Abstract
The incidence of blast attacks and resulting traumatic brain injuries has been on the rise in recent years. Primary blast is one of the mechanisms in which the blast wave can cause injury to the brain. The aim of this study was to investigate the effects of a single sub-lethal blast over pressure (BOP) exposure of either 48.9 kPa (7.1 psi) or 77.3 kPa (11.3 psi) to rodents in an open-field setting. Brain tissue from these rats was harvested for microarray and histopathological analyses. Gross histopathology of the brains showed that cortical neurons were “darkened” and shrunken with narrowed vasculature in the cerebral cortex day 1 after blast with signs of recovery at day 4 and day 7 after blast. TUNEL-positive cells were predominant in the white matter of the brain at day 1 after blast and double-labeling of brain tissue showed that these DNA-damaged cells were both oligodendrocytes and astrocytes but were mainly not apoptotic due to the low caspase-3 immunopositivity. There was also an increase in amyloid precursor protein immunoreactive cells in the white matter which suggests acute axonal damage. In contrast, Iba-1 staining for macrophages or microglia was not different from control post-blast. Blast exposure altered the expression of over 5786 genes in the brain which occurred mostly at day 1 and day 4 post-blast. These genes were narrowed down to 10 overlapping genes after time-course evaluation and functional analyses. These genes pointed toward signs of repair at day 4 and day 7 post-blast. Our findings suggest that the BOP levels in the study resulted in mild cellular injury to the brain as evidenced by acute neuronal, cerebrovascular, and white matter perturbations that showed signs of resolution. It is unclear whether these perturbations exist at a milder level or normalize completely and will need more investigation. Specific changes in gene expression may be further evaluated to understand the mechanism of blast-induced neurotrauma.
Keywords: primary blast injury, central nervous system, histopathology, immunohistochemistry, gene expression
Introduction
Blast attacks involving weapons such as roadside bombs, grenades, and improvised explosive devices (IEDs) are an increasingly common feature of terrorist attacks, with as many as 1513 such attacks recorded in the period of January to November 2007, affecting both civilian and military populations and resulting in over 16,000 casualties (Lawson Terrorism Information Centre, 2009). In particular, blast-induced neurotrauma (BINT) is an increasing problem for which mild traumatic brain injury (MTBI) forms the majority of these injuries (Ling et al., 2009; Cernak and Noble-Haeusslein, 2010). Despite the pressing urgency for accurate and effective diagnostic, prognostic, and therapeutic approaches to blast injuries, there remain significant gaps in our knowledge of this condition (Kochanek et al., 2009).
Primary neurotrauma occurs when the insult delivers a direct blow to the head which may be penetrating or non-penetrating (closed head). In a blast injury, primary injury is a result of the directs effects of the blast wave to the head compared to other forms of blast injuries such as secondary (e.g., victim is hit on head by an object propelled by the blast wave) and tertiary (e.g., victim is flung by the blast wave against an object and injures his head) injuries. The most commonly assessed blast wave parameter for primary blast injury is usually the peak blast over pressure (BOP), duration of the positive phase and impulse. The effects of primary blast injury on air-containing organs such as the lungs have been widely investigated and characterized (Kirkman and Watts, 2011). Blast-induced pulmonary injury thresholds have also been elucidated and refined (Bowen et al., 1968; Rafaels et al., 2010). Advancement in body armor material and protection has been able to mitigate in part, the vulnerability of pulmonary injuries to blast (Phillips et al., 1988) though not totally. Together with improved efficiencies in medical evacuations and advances in medical care which contribute to increased survival rate, incidences of BINT are on the rise in modern warfare.
Given the prevalence of BINT, the mechanism of primary blast injury to the central nervous system (CNS) is less well characterized and especially so for blast-induced MTBI. To date, most primary blast injury rodent CNS research has focused on peak BOPs > 110 kPa. However, it has been reported that BOPs > 110 kPa can also cause concomitant pulmonary injury in animals with high incidence of mortality (Bauman et al., 1997; Gorbunov et al., 2004; Chavko et al., 2006; Long et al., 2009). Hence, we were particularly interested in the effects of low-intensity blast on the brain at peak BOPS < 110 kPa without causing overt pulmonary damage and mortality.
Previous studies investigating BINT in rodents have reported a wide spectrum of perturbations post-blast that encompass cerebrovascular changes, white matter damage, neuronal changes in the hippocampus, oxidative stress, and increased blood–brain barrier permeability (Bauman et al., 1997; Cernak et al., 2001a,b; Long et al., 2009; Cernak, 2010; Readnower et al., 2010; Risling et al., 2011). Recent literature has also pointed toward the presence of cerebral inflammation that could be mediated by systemic inflammation due to the CNS effects of the primary blast wave through the unprotected torso (Cernak, 2010). Hence, in this low level blast study, we aimed to profile the acute changes post-blast especially with regards to regions vulnerable to apoptotic cell death and inflammation through the activation of microglial cells which are the major inflammatory cells in the CNS.
Furthermore, we also sought to profile changes in gene expression post-blast for the identification of broad functional changes through clustering and to provide a platform for biomarker discovery. Biomarkers should be definitive indicators of pathogenic processes (Biomarkers Definitions Working Group, 2001) which are sorely lacking for MTBI for which better experimental designs into underlying molecular mechanisms are required (Svetlov et al., 2009). A proteomics approach to identifying relevant molecules has previously been suggested (Agoston et al., 2009). We present here, a microarray technique that can be applied to low level primary blast research and also venture to provide a conceptual model of an alternative and complementary genomics-based approach.
Materials and Methods
Animals and blast exposure
Animal experiments were approved by the DSO Institutional Animal Care and Use Committee (DSO IACUC). Male Sprague-Dawley rats (250–350 g) were used for this study. Rats were anesthetized prior to blast exposure with an intraperitoneal injection of 75 mg/kg ketamine and 10 mg/kg xylazine. The animals were then secured with Velcro straps in metal cages that were anchored to the ground at the blast site. The source of BOP was 120 kg of 2,4,6-trinitrotoluene (TNT). Blast sensors (seven side-on pressure gages and three stagnation pressure gages) were used to monitor intensity and duration of BOP exposure during test and actual blast trials. Animals were placed at either 24 or 30 m away from the TNT source and were exposed to different sub-lethal BOP intensities (i.e., low, high). Six to eight animals were strapped loosely using Velcro to a metal mesh cage at the specified distances and doused with water to minimize dehydration and singeing of fur. A 0.4 m × 0.4 m concrete block was placed between the animals and the explosive source at a distance of 1.5 m from the animals. This block served as a shield against debris from the explosion, thus protecting animals against secondary blast injuries due to the projectiles. A schematic of the blast set-up is given in Figure 1A. Preliminary trials and simulations revealed no influence of the block on the blast wave at the position of the animals (data not shown). Control animals were transported to the blast site then anesthetized as with the blast-exposed animals, but were not exposed to the actual blast. After blast exposure, the animals were returned to the animal holding facility and allowed to recover from the effects of anesthesia. Access to food and drinks was ad libitum. The animals were sacrificed at day 1, day 4, and day 7 after the blast.
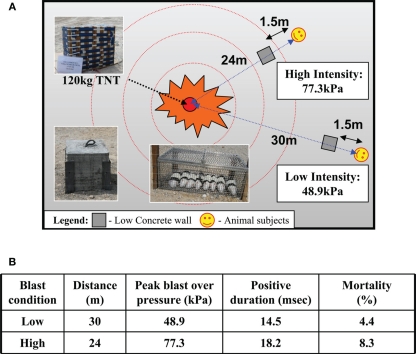
Figure 1.
(A) Schematic of blast set-up for six to eight rodents placed in a mesh metal cage at 24 m (high intensity) and 30 m (low intensity) from blast source (120 kg TNT) shielded from debris with a concrete block (0.4 m × 0.4 m) at 1.5 m from the animals. (B) Actual blast parameters: pressure (kPa) and duration (milliseconds) and mortality (%) at low and high blast exposure.
