 4 °C overnight, to seal the two nicks. Denaturing polyacrylamide gel electrophoresis (PAGE) purification was utilized to obtain the full-length strand (Figure S2 in Supporting Information).
Table 1.
Sequences of Component Strands Used to Synthesize the Full-Length (286-nt) ssDNA
| component strand 1 | AGACGTGCGTTAGATATGCTGTACAAGCGCGATCGTGACGACTGCAGAAGTGCTTCACGCATTTCATGATACGAGCTACGCACGTCTACTCTAGGGCGTGGGTGC |
| component strand 2a | /Phos/GGAGCGCTGGCCGAATTCGCGCTTGTACAGCATATCTTGCTCGTATCATGAAA |
| component strand 3a | /Phos/TGCGTGTGCGACTCTCGTGCCGGCTTGCGTCCGCGTCGCTAGCACTTCTGCAGTCGTCACGTTTCGGCCAGCGCTCCGCACCCTGCGGCCCGGCACGAGAGCGGACGCAAGGCCGCTCGCCCTAGAGT |
The 5′ end of the strand is phosphorylated.
Restriction Enzyme Digestion
The purified, full-length DNA strand was annealed in a water bath from 95 °C to room temperature for about 48 h to facilitate the folding of the single strand into the desired tetrahedron, and the annealed DNA sample was then digested by a restriction enzyme (PstI or BsrGI or AfeI or BspHI). Two picomoles of DNA was digested by 10 units of enzyme in 40 μL of 1× NE buffer 1 at 37 °C for 3 h. The digested products were analyzed by denaturing 10% PAGE.
Ferguson Analysis
The preannealed, single-stranded DNA tetrahedron, the DNA tetrahedron assembled from four oligonucleotides as described by Goodman et al.,30 and a 25-bp DNA ladder were loaded into separate lanes of nondenaturing 6%, 8%, 10%, and 12% polyacrylamide gels (Figure S3 in Supporting Information). The four gels were simultaneously run for 3 h at a constant voltage of 10 V/cm. After staining, the mobilities of corresponding bands were measured from the gel images manually, by use of a millimeter-scale ruler.
AFM Imaging
The DNA tetrahedron samples (2 μL, 10 nM) were deposited onto freshly cleaved mica (Ted Pella, Inc.) and left to adsorb for 3 min. Buffer (1× TAE/Mg2+, 30 μL) was added to the liquid cell and the sample was scanned in tapping mode on a Multimode-V AFM (Veeco. Inc.) with NP-S tips (Veeco, Inc.).
In Vivo Cloning
The single-stranded DNA tetrahedron was extended at both the 5′ and 3′ ends and hybridized to its Watson–Crick complement to form a double strand with the proper sticky end sequence (PstI and SacI) for insertion into a plasmid. To avoid undesired digestion products, the PstI cleavage site [d(CTGCAG)] on one edge of the tetrahedron was changed to d(CTGTAG). The in vivo cloning procedures were adapted from a protocol previously reported by Lin et al.36 (see Supporting Information for additional details). Restriction enzyme digestion and Ferguson analysis were used to characterize the replicated product, as described above.
Rolling-Circle Amplification of the Tetrahedron
RCA was initially attempted to amplify this strand (see Supporting Information for details).
Results and Discussion
Synthesis of the single-stranded tetrahedron began with ligation of the three component strands (105-nt, 53-nt, and 128-nt) to yield the full-length 286-mer oligonucleotide. First, the three component strands were mixed in stoichiometric ratios and annealed to allow intermolecular self-assembly, and T4 DNA ligase was subsequently added to seal the phosphorylated nicks. From the denaturing PAGE assay (Figure S2 in Supporting Information), the yield of the ligation reaction was estimated to be ~50%. The relatively high yield of ligation suggested that the self-assembly of the three component strands formed a discrete nanostructure as expected. The full-length 286-nt ssDNA molecule extracted from the gel was then annealed to fold into the desired tetrahedron. Since the self-assembly process involved only a single DNA strand, experimental uncertainties such as pipetting errors that could lead to stoichiometry problems were minimized. It is worth noting that the annealing process was carried out at a relatively low DNA concentration (50 nM), to minimize undesired interstrand associations and achieve optimal assembly yield.
