Abstract
Purpose
Epigenetic alterations in tissues targeted for cancer play a causal role in carcinogenesis. Changes in DNA methylation in nontarget tissues, specifically peripheral blood, can also affect risk of malignant disease. We sought to identify specific profiles of DNA methylation in peripheral blood that are associated with bladder cancer risk and therefore serve as an epigenetic marker of disease susceptibility.
Methods
We performed genome-wide DNA methylation profiling on participants involved in a population-based incident case-control study of bladder cancer.
Results
In a training set of 112 cases and 118 controls, we identified a panel of 9 CpG loci whose profile of DNA methylation was significantly associated with bladder cancer in a masked, independent testing series of 111 cases and 119 controls (P < .0001). Membership in three of the most methylated classes was associated with a 5.2-fold increased risk of bladder cancer (95% CI, 2.8 to 9.7), and a model that included the methylation classification, participant age, sex, smoking status, and family history of bladder cancer was a significant predictor of bladder cancer (area under the curve, 0.76; 95% CI, 0.70 to 0.82). CpG loci associated with bladder cancer and aging had neighboring sequences enriched for transcription-factor binding sites related to immune modulation and forkhead family members.
Conclusion
These results indicate that profiles of epigenetic states in blood are associated with risk of bladder cancer and signal the potential utility of epigenetic profiles in peripheral blood as novel markers of susceptibility to this and other malignancies.
INTRODUCTION
The incidence of transitional-cell carcinoma of the urinary bladder (ie, bladder cancer) in the United States in 2009 was predicted to be almost 71,000 new cases.1 Worldwide, almost 360,000 cases of the disease were diagnosed in 2009.2 In addition, 16% of individuals initially diagnosed with bladder cancer will, in their lifetime, be diagnosed with additional primary tumors.3 The highly successful treatment of bladder cancer comes at great economic burden to the health care system, with lifetime monitoring and treatment making bladder cancer one of the most expensive of all cancers, with diagnosis to death per patient costs ranging from $96,000 to $187,000, accounting for almost 3.7 billion US dollars (2001 dollars) in direct costs to the US medical system each year.4
Tobacco carcinogen exposure through active smoking is the main established risk factor for bladder cancer; but the attributable risk is far less than for lung cancer, and much of the etiology of bladder cancer remains unclear.5 Other major risk factors for bladder cancer include occupational exposures, particularly aromatic amine and polycyclic aromatic hydrocarbon exposures,6 inorganic arsenic7–10,11 use of certain hair dyes, exposure to chlorination byproducts, individual fluid intake, and dietary factors.12,13 Of course, host susceptibility also plays an important role in bladder carcinogenesis, and family history of bladder cancer confers an almost two-fold risk of disease. Polymorphisms in genes related to environmental toxicant metabolism such as NAT2 and GSTM1 have been clearly linked to bladder cancer risk.14,15 Genome-wide association studies of bladder cancer identified single-nucleotide polymorphisms (SNPs) on chromosome 8q24, upstream of the MYC oncogene, on chromosome 3q28 near the TP63 tumor suppressor gene,16 and in the PSCA gene to be associated with bladder cancer risk.17 Although these SNP studies may point to novel mechanisms of susceptibility to the disease, it is becoming increasingly clear that their contribution to the attributable risk for the disease is minor. In fact, there is great controversy over whether common genetic variants will play a major role in defining disease susceptibility.18
It is now widely accepted that epigenetic alterations in target tissues are causal to the development of malignancy.19,20 The extent of variability of the cellular epigenome, and specifically DNA methylation at gene promoter regions, remains a critical question; the amount of variation in genomic methylation across the population is not currently known. Further, individual variation in the epigenome is likely to have multiple characteristics, with a component that is tissue-specific and a component that is common to all tissues. We know that some of this variability, particularly in blood, is associated with aging and exposures encountered throughout life,21,22 and the data now suggest that it is, in fact, associated with risk of breast,23 ovarian,24 and small-cell lung cancer.25 The profiles of epigenetic change that are found to be associated with disease may reflect genetic or environmental factors (or their interaction) that establish these gene regulatory marks in a fashion that results in disease susceptibility. Therefore, we examined the genome-wide DNA methylation profiles of peripheral blood from a population-based case-control study of bladder cancer to identify profiles of DNA methylation in this accessible (but not diseased) tissue that are associated with bladder cancer. By examining the gene pathways involved, as well as the genomic context of the loci with bladder cancer–associated methylation, we provide insight into the functional consequences of these profiles and their genesis, respectively.