Histopathology and immunohistochemistry
By method of transcardial perfusion, the animals were perfused with Ringer’s solution until the liver and lungs were cleared of blood followed by 10% buffered paraformaldehyde. The brains were harvested and post-fixed in 10% buffered formalin. The brains were then dehydrated in an ascending series of alcohol, cleared with xylene, and then embedded in paraffin wax. Paraffin sections of 4 μm thickness were then cut and microwaved in citrate buffer for antigen retrieval and blocked with peroxidase blocking reagent (S2023, DAKO UK Ltd, UK). Brain and lung sections were stained for routine histology using hematoxylin and eosin (H&E) for general morphology analysis. For apoptosis staining, brain sections were stained according to the protocol provided in the ApopTag® Peroxidase In Situ Apoptosis Detection Kit (S7100, Chemicon International, Inc., MA, USA). For the preparation of double-labeled brain sections, a second antibody of rabbit anti-glial fibrillary acidic protein (GFAP) (AB5804, Chemicon International, Inc., MA, USA) diluted 1:1500 in PBS, biotinylated Ricinus communis Agglutinin I (RCA120) anti-lectin (B-1085, Vector Laboratories, Inc., CA, USA) diluted 1:1000 in PBS or rabbit polyclonal anti-myelin basic protein (MBP; AB980, Chemicon International, Inc., MA, USA) diluted 1:200 in PBS was used to detect GFAP, lectin, and MBP respectively. For immunohistochemistry, brain sections were also incubated with rabbit polyclonal anti-caspase-3 (#RB-1197-P, Thermo Fisher Scientific Inc., USA) diluted 1:100 in PBS; rabbit anti-ionized calcium binding adaptor molecule-1 (Iba-1; #019-19741, Wako Pure Chemical) diluted 1:500 in PBS; and rabbit polyclonal amyloid β precursor protein (APP; AB17467, Abcam) diluted 1:100 in PBS; for detection of caspase-3, Iba-1, and APP respectively. Subsequent antibody detection was carried out using either anti-mouse or anti-rabbit IgG (Envision + system-HRP, DAKO UK Ltd, UK) except for lectin which was carried out using horseradish peroxidase streptavidin (SA-5004, Vector Laboratories). All samples were then visualized using 3,3′-diaminobenzidine (DAB) and examined under a light microscope (Olympus, Japan). A cell count of at least three sections at 20 × magnification of TUNEL, Iba-1, and APP positive cells in the white matter was carried out and results are expressed as mean ± standard error of the mean (SEM). Statistical comparison between groups was performed by one-way ANOVA with post hoc Tukey’s HSD test. Significance was accepted at p < 0.05.
Microarray
Brain tissue from animals exposed to the lower BOP were harvested, quick frozen in lqN2 and stored at −80°C for subsequent microarray analyses. RNA was isolated using standard Trizol-based RNA extraction methods. The RNA quality was then determined based on RNA integrity number (RIN) and an electropherogram, both of which were analyzed using the Agilent 2100 Bioanalyzer platform (Agilent Technologies). Only samples with RIN greater than 6 were used (Fleige and Pfaffl, 2006). RNA samples were amplified and labeled with Cy3, hybridized to Agilent Whole Rat Genome Oligo Microarrays 4x44k, and analyzed using a microarray scanner system. All procedures were carried out in duplicates using commercial kits (Agilent Technologies) by a microarray service provider (Miltenyi Biotec GmbH, Germany). Microarray results were analyzed using R/Bioconductor and Partek Genomic Suite (Partek, MO, USA). Two independent analyses were conducted. The first set of analysis compared the expression levels of genes in blast-exposed animals vs. that in controls at each time-point. The second set of analysis investigated the changes in log ratio of blast-exposed vs. control animals [log (blast/control)] over time [e.g., log (blast/control)day 4 vs. log (blast/control)day 1, log (blast/control)day 7 vs. log (blast/control)day 1]. The overall type I error was taken at 0.01, and p-values were corrected for multiple testings using false discovery rates.
Results
Blast exposure and survival
A total of 58 animals were used in this study, of which 11 were controls, 23 were exposed to BOP at 48.9 kPa (or 7.1 psi) and positive over pressure duration of 14.5 ms at 30 m from TNT source, and 24 were exposed to BOP at 77.3 kPa (or 11.3 psi) and positive over pressure duration of 18.2 ms at 24 m (Figure 1B). For the purposes of the current work, we shall refer to the blast exposure conditions employed in simplistic terms as either high (BOP = 77.3 kPa) or low (BOP = 48.9 kPa) intensity. Corresponding mortality rates for the groups (low, high) were 4.4 and 8.3% respectively (Figure 1B). All of these animals died within 30 min of blast exposure. Deaths in the blast groups revealed pulmonary hemorrhage post-mortem. Surviving rats were used for subsequent investigation.
Histology and immunohistochemistry of blast injuries
Lung gross histopathology
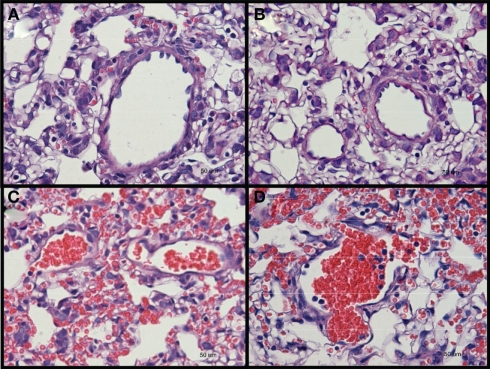
Rats were sacrificed at day 1, day 4, or day 7 after blast for investigation. Two tissues were examined, namely the brain and lungs. Other tissues were not examined as no external hemorrhage was observed. There was no apparent lung injury in both blast-exposed groups on day 1 post-blast. However, a few petechiae and ecchymoses were observed in the periphery of lung tissue upon harvesting at day 4 and day 7 after blast. H&E staining of the lung sections revealed alveolar lesions with accumulation of red blood corpuscles in lung alveolar space at day 4 and day 7 post high-intensity blast (Figure 2).
Figure 2.
Hematoxylin and eosin stained sections of lung tissues from (A) control, and low-intensity blast-exposed animals at (B) day 1 (C) day 4, and (D) day 7, after blast signs of hemorrhage, macrophage infiltration, and thickening of the alveolar septae, were observed at day 4 and day 7 after blast injury.
Brain and lung gross histopathology
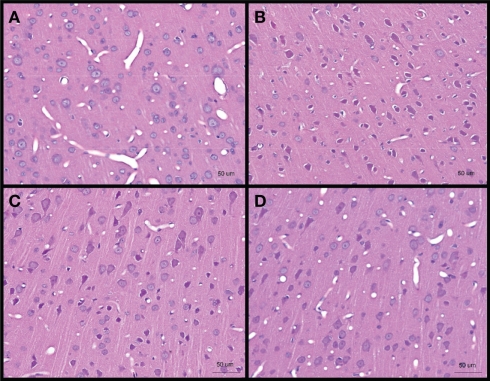
No obvious extra- and/or sub-dural hemorrhage was observed in the brains of all blast-exposed animals relative to the untreated controls H&E staining of brain sections from cerebral cortex showed darkened neurons (identified from the presence of projecting dendrites and polygonal shape of cell body) after high-intensity blast mostly at day 1 post-blast which appeared to abate at day 4 and day 7. These darkened neurons were also shrunken as evidenced by the presence of peri-somal spaces. Furthermore, the vasculature appeared to be narrowed at day 1 and day 4 post-blast compared to control (Figure 3).
Figure 3.
Hematoxylin and eosin stained sections of brain cerebral cortex from (A) control, and low-intensity blast-exposed animals at (B) day 1 (C) day 4, and (D) day 7 after blast. Darkened and shrunken neurons evidenced by the presence of peri-somal spaces at day 1 post-blast compared to control and in lesser quantities at day 4 and day 7 post-blast. Vasculature appears to be narrowed in day 1 and day 4 post-blast compared to control.
White matter damage
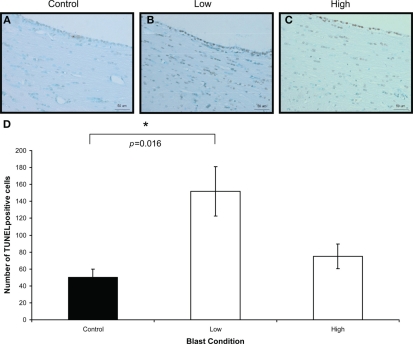
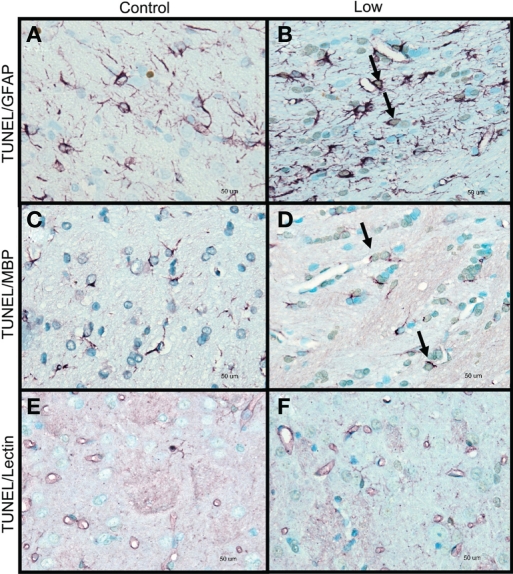
To confirm the presence of injury in the brains of rats exposed to high- and low-intensity BOP, TUNEL and caspase-3 staining was carried out to identify apoptotic cells in the brains. There were only a few cells positive for caspase-3 (not shown) compared to TUNEL-positive cells in the white matter. There was significantly more TUNEL-positive cells in the white matter of blast-exposed rats at high- and low-intensity relative to control on day 1 after blast (Figure 4). By day 7, however, there appeared to be no major difference between controls and blast-treated rats (data not shown). Double-staining for TUNEL and non-neuronal cells (GFAP, lectin, or MBP) revealed that the cells with DNA fragmentation were mainly oligodendrocytes and astrocytes, but not microglial cells (Figure 5). Iba-1 staining for CNS macrophages/microglia also showed no changes in microglia density between control and both blast-groups at day 1 post-blast (Figure 6). In addition to the presence of apoptotic astrocytes and oligodendrocytes, APP immunostaining was also significantly increased in both blast conditions compared to control on day 1 after blast (Figure 7).