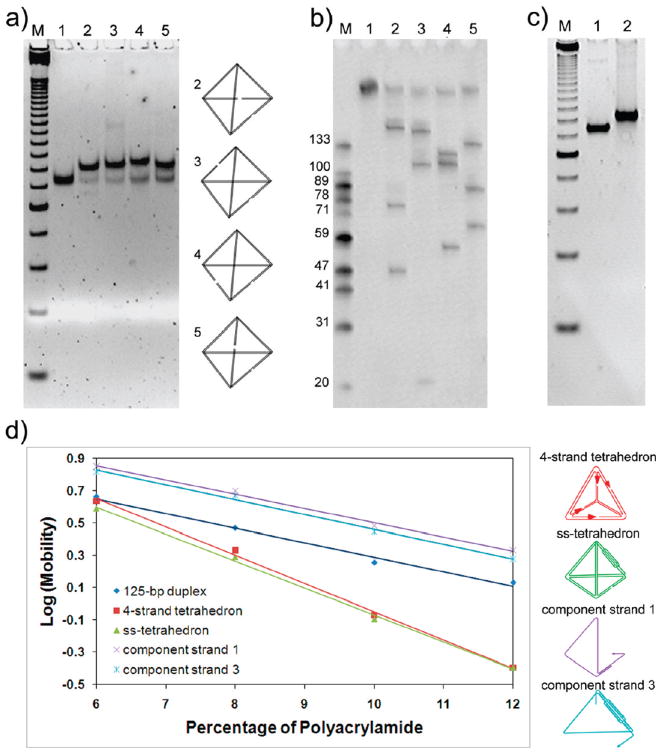
To confirm the correct formation of the tetrahedron after annealing, three experiments were performed: restriction enzyme digestion, Ferguson analysis, and AFM imaging.
According to the design illustrated in Figure 1a, each of the four restriction enzymes will digest the tetrahedron into three fragments with specific lengths (Table 2). Following ref 28, we analyzed the restriction-digested samples by nondenaturing PAGE (Figure 2a). After cleavage by each enzyme, a shift of the mobility of the original band was observed without fragmentation, which suggested that the major structure was assembled from ssDNA rather than from multiple strands. The slightly lower mobility of the digested samples was expected, due to their higher flexibility than the uncut structure. Moreover, a denaturing PAGE assay (Figure 2b) revealed that, after restriction cleavage, the major DNA fragments that resulted were in perfect agreement with the expected enzyme digestion patterns, indicating correct folding of the tetrahedron. A few side products were also observed as faint bands in the gel image in Figure 2b. These are attributed to the products of star reactions of the enzymes or the cleavage of other DNA nanostructures. For example, although the single-stranded tetrahedron represented the major self-assembly product, dimers, trimers, or even higher order aggregates of the ssDNA molecules could form through intermolecular base-pairing, which may have led to the observed side products upon treatment with the restriction enzymes. This assumption was supported by the nondenaturing PAGE assay (Figure 2c), which shows a few minor bands with reduced mobility as compared to the major band of the tetrahedron. These minor bands can be assigned to some multimolecular aggregates. From the gel images, the yield of the correct tetrahedron structure is estimated to be >90%. On the basis of the results above, including one denaturing gel and two nondenaturing gels, we concluded that the assembled structure was formed from ssDNA and folded as designed.
Table 2.
Expected DNA Fragment Lengths after the Tetrahedron was Restriction-Digested
| restriction enzyme | PstI | BsrGI | AfeI | BspHI |
| fragment lengths (nt) | 46, 76, 164 | 20, 109, 157 | 56, 110, 120 | 64, 87, 135 |
| lane in Figure 2a | 2 | 3 | 4 | 5 |
Figure 2.