METHODS
Study Population
The study population has been previously described,26,27 and additional details are provided in the Data Supplement. Briefly, cases of incident bladder cancer were identified from the New Hampshire state cancer registry from July 1, 1994, until June 30, 1998, and a standardized histopathologic review was conducted by a single study pathologist to verify the diagnosis and histopathology of the cases. The case group in this study is limited to white patients because of a limited number of nonwhite participants in the study population. For cases, blood samples were collected on average within 1 year after diagnosis. All controls younger than 65 years of age were selected from records obtained from the New Hampshire Department of Transportation, and controls older than 65 years of age were chosen from records obtained from the Health Care Financing Administration's Medicare Program. Informed consent was obtained from each participant, and all procedures and study materials were approved by the Committee for the Protection of Human Subjects at Dartmouth College and Brown University. Consenting participants underwent a detailed in-person interview covering sociodemographic information and lifestyle factors such as the use of tobacco.
DNA Methylation and Statistical Analysis
DNA was extracted from peripheral-blood buffy coats using the QIAmp DNA mini kit according to the manufacturer's protocol (Qiagen, Valencia, CA) and was subjected to sodium bisulfite modification using the EZ DNA Methylation Kit (Zymo Research, Orange, CA) following the manufacturer's protocol. Methylation profiling was performed using the Illumina Infinium Methylation27 Bead Array at the University of California, San Francisco Institute for Human Genetics Genomic Core Facility.
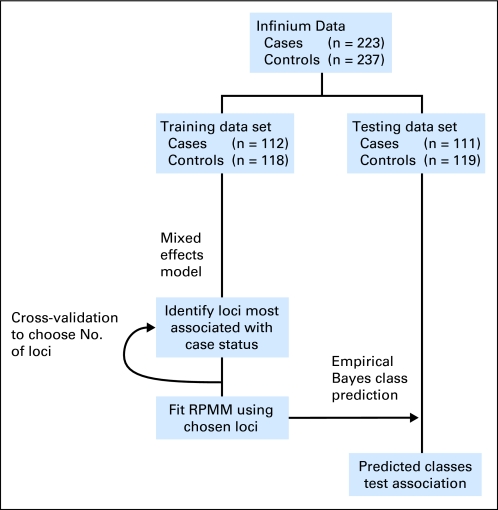
The scheme of our analysis strategy aimed at identifying and validating novel epigenetic biomarkers of bladder cancer in peripheral blood is depicted in Figure 1. We used the methods of Houseman et al,28 the recursively partitioned mixture model (RPMM), because this model-based clustering strategy has been demonstrated to perform effectively and efficiently for methylation data derived from the Illumina array technologies, and it allows for inference in addressing the associations between the methylation-based clusters and covariates. Training and testing sets were obtained by randomly sampling within bladder cancer case-control status. We used a procedure called Semi-Supervised Recursively Partitioned Mixture Models (SS-RPMM),29 similar in spirit to the semi-supervised methodologies proposed by Bair and Tibshirani30 for identifying methylation profiles that are associated with case-control status. To examine the robustness of the association identified in the SS-RPMM, we also used a least absolute shrinkage and selection operator approach for modeling the association between methylation profile and bladder cancer status, using the same training and testing data sets. Details of these methods and results are included in the Data Supplement.
Fig 1.
Diagram of the analysis strategy used in defining the methylation profiles in the training set and applying those profiles for classification in the testing set. RPMM, recursively partitioned mixture model.
Gene set enrichment analysis (GSEA31) was used to explore the biologic relevance of blood-based alterations in DNA methylation for distinguishing bladder cancer cases from controls and also as a result of aging.
RESULTS
The profile of DNA methylation was obtained for 460 peripheral-blood samples using the Human Methylation27 Beadarray. We used a semi-supervised strategy to identify profiles of DNA methylation associated with bladder cancer and to examine whether the identified profiles can predict case status in a series of blinded test samples (Fig 1). Following quality assurance procedures, the data set was split into training and testing series. Characteristics of the cases and controls are shown in Table 1, and do not differ significantly between training and testing sets (Data Supplement).
Table 1.