Figure 4.
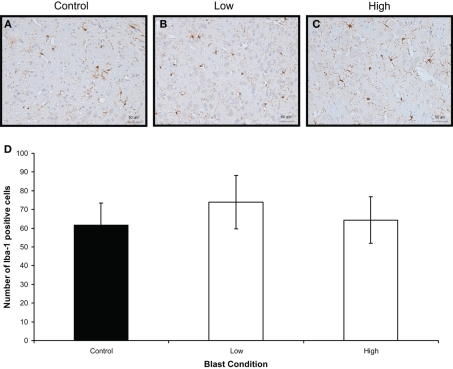
Iba-1 immunostaining (brown) of (A) control (B) low, and (C) high, blast exposure 1 day after blast. Scale bar = 50 μm. (D) Mean number of Iba-1 positive cells per field view in the corpus callosum of control, low (p = 0.26 vs. control) and high (p = 0.019 vs. control) blast-exposed animals 1 day after blast at × 20 magnification. No difference in Iba-1 positive cells was found between blast exposure groups (low and high) and control. *p < 0.05.
Figure 5.
Double-labeling of non-neuronal cells with TUNEL for apoptotic cells (light brown staining, indicated by small arrows) and glia markers (purple staining) for (A,B) apoptotic astrocytes, indicated by thick arrows (TUNEL and GFAP double-labeling). (C,D) Apoptotic oligodendrocytes, indicated by thick arrows (TUNEL and MBP double-labeling). (E,F) Lectin-positive microglia and TUNEL-labeled cells (TUNEL and lectin double-labeling). Control animals (A,C,E), low-intensity blast-exposed animals (B,D,F). Note lack of overlapping of labeling in apoptotic cells and microglia in low-intensity blast-exposed animals. Scale bar = 50 μm.
Figure 6.
Iba-1 immunostaining (brown) of (A) control (B) low, and (C) high, blast exposure 1 day after blast. Scale bar = 50 μm. (D) Mean number of Iba-1 positive cells per field view in the corpus callosum of control, low (p = 0.26 vs. control), and high (p = 0.019 vs. control) blast-exposed animals 1 day after blast at × 20 magnification. No difference in Iba-1 positive cells was found between blast exposure groups (low and high) and control. *p < 0.05.
Figure 7.
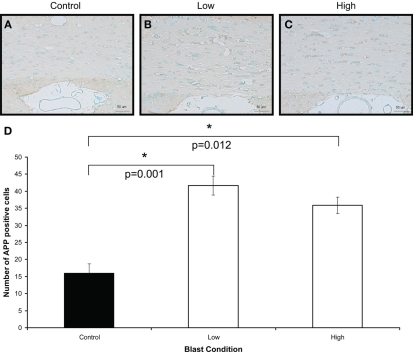
Amyloid β precursor protein immunostaining (brown) of (A) control (B) low, and (C) high, blast exposure 1 day after blast. Scale bar = 50 μm. (D) Mean number of APP positive cells per field view in the white matter of control, low (p = 0.26 vs. control) and high (p = 0.019 vs. control) blast-exposed animals 1 day after blast at × 20 magnification. Both low and high blast exposure animals had significantly higher APP+ cells than control. *p < 0.05.
Gene expression changes in mild traumatic brain injury
Microarray analyses of brain RNA samples from both control and rats exposed to low-intensity blast was carried out to determine gene expression changes. For the purpose of analysis, we hypothesized that functionally relevant genes are likely to include those whose expressions are significantly altered by blast exposure and/or show a significant time evolution after blast.
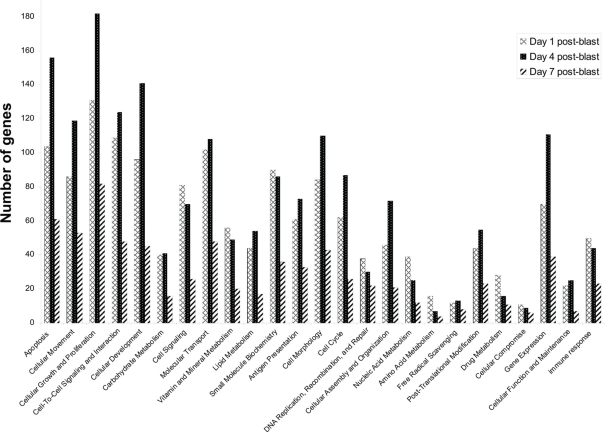
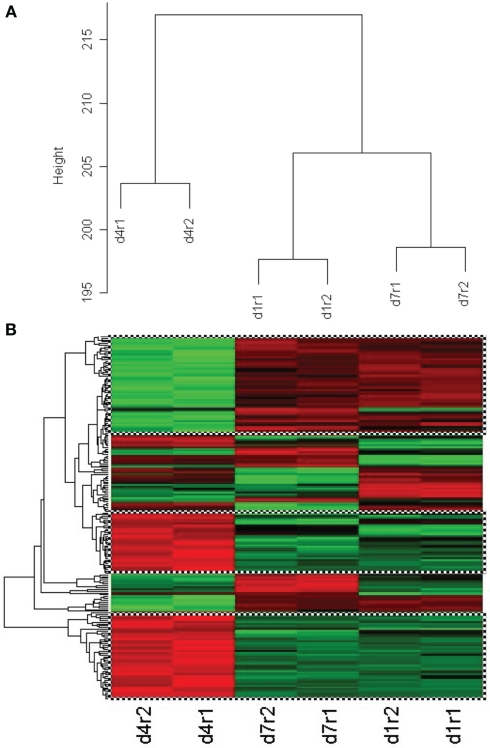
In our first set of analyses, we found 5786 probe sets which showed significant changes in the blast-exposed group relative to the controls at least one time-point. Most of these probe sets either had no confirmed identities or no known biological functions. Only 676 were established genes with well-defined functions (Table A1 in Appendix). We grouped these genes according to their functions and the results are shown in Figure 8. It appears that most changes took place at day 1 and day 4 after blast, with far fewer alterations observed at day 7. Clustering of the genes whose expressions varied most between arrays revealed high concordance between two replicates under the same condition which cluster together. In particular, day 1 and day 7 replicates clusters seemed similar while day 4 is different (Figure 9).
Figure 8.
Functional categorization of genes whose expression levels are altered by blast. Genes were categorized based on their biological functions. Probe sets with unknown functions were ignored in this analysis. A total of 1200 functionally characterized genes were identified in our microarray experiments using brain tissue from rats exposed to a BOP of 48.9 kPa. As each gene can have multiple functions, every gene may contribute more than once to the gene count shown here.
Figure 9.
(A) Cluster dendrogram of genes whose expression varied most from control between arrays for two replicates (r1, r2) of the same condition at day 4, day 1, and day 7 after low-intensity blast. Replicates are observed to cluster at the same level whilst clusters for day 4 appear different from day 1 and day 7. (B) Clustering of 203 genes that show similar behavior between replicates of day 4 (column 1), day 7 (column 2), and day 1 (column 3) after light intensity blast. Green color represents upregulation whilst red color represents downregulation from control. The topmost cluster (checked box) shows genes that are up-regulated at day 4 but down-regulated at day 1 and day 7, whilst the middle and bottom clusters (checked box) show the opposite trend.
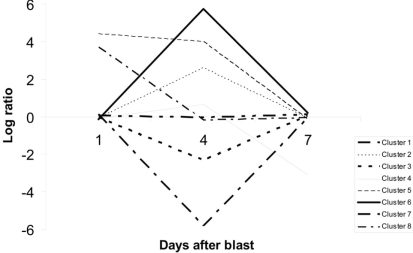
Our second set of analyses revealed 203 probe sets that showed significant time-course evolution, i.e., the log (blast/control)day 4 or day 7 was different relative to that at day 1. Based upon their time-evolution pattern, these 203 probe sets were grouped into eight clusters based on similar evolution (Figure 10). The biological functions of genes in each cluster, except clusters 5 and 8 which do not have genes with information, are given in Table A2 of Appendix. Out of the 203, only 34 are known genes with established functions (Table 1). Between our two sets of microarray analyses, there was an overlap of 10 genes (Table 2).