Characterization of the single-stranded DNA tetrahedron. (a) Result of restriction enzyme digestion of the ss-tetrahedron on a nondenaturing PAGE (8% polyacrylamide gel). A 125-bp DNA marker was loaded in lane M. AfeI-, PstI-, BsrGI-, and BspHI-digested samples were loaded in lanes 2, 3, 4, and 5, respectively. The cutting sites are illustrated on the right. (b) Denaturing PAGE showing the result of restriction enzyme digestion. Single-stranded DNA markers were loaded in lane M with the lengths shown on the left of the corresponding marker band. Lane 1 was loaded with the undigested 286-nt ssDNA. PstI-, BsrGI-, AfeI-, and BspHI-digested samples were loaded in lanes 2, 3, 4, and 5, respectively. Note that the lengths of the corresponding fragments were in perfect agreement with the expected digestion product lengths as listed in Table 2. (c) Nondenaturing PAGE (8% polyacrylamide gel) comparing the mobility of the four-stranded tetrahedron (lane 1) and single-stranded tetrahedron (lane 2). Lane M contains 25-bp dsDNA marker as a reference. (d) Ferguson analysis of the ss-tetrahedron (137 bp, green), a four-stranded tetrahedron (120 bp, red), a 125-bp dsDNA (black), the structure formed by component strand 1 (purple), and the structure formed by component strand 3 (cyan). The two tetrahedron molecules displayed similar Ferguson slopes; both were significantly different from that of a 125-bp DNA duplex and partially formed structures.
Ferguson analysis (Figure 2d) was also utilized to characterize the conformation of the DNA molecules using nondenaturing gel electrophoresis. By measuring the mobility of the DNA nanostructure at different gel concentrations, the friction constant of the DNA nanostructure is obtained, which is related to its surface area and shape. The single-stranded tetrahedron was run together with a previously reported tetrahedron assembled from four individual strands, as a positive control (Figure 2c). The one-stranded tetrahedron has the same geometry as the four-stranded tetrahedron, with a wider edge containing the twin double-helical component and fewer nicks. As expected, it ran slightly slower than the four-stranded tetrahedron because of its higher molecular weight (137 bp versus 120 bp). Most importantly, the two tetrahedral molecules displayed very similar slopes in the Ferguson plot (Figure 2d). In contrast, the negative controls, a 125-bp double-stranded DNA (dsDNA) molecule and the partial structures formed from component strands 1 and 3, respectively, showed significantly different slopes from the two tetrahedron structures. These results strongly suggested that the 286-nt single-stranded DNA folded into the desired tetrahedral nanostructure.
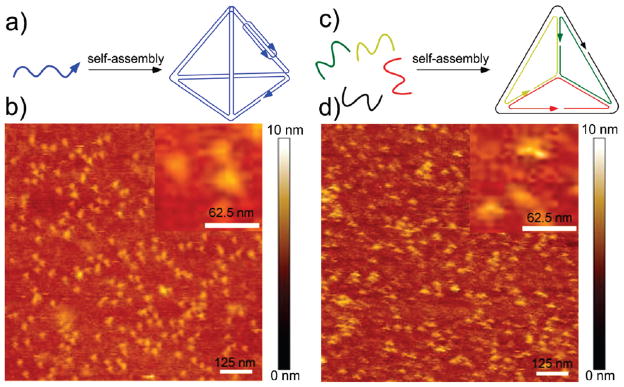
AFM imaging was further used to visualize the assembled structure. We compared our structure assembled by the one-strand strategy with Turberfield’s tetrahedron structure30 formed by the four-strand method. The AFM images shown in Figure 3b,d demonstrate that the particles deposited on the mica surface feature similar morphology with a triangular starlike shape. The sample containing the tetrahedron assembled by the one-strand strategy is more monodisperse, both in size and in shape, as compared to the four-strand tetrahedron sample. This is likely because the tetrahedron composed of four strands has more nick points and is thus more prone to deformation by scanning with an AFM tip. Both structures measure about the same height of ~2 nm, which is consistent with previous observations of tetrahedral structures by the Mao group.31 The height is slightly higher than a DNA duplex, which commonly measures about 1.4 nm on a mica surface via AFM. A height of ~2 nm corresponds to a tetrahedron that has been flattened on the mica surface and squashed by the AFM tip. The lateral dimension of the individual particles measures ~20 nm, larger than the expected ~7 nm, due to resolution that is limited laterally by the tip diameter. This enlargement effect has also been observed by Mao’s group with their tetrahedral DNA structures.31 Overall, side-by-side AFM comparison of our one-strand tetrahedron with the four-stranded tetrahedron, combined with the Ferguson analysis, strongly suggests the correct formation of our designed structures.
Figure 3.
Schematics and direct comparison of AFM images of tetrahedron DNA structures formed by one-strand strategy (a, b) and four-strand strategy (c, d). Scale bars are labeled in each image and inset.