Characteristics of the Participants Used in the Analysis
| Characteristic | Controls |
Cases |
||
|---|---|---|---|---|
| No. | % | No. | % | |
| Total No. | 237 | 223 | ||
| Age, years | ||||
| Median | 65 | 66 | ||
| Range | 28-74 | 25-74 | ||
| Sex | ||||
| Male | 158 | 48 | 171 | 52 |
| Female | 79 | 60 | 52 | 40 |
| Family history of bladder cancer* | ||||
| No | 224 | 53 | 199 | 47 |
| Yes | 7 | 44 | 9 | 56 |
| Smoking history | ||||
| Never | 72 | 64 | 40 | 36 |
| Former | 126 | 53 | 111 | 47 |
| Current | 39 | 35 | 72 | 65 |
| Tumor stage/grade designation | ||||
| Carcinoma in situ | — | 6 | 3 | |
| Noninvasive low grade (grade 1-2) | — | 140 | 63 | |
| Noninvasive high grade (grade 3) | — | 17 | 7 | |
| Invasive | — | 60 | 27 | |
Data on family history were not available for 13 participants.
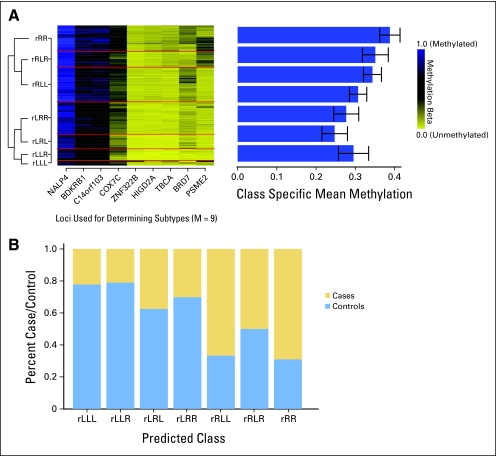
The first step of our semi-supervised strategy was to identify those CpG loci whose methylation state was most significantly associated with being a bladder cancer case rather than control. To do this, we fit a series of linear mixed-effects models using the training data only for each of the 26,486 CpGs in the data set. This allowed us to model each methylation value as the dependent variable, with a random effect for plate (to allow for inter-plate normalization) based on a single normalization sample run on all plates and a fixed effect for case-control status. CpG loci were ranked based on the absolute value of the t statistic derived from the model, and the top nine loci were chosen on the basis of a nested cross-validation procedure (Fig 1) for inclusion in the RPMM, which clustered the samples on the basis of the methylation profile of these nine loci in the training data. To predict class membership in the testing data using only the methylation status of these nine loci, the latent class structure from the RPMM solution fit to the training data was used in conjunction with an empirical Bayes procedure. The methylation profile of these nine loci in the testing data is depicted in Figure 2A, which also shows the mean methylation across loci within a given class and the relationships among the classes through the dendrogram. The right branch classes (those beginning with the letter R) had overall mean methylation that was significantly greater than that of the left branch classes (P < .0001). The distribution of the methylation values for each of the nine loci, across classes, is depicted (Data Supplement).
Fig 2.
DNA methylation profiles defined by a panel of nine loci are significantly associated with bladder cancer. (A) The recursively partitioned mixture model–based classification of methylation of nine loci (columns) in the peripheral blood–derived DNA of the 230 participants (rows) in the testing data set is depicted in the heat map, with the seven classes separated by red lines. The overall mean methylation and CIs (error bars) by class are depicted in the bar graph on the right. (B) The prevalence of cases and controls (y-axis) in each of the predicted classes (x-axis). A permutation-based χ2 test suggests that case-control prevalence is significantly different by methylation class (P < .0001).
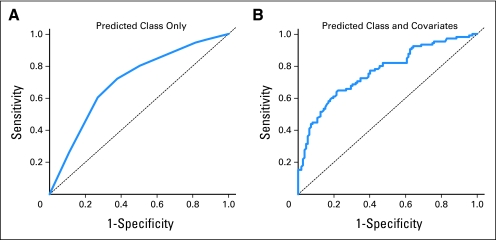
In the test set, we observed that class membership was significantly associated with case-control status (P < .0001, permutation-based χ2 test, Fig 2B), with the right branch classes (those beginning with R) containing a higher proportion of bladder cases than controls compared with the left branch classes. The methylation beta values for cases compared with controls for each of the loci in the testing set are shown (Data Supplement). Each of the nine CpG loci used in the classifier had greater methylation among cases than controls. We assessed performance of the classifier by using receiver operating characteristic curves and calculating the area under the curve (AUC). Using methylation class alone, the AUC was 0.70 (95% bootstrap CI, 0.63 to 0.77). After adjustment for participant age, sex, smoking status (never, former, current), and family history of bladder cancer, the AUC increased to 0.76 (95% bootstrap CI, 0.70 to 0.82; Figs 3A and 3B). To identify whether the association between methylation profiles and bladder cancer is sensitive to the statistical methodology used in the examination, we also performed our analysis using a LASSO approach, using the same training and testing data sets. The methods and results of these analyses are described (Data Supplement) and suggest that our identification of bladder cancer–associated methylation classes is robust to the statistical method used.