Figure 10.
Clustering of probe sets with significant time-course evolution. A total of 203 probe sets showing such evolution were identified, and grouped into eight clusters based on their time-evolution profiles. Probe sets in cluster 1 showed no major variation over time. They are consistently and weakly up-regulated or down-regulated across all time-points. Those in clusters 2 and 6 were up-regulated while those in clusters 3 and 7 were down-regulated at day 4. Probe sets in clusters 4, 5, and 8 were respectively down-regulated at day 7, up-regulated at day 1 and day 4, and up-regulated at day 1 only.
Table 1.
Genes whose expressions show significant time-course evolution after blast in the brains of rats exposed to a BOP of 48.9 kPa.
| AR | FCER1A | ITGB5 | PPIB | SH3GLB1 |
| ARHGAP4 | FLT1 | KIF11 | PRKACB | SLC40A1 |
| CETN1 | FOLR1 | KLHL10 | PRKCH | TFG |
| COQ6 | GADAY 1 | MMP11 | PTTG1 | TPM1 |
| CRYAB | GHR | NFKBIA | PYY | UCP1 |
| DNAJB11 | GTF2F2 | PARP1 | RALBP1 | VEGFA |
| F2RL3 | HMGA1 | PLG | RASA3 |
Table 2.
List of genes whose expression levels are significantly altered in blast-exposed animals and which also show a time-course evolution pattern between day 1, day 4, and day 7 after blast.
| FLT1 | GADAY 1 | KIF11 | PARP1 | PRKCH |
| FOLR1 | HMGA1 | NFKBIA | PRKACB | SLC40A1 |
Discussion
Blast-induced neurotrauma is the signature of the modern war (Elder et al., 2010). We were particularly interested in mild TBI as it accounts for over 77.8% of all TBI injuries sustained during combat (Defense and Veterans Brain Injury Center, 2010). In our open field blast test, we set out to investigate the effects of two relatively low BOP exposures of 77.3 and 48.9 kPa to cause mild BINT and to determine its effects on pulmonary injury in rodents with no body armor. The low BOP exposure was determined from the Bowen’s curve to determine if these BOP values could cause mild BINT without any overt pulmonary damage and also to minimize mortality in the animals. At the time of our study, there was only one report investigating the effects of peak BOP < 100 kPa on the effects in the rodent CNS (Saljo et al., 2009). Our findings showed the appearance of hemorrhagic lesions in the lung which is a feature of pulmonary blast injury even at the test levels of BOP of 77.3 and 48.9 kPa compared to previous studies in which lung injury was reported at higher BOP exposure of 118 kPa (Chavko et al., 2006; Gorbunov et al., 2006). The late appearance of lung petechiae and alveolar hemorrhage at day 4 and day 7 after blast suggests a delayed response as opposed to immediate (as early as 2 h) pulmonary injury that could occur at higher BOPs (Gorbunov et al., 2006). Recent evidence investigating a BOP model <110 kPa also demonstrated that a shockwave of 11.5 kPa resulted in no evidence of lung injury but was evident at 66 kPa (Park et al., 2010). Hence, it may be suggested that the BOP range of 48.9–77.3 kPa represents the threshold for blast-induced lung injury in the unprotected rodent. The use of improved protective body armor in combat situations has largely mitigated against pulmonary injury and mortality (Phillips et al., 1988). It is now generally accepted that the threshold for BINT is higher than that of pulmonary injury. Pulmonary blast injury has been reported to be due to the pressure changes at the tissue-density interface (DePalma et al., 2005). The Bowen’s curve for which the study’s sub-lethal BOP levels were chosen was based on this theory. However, other factors such as the viscoelasticity of the tissue (Stuhmiller, 1997) and internal spalling and implosion (Treadwell, 1989) may also contribute to pulmonary blast injury.
The BOP exposure to animals in our study resulted in the “darkening” or enhanced H&E staining of cortical neurons in the gray matter which may be due to the condensation of the neuronal cytoplasm which has also been previously reported to occur in a global cerebral ischemic condition (Kawai et al., 1992). These cortical changes is also consistent with our previous study investigating BOP of 20 kPa in rodents 1 day post-blast (Moochhala et al., 2004). Furthermore, the changes in vascular profiles post-blast suggest the occurrence of vasospasm which has also been reported as a feature of blast injury (Armonda et al., 2006). The alleviation in darkening at day 4 and day 7 post-blast and a rescue in vascular morphology at day 7 points to the existence of an acute transient ischemic cerebral environment that can recover with time after blast. Interestingly, cerebrovascular changes such as microvascular density and vasospasm have also been reported in studies in blast and impact TBI (Armonda et al., 2006; Park et al., 2010; Svetlov et al., 2010). However, it is unclear whether there is a complete recovery to the original state and whether the mild changes persist. Despite these cortical histopathological changes, TUNEL-staining was not observed in the white matter. This suggests that cortical gray matter and vasculature is affected by the blast wave in a differential manner from white matter.
Our study also showed that low BOP exposures at <110 kPa predominantly caused DNA fragmentation in the glial cells of the white matter with corresponding accumulation of APP probably due to axonal damage which is apparent at day 1 post-blast. The presence of white matter damage post-blast is becoming well-documented and the presence of this damage at low BOP levels suggest that primary injury from the shockwave can act to disrupt axonal transport and to cause cell death in oligodendrocytes and astrocytes which play important supportive functions in the white matter. This axonal pathology is further corroborated by findings of early increases in α-II spectrin, an axonal cytoskeletal protein, and sustained expression of NF200, an axonal neurofilament, in a mild BINT rodent model (Park et al., 2010).
Our study was only focused on the head-on exposure of the shockwave to the skull of the animal although it has been suggested that different orientations of the animal can have different pressure transmission, whether direct or reflected off the skull (Chavko et al., 2011), and the effects of different wave propagation on the CNS due to orientation remains to be investigated. Despite reports of blood–brain barrier permeability changes and inflammation post-blast (Bauman et al., 1997; Cernak et al., 2001a,b; Long et al., 2009; Cernak, 2010; Readnower et al., 2010; Risling et al., 2011), we did not find any changes in AQP-4 expression on S100B+ astrocytes (results not shown) as well as changes in Iba-1 immunoreactivity for microglia, the main inflammatory macrophages in the CNS, as well as systemic cytokines (results not shown). These negative findings may be explained by the lower BOPs used in our study or that the animals were not observed for longer periods of time post-blast.
Given that the lethal threshold is lowest for blast lung injury than other organ systems, no significant mortality was expected in our blast model used here. This was confirmed by our findings (4.4% low BOP, 8.3% high BOP). Although two different BOPs were investigated in this study, both lie on the lower limit of the BOP range tested across many experimental blast studies. This may explain the non-significant differences in mortality, histopathological, and immunohistochemical changes between the low and high BOP. However, the blast set-up employed in this study can be used to establish a blast pressure-dependent mortality or morbidity response curve by placing animals at varying distances from the TNT source and by varying the amount of TNT.
Besides the low BOP, the animals were also exposed to a long positive duration of 14.5 and 18.2 ms in the low and high intensity settings respectively due to the distance that the subjects were placed away from the TNT explosive as opposed to other models of blast injury using compressed air, helium, oxyhydrogen or cyclotrimethylenetrinitramine (Reneer et al., 2011). The duration of the overpressure is thought to be of much significance in causing damage (de Candole, 1967) as the length of this positive duration would also affect the impulse at which the animals are subjected to. This longer positive overpressure duration also reflects the increasing use of thermobaric and other novel explosives in the modern war (Rafaels et al., 2010) and could have different mechanisms from other models investigating short positive overpressure durations (Cooper and Jonsson, 1997). However, it is unclear whether the long positive duration in our study had any impact on CNS injury and is a component of the blast wave that will require a more thorough examination and comparison against other blast models of the same BOP but of different duration.
Separately, the blast set-up and exposure in our study also provides a platform for scaling up to other animal species and to allow comparison between species on BINT thresholds and injury presentation. In a separate study investigating effects of sub-lethal BOP on non-human primates (NHPs) in the same blast set-up, increases in TUNEL-positive cells and APP immunoreactivity in white matter, together with the darkening of neurons were also observed in NHPs exposed to 80 kPa BOP as in our rodent study (unpublished). Further investigations with different small and large animal species using the same blast set-up but with additional strain gages in the body and brains will provide useful information in how blast waves of the same pressure transmit differently through skull and brain tissue properties between species.