After confirmation of the successful assembly of the single-stranded tetrahedron, we sought to scale up the synthesis and replicate the nanostructure by a biological approach. RCA was first used to replicate the structure (see Supporting Information for experimental details and results). However, the replication efficiency was not satisfactory, most likely a result of the complicated 3D structure of the tetrahedron, preventing efficient strand displacement in the RCA reaction.
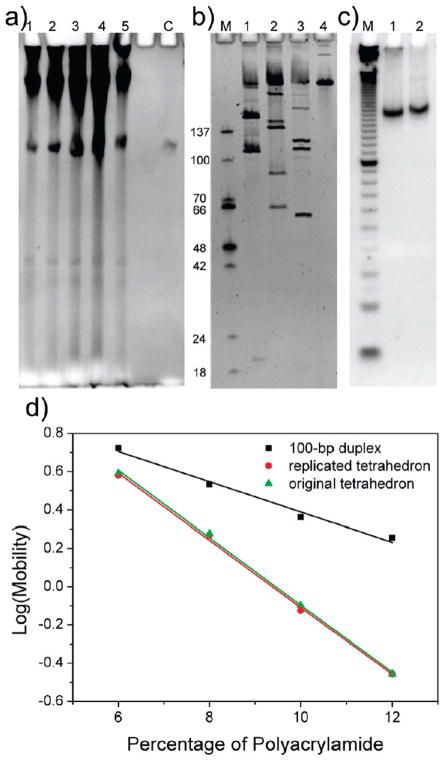
Encouraged by recent findings that artificial DNA nanostructures, such as a Holliday junction-like structure and a paranemic DNA crossover (PX) molecule, can be replicated in viruses and bacteria,36 we exploited the in vivo cloning protocol to amplify the single-stranded tetrahedron (see Figure S5 in Supporting Information for replication scheme). Briefly, the single-stranded tetrahedron (sense strand, 292 nt including the core structure and terminal sticky-end extensions) was inserted into a phagemid, transformed into XL1-Blue cells, and amplified in vivo in the presence of helper phages. The replicated tetrahedrons were recovered by restriction digestion of the single-stranded phagemid extracted from the viral particles. Denaturing PAGE (Figure 4a) was used to evaluate the replication efficiency. The results clearly showed that the replication product had the same molecular weight as the 292-nt sense strand (with the sticky ends added). Approximately 50 pmol of tetrahedron was produced (calculated from the OD260 value of purified DNA) from 250 mL of culture medium. It is very important to point out that this amplification is fully scalable. The final yield of nanostructure is proportional to the volume of the culture medium used. The yield could be improved further by optimizing digestion conditions and the purification process.
Figure 4.
In vivo replication of the single-stranded DNA tetrahedron. (a) Denaturing PAGE showing the final replication product. Lane C, 292-nt sense strand; lanes 1–5, replication products. The DNA species at the top of the gel image represents digested and undigested phagemid vectors; the bands that migrate faster than the complete tetrahedron are truncated nanostructures that may result from incomplete replication. (b) Restriction enzyme digestion assay performed on the replicated tetrahedron. Lane M was loaded with ss-markers with the lengths shown on the left of the corresponding marker band. Lane 4 was loaded with the undigested 292-nt tetrahedron strand. Lanes 1, 2, and 3 are BsrGI-, BspHI-, and AfeI-digested samples, respectively. (c) Nondenaturing PAGE assay showing the mobility of the replicated tetrahedron. Lane M, 10-bp double-stranded DNA ladder; lane 1, annealed original 286-nt tetrahedron; lane 2, annealed replicated tetrahedron (292 nt). (d) Ferguson analysis of the tetrahedron after replication (red circles), the tetrahedron assembled from the original 286-nt strand (green triangles), and a 100-bp dsDNA (black squares).
