Abstract
STAT (Signal transducer and activator of transcription) is a potent transcription factor and its aberrant activation by phosphorylation is associated with human cancers1–4. We have shown previously that overactivation of JAK, which phosphorylates STAT5,6, disrupts heterochromatin formation globally in Drosophila melanogaster7. However, it remains unclear how this effect is mediated and whether STAT is involved. Here, we demonstrate that Drosophila STAT (STAT92E) is involved in controlling heterochromatin protein 1 (HP1) distribution and heterochromatin stability. We found, unexpectedly, that loss of STAT92E, had the same effects as overactivation of JAK in disrupting heterochromatin formation and heterochromatic gene silencing, whereas overexpression of STAT92E had the opposite effects. We have further shown that the unphosphorylated or ‘transcriptionally inactive’ form of STAT92E is localized on heterochromatin in association with HP1, and is required for stabilizing HP1 localization and histone H3 Lys 9 methylation (H3mK9). However, activation by phosphorylation reduces heterochromatin-associated STAT92E, causing HP1 displacement and heterochromatin destabilization. Thus, reducing levels of unphosphorylated STAT92E, either by loss of STAT92E or increased phosphorylation, causes heterochromatin instability. These results suggest that activation of STAT by phosphorylation controls both access to chromatin and activity of the transcription machinery.
To understand the molecular mechanism underlying JAK/STAT-mediated tumour formation, we have previously investigated the role of JAK in a Drosophila leukaemia model, in which a hyperactive mutant form of JAK (Tum-1) causes leukaemia-like overproliferation of blood cells5,8. We have demonstrated that oncogenic JAK disrupts heterochromatin formation globally, allowing transcriptional activation of genes that are not necessarily direct targets of STAT7,9. The molecular mechanism underlying the effects of Hopscotch (Hop, Drosophila JAK) on heterochromatin remains unclear. It may be mediated by phosphorylation of STAT92E, as in the canonical JAK/STAT pathway10,11. Alternatively, Hop may activate cellular targets other than STAT92E9.
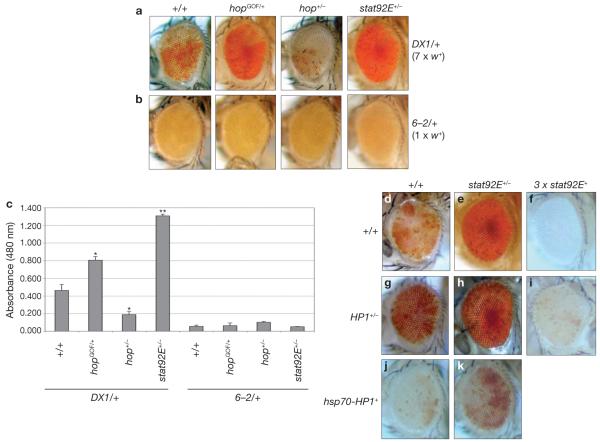
To investigate whether disruption of heterochromatin induced by Hop-activation7 is mediated by STAT92E, we examined the effects of reducing stat92E+ dosage on heterochromatic gene silencing, which can be measured by position-effect variegation (PEV)12. As shown previously7, Hop gain- and loss-of-function mutations suppress and enhance PEV, respectively (Fig. 1). We found that reducing stat92E+ dosage had the opposite effect of reducing Hop and strongly suppressed PEV, causing marked derepression of white+ in two independent variegated lines DX1 and In(1)wm4, resulting in a substantial increase in red eye-pigmentation (Fig. 1a, c, d–k). Heterozygosity of stat92E did not affect expression in a control P[white+] (6–2 mini-white+) element, which does not induce heterochromatin formation (Fig. 1b, c) or the white+ gene in its original locus (data not shown; also see ref. 7), suggesting that P[white+] or white+ is not simply a transcriptional target of STAT92E. Thus, in contrast to Hop, which counteracts heterochromatic gene silencing7, STAT92E seems to promote this silencing process.
Figure 1.
STAT92E is required for heterochromatic gene silencing. (a–c) Effects of mutations in hop and stat92E on white+ variegation induced by transgene repeats, DX1 (a) or a control transgene (b) are shown as changes in red eye-pigmentation in representative images. DX1 consists of seven tandem copies of a P[white+] reporter transgene inserted in a euchromatic region, which induce heterochromatin formation at the insertion locus26. In(1)wm4 is caused by X chromosomal inversion, juxtaposing the white+ gene to centromeric heterochromatin12. (c) Red eye-pigment levels were determined by measuring absorbance at a wavelength of 480 nm. Data are mean ± s. d, **P < 0.001 when compared with +/+ control, Student’s t-test (n = 40 for each group). 6–2 is a single P[white+] element inserted at the same chromosomal location as DX1 but that does not induce heterochromatin formation26,27. (d–k) Effects of altering dosages of stat92E+ and/or HP1+ on white+ variegation induced by chromosomal inversion (In(1)wm4) are shown as changes in red eye-pigmentation in representative images. (d) In(1)wm4 in otherwise wild-type background. Note increased white+ expression (more red pigments) in stat92E (e, h, k) or HP1 heterozygotes (g–i) and decreased white+ expression when stat92E+ (f, i) or HP1+ is overexpressed (j, k). The combination of hsp70–HP1+ and 3 × stat92E+ was not viable. Flies bearing hsp70–HP1+ were raised at 29°C. All others were raised at 25°C. All genotypes are in In(1)wm4/+ or w−/− backgrounds. (hopGOF=hopTum–l, hop−=hop3, stat92E−=stat92E6346, HP1−=Su(var)20505, 3 × stat92E+=Dp(3;3)MRS/+). Similar results were observed when another chromosomal duplication, Dp(3;3)M95A[+]13, that duplicates stat92E+ was used (data not shown), or by a stat92E+ transgene (Supplementary Information, Fig. S1).