Our microarray studies focused on brain samples from animals exposed at the lower BOP of 48.9 kPa vs. the controls. We found 676 genes whose expression profiles were significantly altered by blast. A common feature of trauma to the CNS involves pronounced changes in the expressions of cell proliferation and apoptotic genes (Byrnes and Faden, 2007). Accordingly, we found the highest number of blast-affected genes to belong to these functional groups, suggesting that our model was fundamentally sound in approach. We also observed that the number of genes affected in all functional groups decreased from day 4 to day 7 post-blast. Based on our H&E and TUNEL observations, this corresponds to a period during which there was almost complete recovery of the brain from blast injury, suggesting that recovery from blast injury was associated with a return to baseline of the expression of most genes. However, we did not observe a similar correspondence from day 1 to day 4 post-blast, which suggests that the injury-gene expression association may be time-dependent and differ between the immediate (<day 4) and the short (day 4 to day 7) terms after blast. The difference in the expression levels at day compared to day 1 and day 7 points toward a quadratic expression profile post-blast, i.e., cluster of genes up-regulated or down-regulated at day 4 as opposed to day 1 and day 7 expression. The lack of significance may be due to the small sample size used and the result should be followed up with more extensive sampling and further analysis.
In this study, we also took a multi-pronged approach using a strategy based upon the hypothesis that any gene whose expression is significantly altered by blast exposure is more likely than any other random gene to be critically involved in blast. Likewise, genes whose expression patterns show significant time evolution following blast have higher probability than others to be functionally relevant. Note that genes in the two groups may overlap or be distinct. Based upon our strategy, it follows then that genes in the overlapping group are the more likely genes to influence clinical outcome in blast injuries. The identities of functionally relevant genes will be especially important in the design of novel therapeutics or treatment approaches in blast victims. In terms of diagnostics, however, there could be an additional level of approach besides identifying specific genes. This would involve studying the overall pattern of functional changes in gene expressions based upon the functional categorization of genes whose expressions are modulated following blast exposure. Thus, our model enables the identification of four potential markers for diagnostics and/or treatment design as summarized in Table 3.
Table 3.
Table of illustrative examples of how genes may be classified into non-target and target groups.
| Type of gene | Gene expression ratios | |||
|---|---|---|---|---|
| Day 1 | Day 4 | Day 7 | Linear/quadratic time trend | |
| Gene expression is not significantly altered at any time-point and shows no time-course evolution | 1.1 (p > 0.05) | 1.1 (p > 0.05) | 1.1 (p > 0.05) | p > 0.05 |
| Gene expression is significantly altered by blast in some or all time points, but no | 1.1 (p < 0.05) | 1.1 (p < 0.05) | 1.1 (p < 0.05) | p > 0.05 |
| significant time-course evolution | 1.1 (p < 0.05) | 1.1 (p > 0.05) | 1.1 (p > 0.05) | |
| Gene expression is not significantly altered by blast but overall show significant time-course evolution | 1.1 (p > 0.05) | 1.2 (p > 0.05) | 2 (p > 0.05) | p < 0.05 |
| Gene expression is significantly altered at least one time-point, and shows | 1.1 (p < 0.05) | 5 (p < 0.05) | 1.1 (p < 0.05) | p < 0.05 |
| time-course evolution (overlapping genes) | 1.1 (p > 0.05) | 5 (p < 0.05) | 1.1 (p > 0.05) | |
p-values represent the statistical significance of comparisons between blast-exposed and control samples at each time-point.
We separately found 34 genes to show a time-course evolution over time after blast exposure. Ten genes were common to both our analyses (i.e., blast exposure significantly modulated their expression profiles which also changed over time). It is unclear whether these 10 identified genes and/or their protein products critically affects injury outcome in blast victims and its reliability to form a consistent “fingerprint” of blast-induced mild TBI. Considering the 10 overlapping genes in which the involvement of some such as FLT1 (involved in cell proliferation and angiogenesis) and PARP1 (participates in DNA repair) could be said to be instinctive, that of others such as HMGA1 (commonly found in prostate tumors and thought to be involved in transformation) may be less so. It is possible that genes such as HMGA1 are also involved in injury repair which have yet to be defined. Further studies would be required to determine if this is so. However, the potential for genes and “fingerprints” identified here to be used as biomarkers and therapeutic targets in blast research cannot be denied.
Overall, we have presented a primary blast injury rodent model exposed to low BOP levels in an open-field setting. Pulmonary injury was mild and delayed whilst neuronal and non-neuronal changes were immediate at day 1 and was found to be alleviated at day 4 and day 7 suggesting the ability of the brain to recover from mild BINT on the histopathological level although it is not clear whether there is complete recovery. Acute CNS changes after low BOP exposure suggest that cortical cerebrovascular changes and white matter changes are key features of acute low level BINT. In the first, ischemia may be a resultant effect whilst predominant white matter damage suggests vulnerability to primary blast injury. Furthermore, the concomitant increases in gene expression at day 1 and day 4 suggests a time-dependent injury response and recovery period. Of the 676 genes that were significantly altered, a framework was derived to narrow these to 10 according to the time-course evolution and functional relevance. Some of these up-regulated functional genes such as FLT1 and PARP1 point toward repair after injury and may contribute to the recovery in the histopathological changes seen at day 4 and day 7. Future work will center upon the validation of our model. This would involve efforts to determine if the “fingerprints” identified here are consistent and reproducible across different animal models of blast injury and in different tissues from blast-exposed animals. Most importantly, a time-course profile of the relationship between changes in gene expression patterns, conditions of blast exposure (e.g., BOP), histopathological changes, and blast injury severity should also be evaluated and for a longer time post-blast to observe for sustained changes or the development of secondary pathobiology.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors express their gratitude to Leonard Heng, Karen Chong, Rick Tan, Chor Boon Ng from Defence Science and Technology Agency; Julie Yeo, Jian Wu and Melissa Teo from DSO National Laboratories.