The replicated strand was then subjected to restriction enzyme digestion and Ferguson analysis to verify that it could still fold into the tetrahedron structure as designed. First, the replication product was separately treated with the restriction enzymes BsrGI, BspHI, and AfeI. It should be noted that the PstI site in the original design was removed to avoid conflicts with the sticky end design for ligation with the plasmid. Denaturing PAGE was used to analyze the digestion results (Figure 4b). Again, all fragment lengths were consistent with the expected pattern summarized in Table 3. Some irregular digestion products were also observed, possibly due to misfolding of the long ssDNA that contained extensive self-complementary sequences and potential for aggregation, similar to the observations in Figure 2c. Second, nondenaturing PAGE (Figure 4c) showed that the replicated tetrahedron (292 nt) exhibited almost the same migration rate as the original 286-nt tetrahedron molecule. The slight difference is a result of the additional sticky ends at the 5′ and 3′ ends of the replicated molecule. Ferguson analysis was then used to compare the friction constant of the replicated tetrahedron to that of the original 286-nt tetrahedron (Figure 4d). The plot of the two molecules nearly overlapped, while the plot for a 100-bp double-stranded DNA showed a dramatically different slope. This observation strongly suggested that the replicated strand correctly folded into the tetrahedron structure, confirming that the single-stranded tetrahedron was replicated with high fidelity by in vivo cloning.
Table 3.
Fragment Lengths of the Replicated DNA Digested by the Three Restriction Enzymes, Respectively
| restriction enzyme | BsrGI | BspHI | AfeI |
| fragment lengths (nt) | 21, 109, 162 | 65, 87, 140 | 61, 111, 120 |
| lane in Figure 4a | 1 | 2 | 3 |
Compared with in vitro enzymatic amplification (RCA), in vivo replication resulted in significantly higher amplification efficiency, demonstrating the power of naturally existing cellular machinery. This is consistent with our former finding36 that in vivo replication yields higher replication efficiency of complicated nanostructures such as a paranemic crossover.
Conclusion
In summary, we have successfully constructed a DNA tetrahedron folded from one ssDNA molecule. To the best of our knowledge, this is the first example of a discrete single-stranded 3D DNA nanostructure experimentally constructed. We expect that our method is highly adaptable for the construction of other polyhedral nanostructures. Compared to the multistrand system, the single-stranded folding strategy features the following advantages: First, it simplifies the assembly process and eliminates stoichiometric dependence, leading to a better assembly yield. Second, it makes the resulting 3D nanostructures readily amplifiable. This is important for scaling up the preparation of DNA nanostructures. Third, the single-stranded nanostructures can easily be circularized to impart exonuclease resistance, resulting in longer life spans in biological systems (e.g., inside living cells). This property is appealing for in vivo applications such as biosensing and drug delivery. Finally, the success in building single-stranded 3D DNA nanostructures prompts us to explore other nucleic acid species, such as RNA, for the construction of 3D molecules. Conceivably, we should be able to synthesize an analogous polyhedron using RNA obtained by transcription.
Supplementary Material
Acknowledgments
This work was supported by the research grant Construction of DNA 3-D Superstructures by DNA Self-Assembly, RGC HKUST 604606, of the earmarked grant from the University Grant Council of the Hong Kong government to Y.M.; by grants from the National Science Foundation (NSF), the Army Research Office (ARO), and the Technology and Research Initiative Fund from Arizona State University to Y.L.; and by grants from NSF, ARO, the Air Force Office of Scientific Research, the Office of Naval Research, the National Institutes of Health, and Alfred P. Sloan Foundation to H.Y.