We next examined the epistatic relationship between HP1 and STAT92E (Fig. 1d–k). HP1, encoded by Su(var)205, is a constitutive component of heterochromatin and is essential for heterochromatic gene silencing12,13. Consistent with the results described above, we found that increasing stat92E+ dosage by a chromosomal duplication (referred to as 3 × stat92E+, Fig. 1f) or a stat92E+ transgene (Supplementary Information, Fig. S1) enhanced heterochromatic gene silencing, resulting in complete silencing of the variegated white+ gene. This effect was antagonized, however, by reducing HP1+ gene dosage (Fig. 1i). It was also evident that the effect of halving HP1+ gene dosage on derepression of the silenced white+ gene was strongly opposed by the presence of 3 × stat92E+ (compare Fig. 1g with 1i). Thus, increasing STAT92E levels promotes gene silencing even when HP1 levels are reduced. Consistent with previous reports14, increasing HP1 levels by expression of an hsp70–HP1+ transgene promoted heterochromatin formation, as shown by increased variegation (Fig. 1j). We found that halving stat92E+ gene dosage counterbalanced the effects of increased HP1, allowing increased white+ expression (Fig. 1k). However, expression levels of white+ in hsp70–HP1+/−;stat92E+/− double-heterozygous flies were much lower that those in stat92E+/− flies (compare Fig. 1e with 1k), suggesting that increasing HP1 levels also antagonizes the effects of reducing stat92E+ dosage. Thus, STAT92E and HP1 are interdependent and both required for heterochromatic gene silencing.
We next investigated the requirement for STAT92E in heterochromatin formation by genetic mosaic analyses (see Methods) using HP1 and dimethylated histone H3mK9 as markers in larval salivary glands7,13. We found that clonal overexpression of stat92E+ resulted in higher levels of heterochromatin (H3mK9 and HP1 accumulation), compared with neighbouring wild-type cells (Fig. 2a, b). An increase in the size of the HP1 foci was also found in cells overexpressing a STAT92E–GFP fusion protein (see below). Conversely, in stat92E−/− cells, heterochromatin levels, as characterized by HP1 foci, were markedly reduced, compared with neighbouring wild-type cells (Fig. 2c). Notably, the effects of STAT92E overexpression were opposite to those of JAK overactivation on heterochromatin shown previously7, consistent with the opposite effects we observed on PEV. Thus, STAT92E levels seem to be important for heterochromatin formation or stability.
Figure 2.
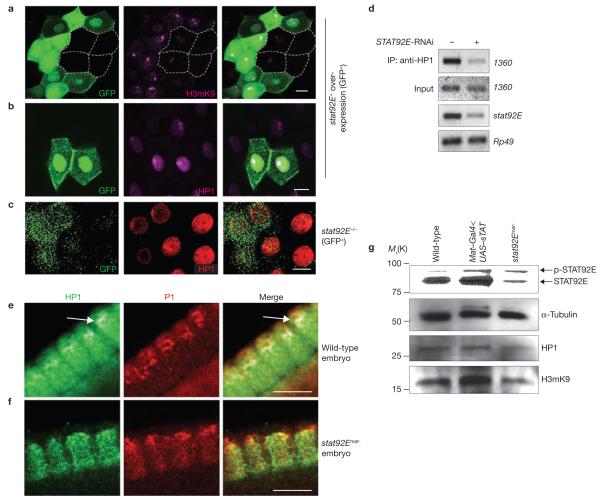
STAT92E levels control heterochromatin abundance and HP1 localization. (a, b) Third-instar salivary glands overexpressing stat92E+ in random cells marked with GFP (green) and stained with anti-H3mK9 (magenta in a) or anti-HP1 (magenta in b) are shown partially. Note the increased levels of H3mK9 or HP1 in GFP+ cells, compared with GFP− cells (four GFP− cells are outlined). (c) A third-instar salivary gland containing stat92E−/− cells marked with GFP (green) and stained with anti-HP1 (red) is shown partially. Note the lack of HP1 foci in the GFP+ cells. (d) Enrichment of HP1 on heterochromatin was detected by ChIP using an anti-HP1 antibody and PCR primers that amplify the 1360 transposon element (heterochromatin). Note the marked reduction in HP1 association with 1360 sequences in the STAT92E RNAi-depleted sample (lane 2), compared with the control (lane 1). (e, f) Cellular blastoderm-stage embryos stained with anti-HP1 (green) and propidium iodide (PI; red) to show DNA. Part of the cortical cell layer is shown. Note the concentrated HP1-staining at the apical region of the wild-type embryo (arrow in e), which is absent in the stat92Emat− embryo (f). (g) Total protein extracts from 0–12 hold embryos of wild-type (lane 1), Nanos–Gal4 × UAS–stat92E+ (lane 2) and stat92Emat− females (lane 3) were subjected to SDS–PAGE and blotted sequentially with anti-STAT92E and anti-α-tubulin antibodies (for full-length gel, see Supplementary Information, Fig. S7), or with anti-H3mK9 and anti-HP1 antibodies, respectively. The membrane was stripped between blots. Note the correlation between H3mK9 and STAT92E levels. Scale bars, 10 μm.