Appendix
Table A1.
Genes whose expressions are significantly altered by blast in the brains of rats exposed to a BOP of 48.9 kPa.
| ABCA2 | CAPZA3 | DLK1 | GPX3 | KIF11 | OMP | RAMP2 | ST3GAL5 |
|---|---|---|---|---|---|---|---|
| ABCC3 | CASR | DNAJA3 | GRIN1 | KIF1C | ONECUT1 | RAP1B | STAG3 |
| ABCG2 | CATSPER2 | DNAJA4 | GRINL1A | KIF2C | OPRL1 | RAPGEF1 | STC1 |
| ABRA | CBARA1 | DNAJB4 | GRK1 | KIT | OPRM1 | RASDAY 1 | STK38 |
| ACCN2 | CBR1 | DNDAY 1 | GRM5 | KLF15 | ORC2L | RASSF2 | STX1A |
| ACHE | CCL2 | DNM1L | GRPR | KLF5 | OXT | RASSF5 | SUFU |
| ACSL1 | CCL3 | DNMT3B | GSPT1 | KLF6 | P2RX2 | RB1 | SULT1B1 |
| ACTB | CCL4 | DPP4 | GUCY1A3 | KNG1 | P2RY2 | RBM17 | SYNJ1 |
| ADAM10 | CCNB2 | DPYSL5 | GUSB | KRT20 | PA2G4 | RBP3 | SYT1 |
| ADAM9 | CCNDAY 1 | DR1 | GZMA | LHB | PAH | RCOR2 | TAAR1 |
| ADCY6 | CCNG1 | DRD2 | GZMB | LITAF | PAQR3 | REG3G | TAC1 |
| ADIPOR1 | CCR4 | DRDAY 4 | H1F0 | LPIN1 | PARG | REST | TACC2 |
| ADM | CCR5 | DSP | HAVCR2 | LPL | PARP1 | RFX3 | TAF5L |
| ADORA2A | CCT6A | DUSP5 | HCLS1 | LRPAP1 | PARVA | RGS10 | TAPBP |
| ADRA2B | CDAY 14 | DUSP9 | HCRT | LRRK2 | PAX4 | RGS19 | TAX1BP1 |
| AES | CD2 | EBF1 | HERPUDAY 1 | LTB4R | PCDHAC2 | RHOA | TBCE |
| AGRP | CD2AP | ECEL1 | HES3 | LTBP1 | PCNA | RHOH | TBX3 |
| AGT | CD320 | ECM1 | HINT1 | LYPD3 | PDCDAY 4 | RIPK2 | TCEA1 |
| AK2 | CD36 | EDNRA | HIST1H1T | LZTS1 | PDCD6IP | RLN1 | TCIRG1 |
| AK3L1 | CD38 | EGF | HIVEP1 | MAEA | PDCL | RNF10 | TDG |
| AKAP13 | CD3G | EGFR | HMGA1 | MAL | PDE4B | RNF14 | TEAD2 |
| AKAP4 | CDAY 44 | EGR1 | HMMR | MAP1B | PDHA2 | ROBO4 | TERF1 |
| ALB | CDC25A | EGR2 | HNF4A | MAP3K10 | PDLIM2 | RPN2 | TGFB1 |
| ALDH2 | CDC25B | EHDAY 1 | HOMER1 | MAPK1 | PDYN | RPS15A | TGFB1I1 |
| ALDOA | CDC2L5 | EIF4B | HOMER2 | MAPK14 | PDZK1IP1 | RRM1 | TGM1 |
| ALG5 | CDC42BPB | ELA2 | HOXA5 | MAPK8 | PELO | RTKN | TH |
| AMDAY 1 | CDCA2 | ENAH | HOXC6 | MAPK9 | PELP1 | S100A8 | THAP1 |
| AMHR2 | CDH16 | ENTPDAY 1 | HP | MAPRE1 | PEMT | SAA4 | THBD |
| ANTXR1 | CDH22 | EP400 | HPS1 | MAT1A | PHGDH | SATB1 | THEM4 |
| ANXA2 | CDK10 | ERBB2 | HPSE | MATK | PIK3C3 | SBDS | THPO |
| AOC3 | CDKN1A | ESM1 | HSDAY 11B2 | MATR3 | PIR | SCAMP2 | TIAM1 |
| APBA1 | CDKN1B | ETS2 | HSPA1A | MBL2 | PKDAY 1 | SCARB1 | TK1 |
| APCS | CDKN1C | ETV6 | HSPA8 | MCF2L | PKNOX1 | SCN10A | TLE4 |
| APH1A | CDX2 | F12 | HSPBP1 | MFN2 | PLA2G4A | SCN4B | TMOD2 |
| APH1B | CEBPE | F2R | HSPDAY 1 | MGAT2 | PLAT | SCN9A | TMOD3 |
| APLN | CES1 | F2RL2 | HTR1B | MINA | PLCG1 | SCNN1B | TNF |
| APOC2 | CFH | F5 | HTR1D | MLH1 | PLD2 | SEMA3D | TNFRSF1A |
| AQP4 | CFTR | FABP7 | HTR2A | MLL | PLEKHF1 | SENP2 | TNNI2 |
| AREG | CGA | FADS1 | HTR2B | MST1 | PLXNA3 | SERINC3 | TNNT1 |
| ARF6 | CHI3L1 | FAIM | IBSP | MSX2 | PMCH | SERPINB2 | TNP2 |
| ARHGEF7 | CHMP5 | FAU | ICAM1 | MTA1 | PNLIP | SERPINI1 | TNR |
| ARL11 | CHRM4 | FCGR3A | ID2 | MTPN | POLA1 | SFRP2 | TOB2 |
| ARL2BP | CHRNA10 | FGF13 | ID3 | MXD3 | PON2 | SFRS2 | TOP2A |
| ASAH2 | CIT | FGF4 | IFNG | MXI1 | POU2F1 | SFTPC | TPH1 |
| ATF3 | CITED2 | FGFR1 | IGF1R | MYCL1 | POU2F2 | SGTB | TPM3 |
| ATG7 | CKAP5 | FGG | IGFBP1 | MYO5A | POU3F1 | SH3BP5 | TPST1 |
| ATP1A1 | CLDN11 | FGL2 | IGFBP4 | NAP1L1 | PPAP2C | SHC1 | TPT1 |
| ATP2A3 | CLU | FGR | IGHMBP2 | NCR3 | PPEF1 | SHH | TRADD |
| ATP2C2 | CNR1 | FHIT | IKBKG | NDFIP1 | PPIL2 | SHMT1 | TRIB3 |
| ATP6V1C1 | CNTF | FKRP | IL12RB2 | NDN | PPM1J | SIP1 | TRIM32 |
| ATP6V1F | CNTN3 | FLT1 | IL13 | NDRG1 | PPP1R1B | SIX3 | TRIM63 |
| ATP7B | CNTN4 | FN1 | IL13RA1 | NEFL | PPP2R2B | SLC16A2 | TRPC3 |
| AVP | COL16A1 | FOLR1 | IL13RA2 | NEO1 | PPP2R3A | SLC16A4 | TRPM6 |
| AZGP1 | COL2A1 | FOXM1 | IL18RAP | NEU1 | PPP2R5B | SLC17A3 | TRPM7 |
| AZI2 | COMT | FSHR | IL1A | NEU3 | PPYR1 | SLC18A2 | TRPV1 |
| B4GALNT1 | CORO1B | FTH1 | IL1B | NEUROG3 | PRDX5 | SLC1A3 | TTN |
| BACE1 | CREB1 | FUBP1 | IL22RA2 | NFIA | PRIM1 | SLC22A2 | TTR |
| BAD | CRKRS | FUT4 | IL4 | NFKBIA | PRKAB1 | SLC24A3 | TUBB2C |
| BAK1 | CRTC2 | FXYD5 | IL8RB | NFKBIB | PRKACB | SLC25A10 | TWIST1 |
| BCAN | CRY1 | GABBR1 | IMPACT | NGFR | PRKCD | SLC25A14 | UBC |
| BCAP31 | CSDA | GADAY 1 | IMPDH2 | NIDAY 1 | PRKCH | SLC25A27 | UBE2D2 |
| BCL2 | CSF3 | GADDAY 45GIP1 | INHBB | NINJ2 | PRKCZ | SLC2A4 | UBE2D3 |
| BCL2L10 | CSNK1A1 | GALNS | INSIG2 | NKX3-1 | PRLR | SLC34A1 | UBTF |
| BDKRB2 | CSNK1G1 | GAP43 | INSRR | NLGN3 | PRM1 | SLC36A2 | UCHL1 |
| BID | CSPG4 | GATA1 | IPPK | NMT1 | PRPF19 | SLC37A4 | UCP2 |
| BMP4 | CSPG5 | GATA6 | IRS1 | NNT | PRPF8 | SLC40A1 | UGCG |
| BNIP3 | CTH | GATAD2A | ITGA1 | NOS3 | PSMB2 | SLC6A3 | USH2A |
| BTRC | CTNNB1 | GFAP | ITGA2 | NOVA1 | PSMD2 | SLC6A4 | USP14 |
| BYSL | CTSB | GGCX | ITGA4 | NPDC1 | PSMDAY 4 | SLC6A5 | VNN1 |
| C3AR1 | CUGBP1 | GHRL | ITGA5 | NPEPPS | PSMD9 | SLC7A2 | VPS4B |
| C9 | CXCL11 | GLI1 | ITGAL | NPFF | PTGDS | SLC7A5 | VTCN1 |
| CA3 | CYB5R4 | GLIPR1 | ITGB2 | NPR1 | PTGER4 | SLC8A1 | WEE1 |
| CABP1 | CYP1A1 | GLP2R | ITPKB | NPY | PTGES | SLC9A1 | WNT2 |
| CACNA1B | CYP2E1 | GLTSCR2 | JAG1 | NR1D2 | PTGS2 | SMAD3 | XRCC1 |
| CACNB2 | CYR61 | GNA14 | JAM2 | NR1I2 | PTHLH | SMO | YBX1 |
| CALCA | DAB2 | GNAL | KCNA1 | NR2C2 | PTK2 | SNAPC2 | ZBTB10 |
| CALCRL | DBH | GNB2 | KCNA6 | NR2F2 | PTMS | SNCA | ZDHHC2 |
| CALDAY 1 | DCC | GNB5 | KCNC1 | NR4A3 | PTPN2 | SOD3 | ZHX2 |
| CAMK2A | DCLK1 | GNG2 | KCNC3 | NR5A2 | PTPN3 | SP2 | ZIC1 |
| CAMK4 | DDR1 | GNG4 | KCNH1 | NRTN | PTPRV | SPG7 | ZMYNDAY 11 |
| CAMKK1 | DEAF1 | GNG5 | KCNJ11 | NRXN2 | PTS | SPP1 | ZP2 |
| CAMKK2 | DGAT1 | GNRH1 | KCNMA1 | NTRK1 | QPRT | SQSTM1 | |
| CAMP | DGAT2 | GPC1 | KCNMB1 | NTRK2 | RABGGTA | SREBF2 | |
| CANT1 | DKC1 | GPR44 | KCNN3 | NUP98 | RAG1 | SST | |
| CAPN2 | DLC1 | GPS1 | KIDINS220 | OMG | RALGDS | ST18 |
Table A2.