Fig 3.
Receiver operating characteristic (ROC) curve analysis of methylation profiles. (A) ROC curve based on methylation class only results in a significant area under the curve (AUC) of 0.70 (95% CI, 0.63 to 0.77). (B) ROC curve including methylation classes, patient sex, age, smoking status (never, former, current), and family history of bladder cancer results in a significant AUC of 0.76 (95% CI, 0.70 to 0.82).
Unconditional logistic regression was used to calculate the magnitude of the association between methylation class and bladder cancer, controlling for potential confounders. The odds ratios (ORs) and 95% CI resulting from each of the pairwise comparisons between the seven predicted classes are shown (Data Supplement). There was a trend of increasing risk of disease moving from the left to right branch of the classification, with the highest risk for members of class RR compared with LLL (OR = 8.7; 95% CI, 1.5 to 55.2). Comparing all the right branch classes with all the left classes, the OR for bladder cancer was 5.2 (95% CI, 2.8 to 9.7), controlled for participant age, sex, smoking status, and family history of bladder cancer. There was no difference in the prevalence of invasive disease across the predicted classes (data not shown).
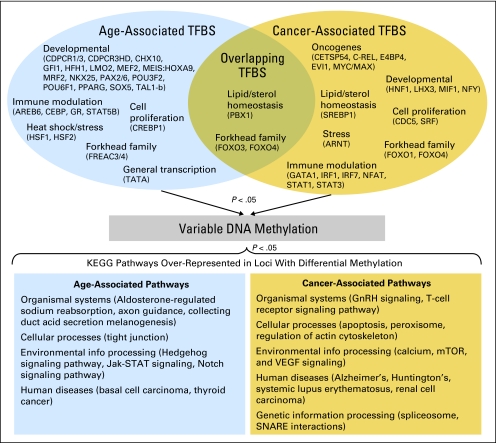
Because previous work has suggested that aging is associated with epigenetic states in peripheral blood and can be related to the alterations associated with cancer, we sought to examine whether there was any overlap in the biologic pathways impacted by differential DNA methylation associated with age or case status. We performed a gene set enrichment analysis (GSEA) based on Kegg-defined pathways using the combined training and testing data and compared pathways over-represented among loci associated with participant age (in controls) with those associated with disease. Pathways with a nominal P < .05 based on the GSEA enrichment statistic are provided in Figure 4, grouped by function. No overlapping pathways based on age- and disease-associated loci were identified. However, similar functional groupings of pathways were identified in both age-associated and bladder cancer–associated loci and are detailed in Figure 4. Genetic information processing pathways were identified exclusively among loci associated with bladder cancer.
Fig 4.
Diagram of the gene-set enrichment analysis on DNA methylation data. The upper panel depicts the transcription factor binding sites (TFBS) within 1 kB of differentially methylated loci associated with aging, bladder cancer, and their overlap grouped by functional role or family. The lower panel depicts the KEGG pathways that are over-represented among the loci with differential methylation associated with aging, bladder cancer, and their overlap, grouped by higher level pathways.
In addition to examining the functional consequences of differential methylation in peripheral blood between cases and controls, we hypothesized that differential methylation profiles may represent a response of the hematopoietic system to a developing tumor (ie, the methylation profiles capture the downstream effects of this response, which may be through differential binding of transcription factors near sites of altered methylation). The top half of Figure 4 depicts the results of this GSEA-based analysis, depicting binding sites of transcription factors over-represented within 1 kB of loci whose DNA methylation was related to age, bladder cancer status, or both, grouped by similar structure or functional response. Binding sites for a forkhead-containing transcription factor and a transcription factor involved in immune modulation (GATA1) overlapped between loci associated with age and disease status. Loci with differential methylation strongly associated with age were nearby binding sites of a large number of transcription factors related to developmental processes, including homeobox-containing transcription factors, as well as factors involved in immune modulation and stress response. Oncogenic transcription factor binding sites as well as immune modulation and development-related transcription factor binding sites were exclusively over-represented near loci whose methylation was associated with bladder cancer.