To further confirm that STAT92E is required for HP1 localization on heterochromatin, we performed chromatin immunoprecipitation (ChIP) using an anti-HP1 antibody before and after stat92E knockdown by RNAi in Drosophila S2 cells. We chose the transposable element 1360 as a heterochromatin marker to detect the co-immunoprecipitated heterochromatin. 1360 is a repetitive sequence found most abundantly in constitutive heterochromatin regions of all Drosophila chromosomes and is believed to be essential for initiating heterochromatin formation15,16. Enrichment of HP1 binding to 1360 sequences has been detected previously by ChIP17. We found that association of HP1 with 1360 was significantly reduced following stat92E RNAi knockdown (Fig. 2d). Thus, STAT92E is essential for the association of HP1 with heterochromatin.
To investigate whether changing STAT92E levels alters HP1 localization in general, we examined additional developmental contexts. First, HP1 localization in heterochromatin can be observed in cellularization-stage (nuclear cycle 14) embryos as foci enriched at the apical region of elongated wild-type nuclei13,18,19 (Fig. 2e). This pattern of HP1 distribution, however, was absent in mutant embryos lacking maternal stat92E+ (referred to as stat92Emat− embryos; see Methods; Fig. 2f). In these stat92Emat− embryos, HP1 protein assumed a punctate and clustered distribution that was no longer restricted to the apical heterochromatin, whereas there were no detectable changes in the total levels of HP1 (compare Fig. 2e with 2f). This is consistent with the notion that STAT92E is required for HP1 localization in centromeric heterochromatin. Second, we examined stat92E mutant clones in imaginal discs. The small size of imaginal disc cells does not allow examination of HP1 sub-nuclear distribution, but does allow assessment of total HP1 levels. We found no detectable changes in HP1 levels in stat92E−/− imaginal disc cells, compared with neighbouring wild-type cells (Supplementary Information, Fig. S2), suggesting that loss of STAT92E does not immediately cause a substantial reduction in HP1 protein levels. However, long-term loss of stat92E+ indeed reduced HP1 levels (Supplementary Information, Fig. S3), suggesting that HP1 transcription may be regulated directly or indirectly by STAT92E. Finally, we examined heterochromatin by performing blots of total protein from embryo extracts. We found that H3mK9 levels were increased significantly in embryos that overexpressed STAT92E and decreased in stat92Emat− embryos, whereas HP1 levels were not changed and moderately reduced, respectively (Fig. 2g). These results indicate that, although the canonical JAK/STAT pathway may regulate HP1 transcription, both loss of STAT92E and overactivation of JAK cause HP1 delocalization and overall loss of H3mK9.
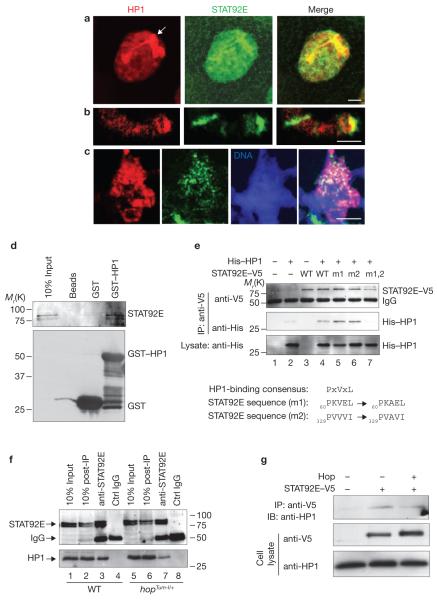
The observation that loss of stat92E+ is equivalent to JAK overactivation and that increasing STAT92E levels has similar effects to loss of hop+ on heterochromatin formation seemed paradoxical, as JAK activates STAT in the canonical JAK/STAT pathway3,4. To understand how STAT92E levels affect heterochromatin formation, we examined STAT92E protein localization by immunostaining with an anti-STAT92E antibody (see Supplementary Information, Figs S4, S6 for specificity). We found that STAT92E was localized mainly in the nucleus in discrete patterns, with the highest levels colocalizing with the HP1 foci (Fig. 3a). Interestingly, it has been shown that unphosphorylated mammalian STAT3 is localized predominantly in the nucleus20, despite the conventional view that it should be in the cytoplasm. STAT92E colocalization with HP1 in heterochromatin regions was also detected in larval fat bodies or using a different anti-STAT92E antibody (data not shown), or in glands overexpressing a STAT92E–GFP transgene, or by ChIP with S2 cells (see below), indicating that STAT92E is indeed localized in heterochromatin. Nuclear localization of STAT92E–GFP is also evident in unstimulated S2 cells21. Moreover, staining of squashed polytene chromosomes from wild-type salivary glands indicated that STAT92E and HP1 colocalize in several regions of heterochromatin, including the chromocentre and telomere (Fig. 3b, c). These results suggest that a significant amount of STAT92E is normally localized on heterochromatin.
Figure 3.