Biological functions of genes in each cluster (clusters 1–4, 6, and 7; except clusters 5 and 8 which do not have genes with information).
| Category | Molecules |
|---|---|
| CLUSTER 1 FUNCTIONS | |
| Amino acid metabolism | GADAY 1 |
| Antigen presentation | PLG, NFKBIA, VEGFA, ITGB5 |
| Carbohydrate metabolism | GPI, F2RL3 |
| Cell cycle | PLG, CETN1, NFKBIA, VEGFA, KIF11, CLIP1, GPI, CRYAB, GHR |
| Apoptosis | LPAR1, NFKBIA, VEGFA |
| Cell morphology | PLG, LPAR1, NFKBIA, VEGFA, CLIP1, ITGB5, GPI, GHR |
| Cell signaling | PLG, LPAR1, VEGFA, FCER1A, PYY, ARHGAP4, PPIB, F2RL3 |
| Cell-to-cell signaling and interaction | PLG, VEGFA, PRKACB, PYY, ITGB5, GPI, GHR |
| Cellular assembly and organization | CETN1, LPAR1, SH3GLB1, VEGFA, FCER1A, ITGB5, F2RL3, CRYAB, PLG, KIF11, CLIP1, ARHGAP4, GPI, GHR |
| Cellular compromise | PLG, VEGFA, KIF11, GPI, GHR |
| Cellular development | PLG, NFKBIA, VEGFA, FCER1A, GHR |
| Cellular function and maintenance | PLG, NFKBIA, KIF11, PYY, ITGB5, ARHGAP4, GPI, F2RL3, GHR |
| Cellular growth and proliferation | LPAR1, VEGFA, C19ORF10, ITGB5, PYY,CRYAB, PLG, TFG, NFKBIA, KIF11, GADAY 1, GPI, WNK1, GHR, SKAP2 |
| Cellular movement | PLG, LPAR1, NFKBIA, VEGFA, PYY, ITGB5, GADAY 1, GPI, PPIB |
| DNA replication, recombination, and repair | VEGFA, PPIB, WNK1, GHR |
| Drug metabolism | VEGFA, PPIB, GHR |
| Energy production | PLG |
| Gene expression | NFKBIA, VEGFA, GHR |
| Lipid metabolism | NFKBIA, VEGFA, PYY, GHR |
| Molecular transport | PLG, LPAR1, NFKBIA, VEGFA, FCER1A, PYY, GPI, PPIB,F2RL3, GHR |
| Nucleic acid metabolism | PLG, VEGFA, PPIB, WNK1, GHR |
| Post-translational modification | VEGFA, GADAY 1 |
| Protein folding | VEGFA |
| Protein synthesis | PLG, VEGFA, PYY |
| Protein trafficking | NFKBIA |
| Small molecule biochemistry | PLG, NFKBIA, VEGFA, PYY, GADAY 1, GPI, PPIB, F2RL3, WNK1, GHR |
| Vitamin and mineral metabolism | PLG, LPAR1, VEGFA, FCER1A, PYY, PPIB, F2RL3 |
| Immune response | PLG, VEGFA, FCER1A, ITGB5 |
| CLUSTER 2 FUNCTIONS | |
| Amino acid metabolism | HIPK2, PRKCH, PPM1D |
| Antigen presentation | PPM1D |
| Carbohydrate metabolism | UCP1 |
| Cell cycle | HIPK2, PRKCH, PPM1D |
| Apoptosis | HIPK2 |
| Cell morphology | PRKCH, UCP1 |
| Cell signaling | HIPK2 |
| Cellular assembly and organization | HIPK2 |
| Cellular compromise | HIPK2, UCP1 |
| Cellular development | HIPK2, PRKCH |
| Cellular function and maintenance | UCP1, PPM1D |
| Cellular growth and proliferation | HIPK2, PRKCH, UCP1 |
| DNA replication, recombination, and repair | PRKCH, UCP1 |
| Drug metabolism | UCP1 |
| Energy production | UCP1 |
| Free radical scavenging | UCP1 |
| Gene expression | PRKCH, UCP1 |
| Lipid metabolism | UCP1 |
| Molecular transport | PRKCH, UCP1 |
| Nucleic acid metabolism | UCP1 |
| Post-translational modification | HIPK2, PRKCH, PPM1D |
| Small molecule biochemistry | HIPK2, PRKCH, UCP1, PPM1D |
| Immnue response | PPM1D |
| CLUSTER 3 FUNCTIONS | |
| Amino acid metabolism | PARP1 |
| Antigen presentation | FLT1, IRF8 |
| Carbohydrate metabolism | PARP1, FLT1, RALBP1 |
| Cell cycle | DDB1, PARP1, FLT1, TPM1, HMGA1 |
| Apoptosis | PARP1, NDRG1, FLT1, RALBP1, TPM1, COQ6, HMGA1, IRF8 |
| Cell morphology | FLT1, CDAY 151, RALBP1, TPM1, HMGA1 |
| Cell signaling | FLT1 |
| Cell-to-cell signaling and interaction | FLT1, CDAY 151, IRF8 |
| Cellular assembly and organization | PARP1, FLT1, TPM1 |
| Cellular compromise | KLHL10, PARP1, NDRG1, FLT1, TPM1, HMGA1 |
| Cellular development | KLHL10, PARP1, NDRG1, FLT1, VSX2, CDAY 151, RALBP1, TPM1, HMGA1, RASA3, IRF8 |
| Cellular function and maintenance | PARP1, FLT1, TPM1, IRF8 |
| Cellular growth and proliferation | NDRG1, FLT1, VSX2, RALBP1, TPM1, HMGA1, IRF8 |
| Cellular movement | PARP1, FLT1, CDAY 151, RALBP1, TPM1 |
| Cellular response to therapeutics | PARP1 |
| DNA replication, recombination, and repair | DDB1, PARP1, HMGA1 |
| Drug metabolism | PARP1, RALBP1 |
| Energy production | PARP1 |
| Gene expression | DDB1, PARP1, GTF2F2, VSX2, RALBP1, HMGA1, IRF8 |
| Lipid metabolism | FDPS, FLT1 |
| Molecular transport | PARP1, RALBP1 |
| Nucleic acid metabolism | DDB1, PARP1, RALBP1 |
| Post-translational modification | PARP1 |
| RNA post-transcriptional modification | PARP1 |
| Small molecule biochemistry | DDB1, FDPS, PARP1, FLT1, RALBP1 |
| Immune response | FLT1, IRF8 |
| CLUSTER 4 FUNCTIONS | |
| Amino acid metabolism | FOLR1 |
| Antigen presentation | MMP11 |
| Carbohydrate metabolism | INPP5K |
| Apoptosis | MMP11 |
| Cellular assembly and organization | INPP5K |
| Cellular function and maintenance | FOLR1, SLC40A1 |
| Cellular growth and proliferation | MMP11, FOLR1 |
| DNA replication, recombination, and repair | FOLR1 |
| Drug metabolism | FOLR1 |
| Lipid metabolism | INPP5K |
| Molecular transport | INPP5K, FOLR1, SLC40A1 |
| Nucleic acid metabolism | FOLR1 |
| Post-translational modification | MMP11 |
| Protein synthesis | FOLR1 |
| Protein trafficking | FOLR1 |
| Small molecule biochemistry | INPP5K, FOLR1, SLC40A1 |
| Vitamin and mineral metabolism | FOLR1 |
| Immune response | MMP11 |
| CLUSTER 6 FUNCTIONS | |
| Cellular assembly and organization | EIF4A1 |
| Gene expression | EIF4A1 |
| Protein synthesis | EIF4A1 |
| RNA post-transcriptional modification | EIF4A1 |
| RNA trafficking | EIF4A1 |
| CLUSTER 7 FUNCTIONS | |
| Carbohydrate metabolism | PTTG1 |
| Cell cycle | AR, PTTG1 |
| Apoptosis | AR, PTTG1 |
| Cell morphology | AR, PTTG1 |
| Cell signaling | AR |
| Cell-to-cell signaling and interaction | AR |
| Cellular assembly and organization | AR, PTTG1 |
| Cellular compromise | AR, PTTG1 |
| Cellular development | AR, PTTG1 |
| Cellular function and maintenance | AR |
| Cellular growth and proliferation | AR, PTTG1 |
| Cellular movement | AR |
| DNA replication, recombination, and repair | AR, PTTG1 |
| Drug metabolism | AR, PTTG1 |
| Gene expression | AR, PTTG1 |
| Lipid metabolism | AR, PTTG1 |
| Molecular transport | AR, PTTG1 |
| Nucleic acid metabolism | AR |
| RNA damage and repair | DNAJB11 |
| RNA post-transcriptional modification | DNAJB11 |
| Small molecule biochemistry | AR, PTTG1 |
| Immune response | AR, PTTG1 |
References
- Agoston D. V., Gyorgy A., Eidelman O., Pollard H. B. (2009). Proteomic biomarkers for blast neurotrauma: targeting cerebral edema, inflammation, and neuronal death cascades. J. Neurotrauma 26, 901–911 10.1089/neu.2008.0724 [DOI] [PubMed] [Google Scholar]
- Armonda R. A., Bell R. S., Vo A. H., Ling G., DeGraba T. J., Crandall B., Ecklund J., Campbell W. W. (2006). Wartime traumatic cerebral vasospasm: recent review of combat casualties. Neurosurgery 59, 1215–1225; discussion 25. [DOI] [PubMed] [Google Scholar]
- Bauman R. A., Elsayed N., Petras J. M., Widholm J. (1997). Exposure to sublethal blast overpressure reduces the food intake and exercise performance of rats. Toxicology 121, 65–79 10.1016/S0300-483X(97)03656-1 [DOI] [PubMed] [Google Scholar]
- Biomarkers Definitions Working Group (2001). Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 69, 89–95 [DOI] [PubMed] [Google Scholar]
- Bowen I. G., Fletcher E. R., Richmond D. R. (1968). Estimae of Man’s Tolerance to the Direct Effects of air Blast. Albuquerque: Lovelace Foundation for Medical Education and Research [Google Scholar]