Footnotes
Supporting Information Available: DNA sequences, materials, detailed methods for in vivo replication, and rolling-circle amplification results. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Seeman NC. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 2.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 3.Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Bruchez MP, Jr, Schultz PG. Nature. 1996;382:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 4.Le JD, Pinto Y, Seeman NC, Musier-Forsyth K, Taton TA, Kiehl RA. Nano Lett. 2004;4:2343–2347. doi: 10.1021/nl0515495. [DOI] [PubMed] [Google Scholar]
- 5.Sharma J, Chhabra R, Liu Y, Ke Y, Yan H. Angew Chem, Int Ed. 2006;45:730–735. doi: 10.1002/anie.200503208. [DOI] [PubMed] [Google Scholar]
- 6.Sharma J, Chhabra R, Andersen CS, Gothelf KV, Yan H, Liu Y. J Am Chem Soc. 2008;130:7820–7821. doi: 10.1021/ja802853r. [DOI] [PubMed] [Google Scholar]
- 7.Li H, Park SH, Reif JH, LaBean TH, Yan H. J Am Chem Soc. 2004;126:418–419. doi: 10.1021/ja0383367. [DOI] [PubMed] [Google Scholar]
- 8.Chhabra R, Sharma J, Ke Y, Liu Y, Rinker S, Lindsay S, Yan H. J Am Chem Soc. 2007;129:10304–10305. doi: 10.1021/ja072410u. [DOI] [PubMed] [Google Scholar]
- 9.Ke Y, Lindsay S, Chang Y, Liu Y, Yan H. Science. 2008;319:180–183. doi: 10.1126/science.1150082. [DOI] [PubMed] [Google Scholar]
- 10.Rinker S, Ke Y, Liu Y, Chhabra R, Yan H. Nat Nanotechnol. 2008;3:418–422. doi: 10.1038/nnano.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winfree E, Liu F, Wenzler LA, Seeman NC. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 12.Mao C, Sun W, Seeman NC. J Am Chem Soc. 1999;121:5437–5443. [Google Scholar]
- 13.LaBean TH, Yan H, Kopatsch J, Liu F, Winfree E, Reif JH, Seeman NC. J Am Chem Soc. 2000;122:1848–1860. [Google Scholar]
- 14.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 15.Liu D, Wang M, Deng Z, Walulu R, Mao C. J Am Chem Soc. 2004;126:2324–2325. doi: 10.1021/ja031754r. [DOI] [PubMed] [Google Scholar]
- 16.Ding B, Sha R, Seeman NC. J Am Chem Soc. 2004;126:10230–10231. doi: 10.1021/ja047486u. [DOI] [PubMed] [Google Scholar]
- 17.Rothemund PWK, Papadakis N, Winfree E. PLoS Biol. 2004;2:2041–2053. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shih WM, Quispe JD, Joyce GF. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 19.Chelyapov N, Brun Y, Gopalkrishnan M, Reishus D, Shaw B, Adleman L. J Am Chem Soc. 2004;126:13924–13925. doi: 10.1021/ja0458120. [DOI] [PubMed] [Google Scholar]
- 20.Malo J, Mitchell JC, Venien-Bryan C, Harris JR, Wille H, Sherratt DJ, Turberfield AJ. Angew Chem, Int Ed. 2005;44:3057–3061. doi: 10.1002/anie.200463027. [DOI] [PubMed] [Google Scholar]
- 21.Mathieu F, Liao S, Kopatsch J, Wang T, Mao C, Seeman NC. Nano Lett. 2005;5:661–665. doi: 10.1021/nl050084f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park SH, Barish R, Li H, Reif JH, Finkelstein G, Yan H, LaBean TH. Nano Lett. 2005;5:693–696. doi: 10.1021/nl050108i. [DOI] [PubMed] [Google Scholar]
- 23.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 24.Ke Y, Liu Y, Zhang J, Yan H. J Am Chem Soc. 2006;128:4414–4421. doi: 10.1021/ja058145z. [DOI] [PubMed] [Google Scholar]
- 25.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Seeman NC. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Seeman NC. J Am Chem Soc. 1994;116:1661–1669. [Google Scholar]
- 28.Goodman RP, Berry RM, Turberfield AJ. Chem Commun. 2004:1372–1373. doi: 10.1039/b402293a. [DOI] [PubMed] [Google Scholar]
- 29.Shih WM, Quispe JD, Joyce GF. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 30.Goodman RP, Schaap IAT, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 31.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 32.Bhatia D, Mehtab S, Krishnan R, Indi SS, Basu A, Krishnan Y. Angew Chem, Int Ed. 2009;121:4198–4201. doi: 10.1002/ange.200806000. [DOI] [PubMed] [Google Scholar]
- 33.Mastroianni AJ, Claridge SA, Alivisatos PA. J Am Chem Soc. 2009;131:8455–8459. doi: 10.1021/ja808570g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson ES, et al. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 35.Ke Y, Sharma J, Liu M, Jahn K, Liu Y, Yan H. Nano Lett. 2009;9:2445–2447. doi: 10.1021/nl901165f. [DOI] [PubMed] [Google Scholar]
- 36.Lin C, Rinker S, Wang X, Liu Y, Seeman NC, Yan H. Proc Natl Acad Sci USA. 2008;105:17626–17631. doi: 10.1073/pnas.0805416105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei B, Wang Z, Mi Y. J Comput Theor Nanosci. 2007;4:133–141. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.