STAT92E colocalizes and physically associates with HP1. (a–c) Third-instar wild-type salivary glands were stained with anti-HP1 (red) and anti-STAT92E (green) antibodies in whole-mount tissues or squashed polytene chromosomes. Representative giant nuclei and squashed chromosomes are shown. (a) In wild-type larvae, the HP1 foci in the salivary gland nuclei, which mark heterochromatin, colocalize with STAT92E. In squashed chromosomes, STAT92E and HP1 colocalization is shown in the telomere (b) and chromocentre (c). (d) Drosophila embryo extracts were incubated with bacterially expressed GST–HP1, which was then purified by glutathione beads, subjected to SDS–PAGE and blotted with an anti-STAT92E antibody. (e) Drosophila S2 cells transfected with or without STAT92E–V5 variants or His–HP1 were immunoprecipitated with an anti-V5 antibody and subjected to SDS–PAGE. The associated His–HP1 was detected with an anti-His antibody. Note the double-mutant STAT92E–V5 (lane 7) was unable to bind to HP1. The HP1-binding consensus motif, the two sequences present in STAT92E and the mutated sequences are shown under the blots. (f) Embryo extracts were immunoprecipitated with an anti-STAT92E antibody, subjected to SDS–PAGE and blotted with anti-HP1. Note that a greater amount of the endogenous HP1 was co-immunoprecipitated with endogenous STAT92E from wild-type than hopTum–l/+ embryos. Also note that depleting STAT92E using an antibody did not seem to affect HP1 levels in the lysate (lanes 2, 6), suggesting that perhaps only a small portion of HP1 is normally associated with STAT92E. (g) S2 cells transfected with or without STAT92E–V5 or Hop (lane 3) were immunoprecipitated with anti-V5 and subjected to SDS–PAGE. The associated endogenous HP1 was detected with an anti-HP1 antibody. Note that HP1 is co-immunoprecipitated with STAT92E–V5 (lane 2) and a reduced amount of HP1 is detected in the presence of Hop (lane 3). Scale bars, 2 μm.
As STAT92E colocalizes with HP1, we performed co-immunoprecipitation studies to explore the possibility that STAT92E may interact physically with HP1. Indeed, we found that bacterially expressed GST–HP1 can pull down the endogenous STAT92E from embryo extracts (Fig. 3d), and that HP1 and STAT92E co-immunoprecipitate when transfected in S2 cells (Fig. 3e, lane 4). STAT92E contains a perfect and an imperfect HP1-binding sequence motif PVL22 (Fig. 3e). We found that mutations of only one or the other site had minimal effects on STAT92E–HP1 binding (Fig. 3e, lanes 5, 6). However, mutations of both sites abolished the STAT92E–HP1 interaction (Fig. 3e, lane 7), suggesting that either of the two sites may be sufficient to mediate STAT92E binding to HP1. Moreover, we found that endogenous HP1 can co-immunoprecipitate with STAT92E (Fig. 3f, left). Despite the presence of higher levels of both HP1 and STAT92E proteins in hop-Tum–l/+ embryos, less HP1 was co-immunoprecipitated with STAT92E from these embryos (Fig. 3f). This suggests that unphosphorylated STAT92E binds to HP1, as the ratio of unphosphorylated-to-phosphorylated STAT92E levels was lower in hopTum–l/+ than in wild-type embryos (caused by increased STAT92E phosphorylation). Finally, increasing STAT92E phosphorylation, by co-transfecting Hop, reduced the amount of HP1 co-immunoprecipitated with transfected STAT92E (Fig. 3g). These results suggest that unphosphorylated STAT92E can associate with HP1.
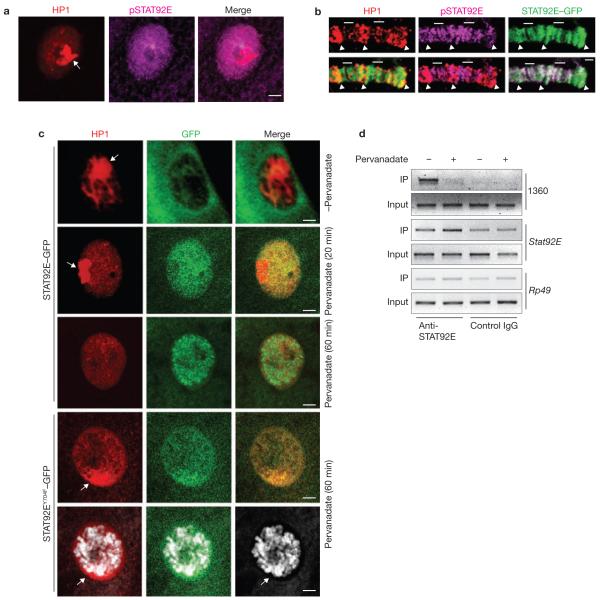
We next determined whether the heterochromatin-associated STAT92E is ‘active’ (that is, tyrosine-phosphorylated) by staining with an anti-phospho-STAT92E antibody23 (see Supplementary Information, Figs S4, S6 for specificity). Compared with total STAT92E, phosphorylated STAT92E seemed to be distributed more uniformly in the nucleus and did not colocalize with HP1 (Fig. 4a). Moreover, we examined squashed polytene chromosomes from larvae expressing STAT92E–GFP by immunostaining and found that STAT92E–GFP was colocalized either with pSTAT92E or HP1 signals, but rarely with both (Fig. 4b). These results indicate that it is the unphosphorylated form of STAT92E that colocalizes with HP1 on heterochromatin.
Figure 4.
Only unphosphorylated STAT92E is localized on heterochromatin. (a) Wild-type larval salivary gland stained with anti-HP1 and anti-pSTAT92E antibodies. Note that the signals do not overlap. (b) In polytene chromosomes from sgs–Gal4<UAS–stat92E–GFP larvae, STAT92E–GFP was colocalized with either pSTAT92E (bars) or HP1 (arrowheads) signals. (c) Third-instar larval salivary glands expressing UAS–STAT92E–GFP or STAT92EY704F–GFP (by arm–Gal4) were treated with pervanadate ex vivo. Distribution of HP1 (red) and STAT92E–GFP or STAT92EY704F–GFP (green) was examined at the indicated time points. Note that following pervanadate stimulation, STAT92E–GFP moves away from heterochromatin (HP1 foci; 20 min) and then binds to chromosomes as distinct bands (60 min), whereas UAS–STAT92EY704F–GFP remains colocalized with HP1 foci (60 min). DNA was stained with Toto-3. (d) Chromatin was immunoprecipitated from S2 cells with an anti-STAT92E antibody (which recognizes both unphosphorylated and phosphorylated STAT92E) before and after pervanadate treatment. Note the enrichment of STAT92E on heterochromatin (1360; row 1) and target-gene promoter (stat92E itself) before (lane 1) and after (lane 2) pervanadate treatment. The stat92E promoter contains multiple STAT92E-binding sites that mediate stat92E positive-feedback autoregulation. Rp49 is a negative control (row 3). A goat anti-human pan SMAD antibody was used as a control IgG for goat anti-STAT92E. Scale bars, 2 μm.