- Byrnes K. R., Faden A. I. (2007). Role of cell cycle proteins in CNS injury. Neurochem. Res. 32, 1799–1807 [DOI] [PubMed] [Google Scholar]
- Cernak I. (2010). The importance of systemic response in the pathobiology of blast-induced neurotrauma. Front. Neurol. 1:151. 10.3389/fneur.2010.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernak I., Noble-Haeusslein L. J. (2010). Traumatic brain injury: an overview of pathobiology with emphasis on military populations. J. Cereb. Blood Flow Metab. 30, 255–266 10.1038/jcbfm.2009.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernak I., Wang Z., Jiang J., Bian X., Savic J. (2001a). Cognitive deficits following blast injury-induced neurotrauma: possible involvement of nitric oxide. Brain Inj. 15, 593–612 10.1080/02699050010009559 [DOI] [PubMed] [Google Scholar]
- Cernak I., Wang Z., Jiang J., Bian X., Savic J. (2001b). Ultrastructural and functional characteristics of blast injury-induced neurotrauma. J. Trauma 50, 695–706 10.1097/00005373-200104000-00017 [DOI] [PubMed] [Google Scholar]
- Chavko M., Prusaczyk W. K., McCarron R. M. (2006). Lung injury and recovery after exposure to blast overpressure. J. Trauma 61, 933–942 10.1097/01.ta.0000233742.75450.47 [DOI] [PubMed] [Google Scholar]
- Chavko M., Watanabe T., Adeeb S., Lankasky J., Ahlers S. T., McCarron R. M. (2011). Relationship between orientation to a blast and pressure wave propagation inside the rat brain. J. Neurosci. Methods 195, 61–66 [DOI] [PubMed] [Google Scholar]
- Cooper G., Jonsson A. (1997). Protection Against Blast Injury. New York: Butterworth- Heinemann [Google Scholar]
- de Candole C. A. (1967). Blast injury. Can. Med. Assoc. J. 96, 207–214 [PMC free article] [PubMed] [Google Scholar]
- Defense and Veterans Brain Injury Center (2010). Department of Defense Numbers for Traumatic Brain Injury. Accessed January 19, 2010, from http://www.dvbic.org/TBI-Numbers.aspx [Google Scholar]
- DePalma R. G., Burris D. G., Champion H. R., Hodgson M. J. (2005). Blast injuries. N. Engl. J. Med. 352, 1335–1342 [DOI] [PubMed] [Google Scholar]
- Elder G. A., Mitsis E. M., Ahlers S. T., Cristian A. (2010). Blast-induced mild traumatic brain injury. Psychiatr. Clin. North Am. 33, 757–781 10.1016/j.psc.2010.08.001 [DOI] [PubMed] [Google Scholar]
- Fleige S., Pfaffl M. W. (2006). RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Aspects Med. 27, 126–139 10.1016/j.mam.2005.12.003 [DOI] [PubMed] [Google Scholar]
- Gorbunov N. V., Asher L. V., Ayyagari V., Atkins J. L. (2006). Inflammatory leukocytes and iron turnover in experimental hemorrhagic lung trauma. Exp. Mol. Pathol. 80, 11–25 [DOI] [PubMed] [Google Scholar]
- Gorbunov N. V., McFaul S. J., Van Albert S., Morrissette C., Zaucha G. M., Nath J. (2004). Assessment of inflammatory response and sequestration of blood iron transferrin complexes in a rat model of lung injury resulting from exposure to low-frequency shock waves. Crit. Care Med. 32, 1028–1034 10.1097/01.CCM.0000120051.79520.B6 [DOI] [PubMed] [Google Scholar]
- Kawai K., Nitecka L., Ruetzler C. A., Nagashima G., Joo F., Mies G., Nowak T. S., Jr., Saito N., Lohr J. M., Klatzo I. (1992). Global cerebral ischemia associated with cardiac arrest in the rat: I. Dynamics of early neuronal changes. J. Cereb. Blood Flow Metab. 12, 238–249 [DOI] [PubMed] [Google Scholar]
- Kirkman E., Watts S. (2011). Characterization of the response to primary blast injury. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 366, 286–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanek P. M., Bauman R. A., Long J. B., Dixon C. R., Jenkins L. W. (2009). A critical problem begging for new insight and new therapies. J. Neurotrauma 26, 813–814 10.1089/neu.2008.0893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson Terrorism Information Centre (2009). Terrorism incidents and significant dates calendar. Accessed May 20, 2009, from http://www.terrorisminfo.mipt.org [Google Scholar]
- Ling G., Bandak F., Armonda R., Grant G., Ecklund J. (2009). Explosive blast neurotrauma. J. Neurotrauma 26, 815–825 10.1089/neu.2007.0484 [DOI] [PubMed] [Google Scholar]
- Long J. B., Bentley T. L., Wessner K. A., Cerone C., Sweeney S., Bauman R. A. (2009). Blast overpressure in rats: recreating a battlefield injury in the laboratory. J. Neurotrauma 26, 827–840 10.1089/neu.2008.0748 [DOI] [PubMed] [Google Scholar]
- Moochhala S. M., Md S., Lu J., Teng C. H., Greengrass C. (2004). Neuroprotective role of aminoguanidine in behavioral changes after blast injury. J. Trauma 56, 393–403 10.1097/01.TA.0000066181.50879.7A [DOI] [PubMed] [Google Scholar]
- Park E., Gottlieb J. J., Cheung B., Shek P. N., Baker A. J. (2010). A model of low-level primary blast exposure results in cytoskeletal proteolysis and chronic functional impairment in the brain in the absence of lung barotrauma. J. Neurotrauma. 28, 343–357 10.1089/neu.2009.1050 [DOI] [PubMed] [Google Scholar]
- Phillips Y. Y., Mundie T. G., Yelverton J. T., Richmond D. R. (1988). Cloth ballistic vest alters response to blast. J. Trauma 28, S149–S152 [DOI] [PubMed] [Google Scholar]
- Rafaels K. A., Bass C. R., Panzer M. B., Salzar R. S. (2010). Pulmonary injury risk assessment for long-duration blasts: a meta-analysis. J. Trauma 69, 368–374 10.1097/TA.0b013e3181e88122 [DOI] [PubMed] [Google Scholar]
- Readnower R. D., Chavko M., Adeeb S., Conroy M. D., Pauly J. R., McCarron R. M., Sullivan P. G. (2010). Increase in blood–brain barrier permeability, oxidative stress, and activated microglia in a rat model of blast-induced traumatic brain injury. J. Neurosci. Res. 88, 3530–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reneer D. V., Hisel R. D., Hoffman J. M., Kryscio R. J., Lusk B. T., Geddes J. W. (2011). A multi-mode shock tube for investigation of blast-induced traumatic brain injury. J. Neurotrauma 28, 95–104 10.1089/neu.2010.1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risling M., Plantman S., Angeria M., Rostami E., Bellander B. M., Kirkegaard M., Arborelius U., Davidsson J. (2011). Mechanisms of blast induced brain injuries, experimental studies in rats. Neuroimage 54(Suppl. 1), S89–S97 [DOI] [PubMed] [Google Scholar]
- Saljo A., Svensson B., Mayorga M., Hamberger A., Bolouri H. (2009). Low-level blasts raise intracranial pressure and impair cognitive function in rats. J. Neurotrauma 26, 1345–1352 10.1089/neu.2008.0856 [DOI] [PubMed] [Google Scholar]
- Stuhmiller J. H. (1997). Biological response to blast overpressure: a summary of modeling. Toxicology 121, 91–103 10.1016/S0300-483X(97)03658-5 [DOI] [PubMed] [Google Scholar]
- Svetlov S. I., Larner S. F., Kirk D. R., Atkinson J., Hayes R. L., Wang K. K. (2009). Biomarkers of blast-induced neurotrauma: profiling molecular and cellular mechanisms of blast brain injury. J. Neurotrauma 26, 913–921 10.1089/neu.2008.0609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlov S. I., Prima V., Kirk D. R., Gutierrez H., Curley K. C., Hayes R. L., Wang K. K. (2010). Morphologic and biochemical characterization of brain injury in a model of controlled blast overpressure exposure. J. Trauma 69, 795–804 10.1097/TA.0b013e3181bbd885 [DOI] [PubMed] [Google Scholar]
- Treadwell I. (1989). Effects of blasts on the human body. Nurs. RSA 4, 32–36 [PubMed] [Google Scholar]