DISCUSSION
This study represents a very novel, large-scale examination of the utility of DNA methylation in peripheral blood as a biomarker of bladder cancer risk and suggests that there are epigenetic alterations detectable in accessible, nondiseased tissue that reflect susceptibility to bladder tumorigenesis. It is important to note that we cannot definitively determine whether these altered profiles of DNA methylation are a response of the hematopoietic systems to the presence of the developing tumor or are extant before tumor development and in some way allow for or potentiate the growth of the tumor. At the same time, we have hypothesized that these profiles may represent a directed alteration, and that by looking at the genomic context of those loci whose differential methylation was associated with aging or bladder cancer, we may be able to better define how these pathologic processes are influencing methylation status.
Specifically, we examined the representation of transcription factor binding sites within 1 kB of the loci demonstrating differential methylation with age and with case status. Loci associated with aging and with disease demonstrated over-representation of specific transcription factors involved in immune modulation and proliferation. Specific to those loci associated with bladder cancer, we observed an over-representation of transcription factor binding sites for transcription factors that have been functionally characterized as oncogenes and those involved in lipid/sterol homeostasis, whereas specific to loci associated with aging were a number of transcription factor binding sites critical in developmental processes. The key role of immune modulation in both aging and carcinogenesis, and particularly bladder carcinogenesis,32 highlights how the detected methylation alterations may represent specific changes to the immune system that enable tumorigenesis. For instance, there is a growing literature on the role of regulatory T cells (known as suppressor T cells) (Treg) and their over-abundance in both the peripheral blood as well as in the target epithelial tissues of a developing tumor,33–35 and these methylation alterations may represent changes to the representation of specific lymphocyte subsets in peripheral blood as either a mediator or consequence of bladder tumorigenesis. In fact, recent work has demonstrated that Foxo1 and Foxo3 proteins are critical for the control of Treg cell differentiation and specifying Treg cell lineage.36 It is equally important to consider transcription factor binding sites that were not over-represented in our analyses; these include those involved in angiogenesis/vascular endothelial growth factor signaling and cellular interaction and communication. Taken together, these results are striking, as they suggest that methylation of DNA in the hematopoietic system associated with aging and bladder cancer may be associated with the presence or absence of transcription factor binding that is both specific to the two processes as well as overlapping.
The functional consequence of the differences in methylation between individuals with and without bladder tumors is unclear, although the nine genes identified as harboring signal CpGs that are most associated with bladder cancer represent a wide range of cellular processes. For example, BRD7 is an activator of the WNT signaling pathway, which plays a critical role in stem cell maintenance and a pathway whose alteration has been linked to bladder cancer.37 TBCA encodes a member of the multiprotein complex responsible for appropriate folding of the tubulin protein and may be involved in responding to cellular stress events leading to an unfolded protein response.38 COX7C is one member of the cytochrome c oxidase complex responsible for mitochondrial respiration,39 and changes in its expression have been observed in skin squamous cell carcinoma40 and in response to fluorouracil treatment.41 At a higher level, similar pathways were disrupted in both age-associated and cancer-associated methylation, including those related to organismal systems, cellular processes, human disease, and environmental information processing.
This work, in summary, suggests that there is untapped potential in the use of peripheral blood–based epigenetic profiling for bladder cancer risk prediction or early detection, as well as in understanding the complicated interplay of multiple systems in tumorigenesis. We have demonstrated, with high accuracy, the ability to distinguish bladder cancers from controls using a model containing the DNA methylation profile of nine loci, patient age, sex, smoking status, and family history (AUC = 0.76). Profiles of DNA methylation reflecting increased methylation of these nine loci were associated with a more than five-fold increased risk for bladder cancer compared with profiles with lesser extents of methylation.