To investigate how STAT92E activation by phosphorylation leads to heterochromatin destabilization, we examined STAT92E–GFP and HP1 localization in ex vivo cultured salivary glands at different times after stimulating STAT92E phosphorylation with pervanadate (H2O2/vanadate), which potently and rapidly increases STAT92E phosphorylation (Supplementary Information, Fig. S6a)23–25. Before stimulation, STAT92E–GFP was localized in both cytoplasm and nucleus, with the nuclear portion mostly colocalized with HP1 (Fig. 4c). The higher levels of STAT92E–GFP than endogenous STAT92E in the cytoplasm (compare Fig. 4c with Fig. 3a) may be caused by transgene overexpression. At 20 min post-stimulation, the nuclear STAT92E–GFP was enriched, due to entry of the cytoplasmic protein, and was not colocalized with HP1 (Fig. 4c). This pattern of STAT92E–GFP resembles that of phosphorylated STAT92E (Fig. 4a). We suggest that the change in STAT92E–GFP distribution is caused by nuclear STAT92E–GFP moving away from heterochromatin and cytoplasmic STAT92E–GFP moving into the nucleus following phosphorylation. At 60 min post-stimulation, STAT92E–GFP was mostly bound to euchromatic chromosomes in distinct bands and HP1 had diffused from heterochromatin (Fig. 4c). When an unphosphorylatable mutant stat92EY704F–GFP was overexpressed, however, pervanadate treatment did not cause complete HP1 dispersal and STAT92EY704F–GFP remained colocalized with HP1 (Fig. 4c). Thus, STAT92E dispersal precedes, and is required for, HP1 displacement from heterochromatin. This conclusion is consistent with observations of HP1 and STAT92E distribution in hopTum–l mutant larval salivary glands (Supplementary Information, Fig. S5). It thus seems that unphosphorylated STAT92E is associated with heterochromatin and phosphorylation causes STAT92E dispersal and translocation to euchromatic regions, presumably binding to cognate promoters.
To confirm this observation, we carried out ChIP experiments in Drosophila S2 cells to determine the effects of pervanadate treatment on the abundance of 1360 (heterochromatin) and stat92E (target gene) promoter sequences in ChIP, using an anti-STAT92E antibody. Indeed, we found enrichment of 1360 sequences in STAT92E–chromatin complexes before, but not after, treatment with pervanadate (Fig. 4d). The same treatment caused enrichment of phosphorylated STAT92E in the promoter of stat92E itself (Fig. 4d).
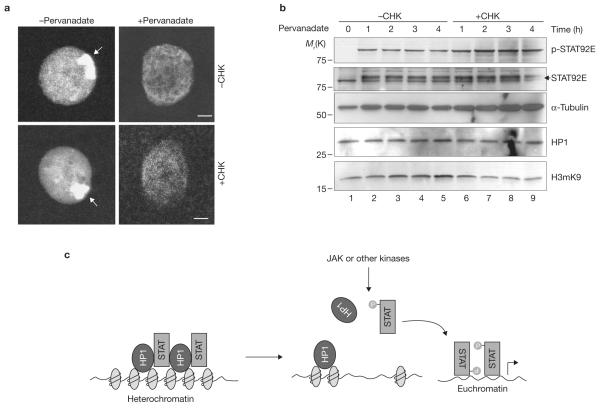
To investigate whether STAT92E activation could affect heterochromatin indirectly by transcriptionally inducing other genes, we blocked protein synthesis by treating salivary gland with cycloheximide before pervanadate stimulation. We found that cycloheximide treatment did not prevent pervanadate-induced HP1 dispersal (Fig. 5a), but rather, accelerated heterochromatin loss (Fig. 5b). This suggests that heterochromatin destabilization induced by STAT92E activation does not require synthesis of new protein and therefore, cannot be mediated by induction of STAT92E transcription targets.
Figure 5.
Protein-synthesis-independent heterochromatin disruption following STAT92E phosphorylation. (a) Wild-type salivary glands were cultured with or without cycloheximide (CHX) for 1 h before addition of pervanadate for 1 h in vitro. Represented nuclei are shown. Note the disappearance of the HP1 foci in treated cells. (b) S2 cells were cultured with or without CHX for 1 h, and then stimulated with pervanadate for the indicated times. Cell extracts were blotted with the indicated antibodies. Note a greater increase in pSTAT92E (phosphorylated) and decrease in STAT92E (unphosphorylated) bands in CHX-treated samples, which is correlated with a greater decrease in H3mK9 levels. (c) A model for the role of STAT in heterochromatin stability. Unphosphorylated STAT is localized on heterochromatin in association with HP1. Increasing STAT phosphorylation (by JAK or other tyrosine kinases) reduces the amount of unphosphorylated STAT localized in heterochromatin. This in turn causes HP1 displacement from heterochromatin and heterochromatin instability. Dispersed phosphorylated STAT binds to cognitive sites in euchromatin to induce target-gene expression. Scale bars, 2 μm.
To further explore the consequences of cycloheximide and pervanadate treatment on STAT92E activation and heterochromatin stability, we examined total protein blots of S2 cell extracts at different times after drug treatment. STAT92E phosphorylation (activation) was evident 5 min after pervanadate treatment and reached a maximum at 1 h in the absence of cycloheximide treatment, but continued to increase in the presence of cycloheximide (Fig. 5b; Supplementary Information, Fig. S6a). The increase in phosphorylated STAT92E levels occurred at the expense of unphosphorylated STAT92E, which was most evident when synthesis of new protein was blocked (Fig. 5b, second panel, lower band), such that after 4 h of drug treatment, unphosphorylated STAT92E levels were significantly reduced (Fig. 5b, second panel, lower band, lane 9). Drug treatment did not significantly affect HP1 levels (Fig. 5b) and cycloheximide treatment alone had no effects on HP1 localization (Fig. 5a), H3mK9 or STAT92E levels or its phosphorylation status (Supplementary Information, Fig. S6b). However, a decrease in the heterochromatin marker H3mK9 was detected 2 h after combined pervanadate and cycloheximide treatment (Fig. 5b).
These results suggest that, in the presence of cycloheximide, pervanadate causes a greater increase in phosphorylated STAT92E levels and a greater decrease in unphosphorylated STAT92E levels, presumably caused by a lack of both positive and negative feedback mechanisms. The negative feedback on STAT activation is mediated by induction of inhibitors such as SOCS36E and PTP61F, which reduce pSTAT92E levels, and the positive feedback by STAT92E autoregulation, which increases STAT92E levels10. Both feedback mechanisms require new protein synthesis, which was blocked by cycloheximide. The resultant loss of unphosphorylated STAT92E after combined drug treatment would thus lead to accelerated heterochromatin destabilization. Taken together, these results suggest that activation of STAT92E by phosphorylation causes STAT92E dispersal, leading to HP1 displacement and heterochromatin destruction in a transcription-independent manner (Fig. 5c).
The results of these genetic and biochemical studies suggest a model in which at least a portion of unphosphorylated STAT is normally localized in heterochromatin, where STAT activation causes its diffusion from heterochromatin and relocalization to euchromatin, leading to HP1 dispersal and heterochromatin destabilization (Fig. 5c). Our results, however, do not rule out the possibility that activation of JAK reduces methylation of histone H3mK9 through a STAT-independent mechanism. We envision that under physiological conditions, a low-intensity signal may be sufficient for activating specific STAT target genes without affecting heterochromatin, whereas high intensity or persistent activation of JAK/STAT may bring about widespread heterochromatin destruction. It remains to be determined whether JAK enters the nucleus to phosphorylate STAT or whether the redistribution of unphosphorylated nuclear STAT is a result of an altered equilibrium between nuclear and cytoplasmic or phosphorylated and unphosphorylated STAT in response to JAK activation.
METHODS
Fly stocks and genetics
All crosses were carried out at 25°C on standard cornmeal/agar medium, unless otherwise specified. Fly stocks of hopTum–l, stat92E6346, Su(var)20505, In(1)wm4, Dp(3;3)MRS/+, Dp(3;3)M95A[+]13, FRT82B ubiq–GFP, FRT82B Gal80, hemese–Gal4, Nanos–Gal4 and hsp70–flp; Act5C>y+>Gal4 UAS–GFP were from the Bloomington Drosophila Stock Center. Fly stocks of DX1 and 6–2 mini-white+ (J. Birchler, University of Missouri), hsp70–HP1 (L. Wallrath, University of Iowa), UAS–stat92E–EGFP and UAS–stat92EY704F–EGFP (M. Zeidler, University of Sheffied) and UAS–stat92E (S. Hou, National Cancer Institute) were gifts.
To express UAS–stat92E+ in random clones, hsp70–flp;Act5C>y+>Gal4 UAS–GFP flies were crossed with UAS–stat92E+ flies. To generate stat92E loss-of-function clones, we crossed hsp70–flp; FRT82B stat92E6346/TM3 females with males of hsp70–flp; FRT82B ubiq–GFP (for clones in imaginal discs), or Act5C>y+>Gal4; FRT82B Gal80 (for GFP+ clones in salivary glands), or hsp70–Flp; FRT82B [ovoD1, w+]/TM3 (for clones in the female germ-line). The progeny were heat-shocked at 37°C for 2 h in late-embryo (for salivary gland clones) or early-larval stages and allowed to develop at 25°C until examination.
Antibodies, drug treatment, imaging and eye-pigmentation measurement
Mouse monoclonal anti-HP1 (C1A9; 1:200, Developmental Hybridoma Bank), rabbit anti-H3(di)mK9 (07–212, 1:200, Upstate Biotechnology), goat anti-STAT92E (sc-15708; affinity purified against the amino-terminus of STAT92E, 1:200, Santa Cruz), rat anti-STAT92E (1:500)24 and anti-pSTAT92E (1:1,000, Cell Signaling Technology) were used as primary antibodies and fluorescent secondary antibodies (1:250; Molecular Probes) were used in whole-mount immunostaining. Stained tissues were photographed with a Leica confocal microscope. Images were cropped and minimally processed with Adobe Photoshop 7.0.
To stimulate JAK/STAT activation in S2 cells, 2 mM H2O2 and 1 mM sodium vanadate (final concentrations; Sigma) were added for 30 min before collection for ChIP experiments or as indicated. For salivary gland ex vivo culture, the final concentrations were 5 mM H2O2 and 0.5 mM vanadate. Alternatively, salivary glands were pretreated with cycloheximide (0.1 mg ml−1; Sigma) for 1 h or as indicated.