The addition of GWAS-based SNPs to bladder cancer prediction models does not seem to significantly improve their performance as compared with models that include risk factors and demographics alone.42 Wu et al recently developed a risk modeling strategy for bladder cancer, including epidemiologic variables, as well as a phenotypic measure of mutagen sensitivity, and this model showed similar performance (AUC = 0.8) to our model and points to the need for including phenotypically relevant data in risk prediction strategies.43 Our use of phenotypically relevant DNA methylation profiles, though, may be more appealing, because measurement of DNA methylation is more amenable to the clinical setting than are those of mutagen sensitivity assays, which are time intensive and laborious, requiring lymphocyte culture and microscopic assessment after exposure to a test mutagen. Although our results are not at the level of accuracy necessary for immediate diagnostic utility, they do point, along with a small but growing number of other studies of other solid tumors,23–25 to the tremendous clinical potential of epigenetic profiling of peripheral-blood DNA. Confirmation of these findings in additional populations is warranted, as is expanded examination of the role of aging and environmental exposures on the production of disease-associated methylation profiles.
Supplementary Material
Glossary Terms
- DNA methylation:
Methylation of bases contained in the DNA double helix, resulting in a loss of gene function. Generally occurring on cytosine residues in the DNA, methylation is important in regulating cell growth and differentiation and has resulted in the testing of DNA methyltransferase inhibitors as anti-cancer agents and differentiation agents.
- Epigenetic:
The transfer of information from one cell to its descendants without the information's being encoded in the nucleotide sequence of the DNA. The methylation of the promoter to inactivate a gene is an example of an epigenetic change. Epigenetic inheritance is typically transmitted in dividing cells. Although rare, it is occasionally seen in traits being transmitted from one generation to another. Epigenetic variants can arise spontaneously and just as spontaneously revert.
- Epigenome:
The overall epigenetic state of a cell.
- Gene Set Enrichment Analysis (GSEA):
GSEA is a computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biologic states.
- KEGG:
Kyoto Encyclopedia of Genes and Genomes is an integrate bioinformatic database resource that is used as a reference knowledge base for biologic interpretation of large-scale data sets generated by sequencing and other high-throughput experimental technologies.
- Population-based:
Study in which the subjects are drawn from a defined population in a manner that is representative of the source population studied. Such a design can avoid bias arising from the selective factors that guide affected individuals to a particular medical facility, allowing for greater generalizability of the findings.
- Recursively partitioned mixture model (RPMM):
A likelihood-based hierarchical clustering procedure that produces classification solutions similar to those of conventional mixture models in a computationally efficient manner and allows for precise inference regarding potential covariates.
- Regulatory T cells (known as suppressor T cells):
are a specialized subpopulation of T cells that act to suppress activation of the immune system and thereby maintain immune system homeostasis and tolerance to self-antigens. This is an important “self-check” built into the immune system so that responses do not go haywire. Regulatory T cells come in many forms, including those that express the CD8 transmembrane glycoprotein (CD8 T cells), those that express CD4, CD25 and Foxp3 (CD4CD25 regulatory T cells or “Tregs”) and other T cell types that have suppressive function. These cells are involved in closing down immune responses after they have successfully tackled invading organisms and also in keeping in check immune responses that may potentially attack one's own tissues (autoimmunity).
Footnotes
Supported by grants from the National Institutes of Health, National Cancer Institute (Grant No. R01CA121147), National Institute of Environmental Health Sciences (Grant No. P42ES007373), and Flight Attendants Medical Research Institute (Young Clinical Scientist Award No. 052341).