To measure eye pigmentation, the heads of 40 female flies (2–3 days old, raised at 25°C) of each genotype were homogenized in methanol (1 ml, acidified with 0.1% HCl). Eye pigmentation was represented as the absorbance of the supernatant at 480 nm.
Cell culture, oligonucleotides and plasmids
V5–STAT92E and Hop expression plasmids in pMT vector were gifts from S. Hou. Oligonucleotides used to amplify the 1360 transposon (forward: TGTATCGTTTTTAAAAAATTGTCAG; reverse: GTGGACCTGTAATATATGCTCT) were as described17. A 500-bp stat92E cDNA fragment was amplified by T7 promoter attached to PCR primers (forward: GAAT TAATACGACTCACTATAGGGAGACTTGCCCAAAACTACAGTTAC; reverse: GAATTAATACGACTCACTATAGGGAGACGACTGTGGGTGGATTGTT) and used for stat92E RNAi synthesis by in vitro transcription with the Fermentas T7 RNA transcription kit, according to the manufacturer’s instruction.
Drosophila Schneider L2 (S2) cells were cultured at 25°C in Drosophila serum-free medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 0.5× antibiotic-antimycotic (Invitrogen) solution. Cells were cultured at 2.5 × 106 ml−1 before transfection, which was performed with Cellfectin (Invitrogen), according to the manufacturer’s instructions. Copper sulphate (Sigma) was added to the medium 16 h after transfection, at a final concentration of 0.5 mM, and cells were collected 48 h of after induction. S2 cells were collected in cell lysis buffer (Cell Signaling Technology). For RNAi treatment, cells were grown in serum-free medium for 12 h before addition of dsRNA (15 μg ml−1). Cells were grown for another 4 days before processing.
ChIP
Analysis was performed according to standard protocols (see Supplementary Information) with the following modifications: 1 × 107 confluent S2 cells were cross-linked with 1% formaldehyde. The cell lysate was sonicated to produce DNA fragments of 500–1000 bps. Chromatin (25 μg; determined by A260nm) was pre-cleared with protein G beads and then precipitated with primary antibody (5 μg in 1 ml total volume) overnight at 4°C. Following purification with protein G beads, samples were heated at 65°C for 5 h to reverse cross-linking. Genomic DNA was purified with QIAquick (Qiagen) and amplified with specific PCR primers.
Supplementary Material
Figure S1 STAT92E enhances repeat-mediated variegation. Eye pigmentation phenotypes of adult flies of indicated genotypes raised at 25°C. The light orange background was due to the presence of mini-w+ in the UAS-Stat92E+ transgene. Note the deceased pigmentation when UAS-Stat92E+ was expressed by the basal level of hsp70-Gal4 transgene (which does not carry mini-w+).
Figure S2 Loss of Stat92E+ does not affect HP1 expression in imaginal discs. A 3rd instar wing imaginal disc bearing Stat92E−/− cells (marked by lack of GFP) is shown for HP1 levels (stained by anti-HP1, red).
Figure S3 Long-term loss of STAT92E reduced HP1. A Stat92E homozygous loss of function cell (marked by GFP) was induced during embryogenesis and examined for HP1 (red) in late 3rd instar larval salivary gland.
Figure S4 Specificity of anti-STAT92E and anti-pSTAT92E. (a) A 3rd instar wing imaginal disc bearing Stat92E−/− cells (marked by lack of GFP) was stained by anti-STAT92E (red). Note that the red signals closely matches STAT92E protein levels, and that no signal is detected in Stat92E−/− cells. (b) S2 cells were transfected with Hop-V5 and/or STAT92E-V5. Total cell lysates were subjected to SDS-PAGE and transferred to nitrocellulose membrane and immunoblotted sequentially with anti-pSTAT92E, anti-V5, and anti-STAT92E, respectively. Note that anti-pSTAT92E does not cross-react with non-phospho-STAT92E (lack of signals in lane 1, 2, left panel), and that anti-STAT92E does not detect other proteins (right panel). Extra bands on the right panel were due to incomplete stripping. Anit-V5 detected non-specific bands (bands across all 3 lanes in middle panel).
Figure S5 HP1 and STAT92E distribution in hopGOF mutant larvae. Salivary glands isolated from hopGOF/+ (hopTum−l/+) larvae were stained with anti-HP1 (red) and anti-STAT92E (green). (a) In glands isolated from hopTum−l/+ larvae raised at 29°C, both HP1 and STAT92E appear diffused, lacking prominent heterochromatin foci. (b) In hopTum−l/+ larvae raised at 25°C, a moderately nonpermissive temperature1, a fraction of the nuclei (32%; n=64/198 nuclei) exhibited nearly normal patterns of HP1 foci (left panel), whereas in nearly all of these nuclei (95%; n=61/64 nuclei), STAT92E appeared dispersed and did not colocalize with HP1 (middle panel). These results suggest that STAT92E disperses in response to HopTum−l phosphorylation, and this process precedes changes in HP1 subnuclear localization.
Figure S6 Effects of H2O2/vanadate and CHX on STAT92E phosphorylation and histone H3 K9 methylation. S2 cells were cultured with or without CHX for 1 h, and then stimulated with H2O2/vanadate for indicated times (a), or incubated in the presence of CHX for indicated times (b). Cell extracts were blotted sequentially with anti-pSTAT92E and anti-STAT92E, or with anti-H3mK9, respectively. Membrane was stripped between blots.
Figure S7 Full-length protein gels. Note that Fig. 2f used a lower percentage gel and longer run, which resulted in a greater separation phospho-STAT92E from STAT92E bands (compared with Fig. 5b).