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Carmen J. Marsit, Margaret R. Karagas, E. Andres Houseman, Karl T. Kelsey
Financial support: Carmen J. Marsit, Margaret R. Karagas, Karl T. Kelsey
Provision of study materials or patients: Margaret R. Karagas
Collection and assembly of data: Carmen J. Marsit, Brock C. Christensen, Margaret R. Karagas, Karl T. Kelsey
Data analysis and interpretation: Carmen J. Marsit, Devin C. Koestler, Brock C. Christensen, Margaret R. Karagas, E. Andres Houseman, Karl T. Kelsey
Manuscript writing: Carmen J. Marsit, Devin C. Koestler, Brock C. Christensen, Margaret R. Karagas, E. Andres Houseman, Karl T. Kelsey
Final approval of manuscript: Carmen J. Marsit, Devin C. Koestler, Brock C. Christensen, Margaret R. Karagas, E. Andres Houseman, Karl T. Kelsey
REFERENCES
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM. The global burden of urinary bladder cancer. Scand J Urol Nephrol Suppl. 2008:12–20. doi: 10.1080/03008880802285032. [DOI] [PubMed] [Google Scholar]
- 3.Hayat MJ, Howlader N, Reichman ME, et al. Cancer statistics, trends, and multiple primary cancer analyses from the Surveillance, Epidemiology, and End Results (SEER) Program. Oncologist. 2007;12:20–37. doi: 10.1634/theoncologist.12-1-20. [DOI] [PubMed] [Google Scholar]
- 4.Botteman MF, Pashos CL, Redaelli A, et al. The health economics of bladder cancer: A comprehensive review of the published literature. Pharmacoeconomics. 2003;21:1315–1330. doi: 10.1007/BF03262330. [DOI] [PubMed] [Google Scholar]
- 5.Kogevinas M, Trichopoulos D. Urinary bladder cancer. In: Adami H, Hunter DJ, Trichopoulos D, editors. Textbook of Cancer Epidemiology. London, England: Oxford University Press; 2002. pp. 446–466. [Google Scholar]
- 6.Colt JS, Baris D, Stewart P, et al. Occupation and bladder cancer risk in a population-based case-control study in New Hampshire. Cancer Causes Control. 2004;15:759–769. doi: 10.1023/B:CACO.0000043426.28741.a2. [DOI] [PubMed] [Google Scholar]
- 7.National Research Council. Arsenic in Drinking Water. Washington, DC: National Academy Press; 2001. [Google Scholar]
- 8.Chen Y, Ahsan H. Cancer burden from arsenic in drinking water in Bangladesh. Am J Public Health. 2004;94:741–744. doi: 10.2105/ajph.94.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith AH, Lingas EO, Rahman M. Contamination of drinking-water by arsenic in Bangladesh: A public health emergency. Bull World Health Organ. 2000;78:1093–1103. [PMC free article] [PubMed] [Google Scholar]
- 10.Bates MN, Smith AH, Cantor KP. Case-control study of bladder cancer and arsenic in drinking water. Am J Epidemiol. 1995;141:523–530. doi: 10.1093/oxfordjournals.aje.a117467. [DOI] [PubMed] [Google Scholar]
- 11.Karagas MR, Tosteson TD, Morris JS, et al. Incidence of transitional cell carcinoma of the bladder and arsenic exposure in New Hampshire. Cancer Causes Control. 2004;15:465–472. doi: 10.1023/B:CACO.0000036452.55199.a3. [DOI] [PubMed] [Google Scholar]
- 12.King WD, Marrett LD. Case-control study of bladder cancer and chlorination by-products in treated water (Ontario, Canada) Cancer Causes Control. 1996;7:596–604. doi: 10.1007/BF00051702. [DOI] [PubMed] [Google Scholar]
- 13.Wilkens LR, Kadir MM, Kolonel LN, et al. Risk factors for lower urinary tract cancer: The role of total fluid consumption, nitrites and nitrosamines, and selected foods. Cancer Epidemiol Biomarkers Prev. 1996;5:161–166. [PubMed] [Google Scholar]
- 14.Mueller CM, Caporaso N, Greene MH. Familial and genetic risk of transitional cell carcinoma of the urinary tract. Urol Oncol. 2008;26:451–464. doi: 10.1016/j.urolonc.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engel LS, Taioli E, Pfeiffer R, et al. Pooled analysis and meta-analysis of glutathione S-transferase M1 and bladder cancer: A HuGE review. Am J Epidemiol. 2002;156:95–109. doi: 10.1093/aje/kwf018. [DOI] [PubMed] [Google Scholar]
- 16.Kiemeney LA, Grotenhuis AJ, Vermeulen SH, et al. Genome-wide association studies in bladder cancer: First results and potential relevance. Curr Opin Urol. 2009;19:540–546. doi: 10.1097/MOU.0b013e32832eb336. [DOI] [PubMed] [Google Scholar]
- 17.Wu X, Ye Y, Kiemeney LA, et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat Genet. 2009;41:991–995. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schork NJ, Murray SS, Frazer KA, et al. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 20.Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 21.Christensen BC, Houseman EA, Marsit CJ, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teschendorff AE, Menon U, Gentry-Maharaj A, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Widschwendter M, Apostolidou S, Raum E, et al. Epigenotyping in peripheral blood cell DNA and breast cancer risk: A proof of principle study. PLoS One. 2008;3:e2656. doi: 10.1371/journal.pone.0002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teschendorff AE, Menon U, Gentry-Maharaj A, et al. An epigenetic signature in peripheral blood predicts active ovarian cancer. PLoS One. 2009;4:e8274. doi: 10.1371/journal.pone.0008274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Aakre JA, Jiang R, et al. Methylation markers for small cell lung cancer in peripheral blood leukocyte DNA. J Thorac Oncol. 2010;5:778–785. doi: 10.1097/JTO.0b013e3181d6e0b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karagas MR, Tosteson TD, Blum J, et al. Design of an epidemiologic study of drinking water arsenic exposure and skin and bladder cancer risk in a U.S. population. Environ Health Perspect. 1998; 106(suppl 4):1047–1050. doi: 10.1289/ehp.98106s41047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace K, Kelsey KT, Schned A, et al. Selenium and risk of bladder cancer: A population-based case-control study. Cancer Prev Res (Phila Pa) 2009;2:70–73. doi: 10.1158/1940-6207.CAPR-08-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houseman EA, Christensen BC, Yeh RF, et al. Model-based clustering of DNA methylation array data: A recursive-partitioning algorithm for high-dimensional data arising as a mixture of beta distributions. BMC Bioinformatics. 2008;9:365. doi: 10.1186/1471-2105-9-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koestler DC, Marsit CJ, Christensen BC, et al. Semi-supervised recursively partitioned mixture models for identifying cancer subtypes. Bioinformatics. 2010;26:2578–2585. doi: 10.1093/bioinformatics/btq470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2:E108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dietrich K, Schned A, Fortuny J, et al. Glucocorticoid therapy and risk of bladder cancer. Br J Cancer. 2009;101:1316–1320. doi: 10.1038/sj.bjc.6605314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loskog A, Ninalga C, Paul-Wetterberg G, et al. Human bladder carcinoma is dominated by T-regulatory cells and Th1 inhibitory cytokines. J Urol. 2007;177:353–358. doi: 10.1016/j.juro.2006.08.078. [DOI] [PubMed] [Google Scholar]
- 34.Wang HY, Wang RF. Regulatory T cells and cancer. Curr Opin Immunol. 2007;19:217–223. doi: 10.1016/j.coi.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Ruffell B, DeNardo DG, Affara NI, et al. Lymphocytes in cancer development: Polarization towards pro-tumor immunity. Cytokine Growth Factor Rev. 2010;21:3–10. doi: 10.1016/j.cytogfr.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouyang W, Beckett O, Ma Q, et al. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol. 2010;11:618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- 37.Marsit CJ, Karagas MR, Andrew A, et al. Epigenetic inactivation of SFRP genes and TP53 alteration act jointly as markers of invasive bladder cancer. Cancer Res. 2005;65:7081–7085. doi: 10.1158/0008-5472.CAN-05-0267. [DOI] [PubMed] [Google Scholar]
- 38.Tian G, Huang Y, Rommelaere H, et al. Pathway leading to correctly folded beta-tubulin. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- 39.Hofmann S, Lichtner P, Schuffenhauer S, et al. Assignment of the human genes coding for cytochrome c oxidase subunits Va (COX5A), VIc (COX6C) and VIIc (COX7C) to chromosome bands 15q25, 8q22–>q23 and 5q14 and of three pseudogenes (COX5AP1, COX6CP1, COX7CP1) to 14q22, 16p12 and 13q14–>q21 by FISH and radiation hybrid mapping. Cytogenet Cell Genet. 1998;83:226–227. doi: 10.1159/000015185. [DOI] [PubMed] [Google Scholar]
- 40.Dang C, Gottschling M, Manning K, et al. Identification of dysregulated genes in cutaneous squamous cell carcinoma. Oncol Rep. 2006;16:513–519. [PubMed] [Google Scholar]
- 41.De Angelis PM, Svendsrud DH, Kravik KL, et al. Cellular response to 5-fluorouracil (5-FU) in 5-FU-resistant colon cancer cell lines during treatment and recovery. Mol Cancer. 2006;5:20. doi: 10.1186/1476-4598-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu X, Hildebrandt MA, Chang DW. Genome-wide association studies of bladder cancer risk: A field synopsis of progress and potential applications. Cancer Metastasis Rev. 2009;28:269–280. doi: 10.1007/s10555-009-9190-y. [DOI] [PubMed] [Google Scholar]
- 43.Wu X, Lin J, Grossman HB, et al. Projecting individualized probabilities of developing bladder cancer in white individuals. J Clin Oncol. 2007;25:4974–4981. doi: 10.1200/JCO.2007.10.7557. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.