ACKNOWLEDGEMENTS
We thank J. Birchler, S. Elgin, S. Hou, L. Wallrath, M. Zeidler, G. Reuter, the Developmental Hybridoma Bank (Iowa), the Bloomington Drosophila Stock Center for Drosophila strains and reagents and H. Land and D. Bohmann for helpful comments on the manuscript. This study was supported, in part, by grants from the National Institutes of Health (R01GM65774; R01GM077046), an American Cancer Society Research Scholar Grant (RSG-06-196-01-TBE) and a Leukemia & Lymphoma Society Research Scholar Grant (1087-08) to W.X.L.
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
References
- 1.Bromberg J. Stat proteins and oncogenesis. J. Clin. Invest. 2002;109:1139–1142. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu H, Jove R. The STATs of cancer – new molecular targets come of age. Nature Rev. Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 3.Aaronson DS, Horvath CM. A road map for those who know JAK-STAT. Science. 2002;296:1653–1655. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 4.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 5.Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo H, Hanratty WP, Dearolf CR. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. EMBO J. 1995;14:1412–1420. doi: 10.1002/j.1460-2075.1995.tb07127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi S, et al. JAK signaling globally counteracts heterochromatic gene silencing. Nature Genet. 2006;38:1071–1076. doi: 10.1038/ng1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanratty WP, Dearolf CR. The Drosophila Tumorous-lethal hematopoietic oncogene is a dominant mutation in the hopscotch locus. Mol. Gen. Genet. 1993;238:33–37. doi: 10.1007/BF00279527. [DOI] [PubMed] [Google Scholar]
- 9.Betz A, Darnell JE., Jr A Hopscotch-chromatin connection. Nature Genet. 2006;38:977–979. doi: 10.1038/ng0906-977. [DOI] [PubMed] [Google Scholar]
- 10.Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605–2616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- 11.Hou SX, Zheng Z, Chen X, Perrimon N. The Jak/STAT pathway in model organisms. Emerging roles in cell movement. Dev. Cell. 2002;3:765–778. doi: 10.1016/s1534-5807(02)00376-3. [DOI] [PubMed] [Google Scholar]
- 12.Grewal SI, Elgin SC. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 2002;12:178–187. doi: 10.1016/s0959-437x(02)00284-8. [DOI] [PubMed] [Google Scholar]
- 13.James TC, et al. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- 14.Li Y, Danzer JR, Alvarez P, Belmont AS, Wallrath LL. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development. 2003;130:1817–1824. doi: 10.1242/dev.00405. [DOI] [PubMed] [Google Scholar]
- 15.Kaminker JS, et al. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002;3:0084.1–0084.20. doi: 10.1186/gb-2002-3-12-research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun FL, et al. cis-Acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four. Mol. Cell Biol. 2004;24:8210–8220. doi: 10.1128/MCB.24.18.8210-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Lucia F, Ni JQ, Vaillant C, Sun FL. HP1 modulates the transcription of cell-cycle regulators in Drosophila melanogaster. Nucleic Acids Res. 2005;33:2852–2858. doi: 10.1093/nar/gki584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kellum R, Raff JW, Alberts BM. Heterochromatin protein 1 distribution during development and during the cell cycle in Drosophila embryos. J. Cell Sci. 1995;108:1407–1418. doi: 10.1242/jcs.108.4.1407. [DOI] [PubMed] [Google Scholar]
- 19.Shareef MM, et al. Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Mol. Biol. Cell. 2001;12:1671–1685. doi: 10.1091/mbc.12.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L, McBride KM, Reich NC. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-α3. Proc. Natl Acad. Sci. USA. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karsten P, Plischke I, Perrimon N, Zeidler MP. Mutational analysis reveals separable DNA binding and trans-activation of Drosophila STAT92E. Cell Signal. 2006;18:819–829. doi: 10.1016/j.cellsig.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Thiru A, et al. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004;23:489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, et al. Patterns and functions of STAT activation during Drosophila embryogenesis. Mech. Dev. 2003;120:1455–1468. doi: 10.1016/j.mod.2003.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li WX, Agaisse H, Mathey-Prevot B, Perrimon N. Differential requirement for STAT by gain-of-function and wild-type receptor tyrosine kinase Torso in Drosophila. Development. 2002;129:4241–4248. doi: 10.1242/dev.129.18.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sweitzer SM, Calvo S, Kraus MH, Finbloom DS, Larner AC. Characterization of a Stat-like DNA binding activity in Drosophila melanogaster. J. Biol. Chem. 1995;270:16510–16513. doi: 10.1074/jbc.270.28.16510. [DOI] [PubMed] [Google Scholar]
- 26.Dorer DR, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- 27.Ronsseray S, Boivin A, Anxolabehere D. P-Element repression in Drosophila melanogaster by variegating clusters of P-lacZ-white transgenes. Genetics. 2001;159:1631–1642. doi: 10.1093/genetics/159.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 STAT92E enhances repeat-mediated variegation. Eye pigmentation phenotypes of adult flies of indicated genotypes raised at 25°C. The light orange background was due to the presence of mini-w+ in the UAS-Stat92E+ transgene. Note the deceased pigmentation when UAS-Stat92E+ was expressed by the basal level of hsp70-Gal4 transgene (which does not carry mini-w+).
Figure S2 Loss of Stat92E+ does not affect HP1 expression in imaginal discs. A 3rd instar wing imaginal disc bearing Stat92E−/− cells (marked by lack of GFP) is shown for HP1 levels (stained by anti-HP1, red).
Figure S3 Long-term loss of STAT92E reduced HP1. A Stat92E homozygous loss of function cell (marked by GFP) was induced during embryogenesis and examined for HP1 (red) in late 3rd instar larval salivary gland.
Figure S4 Specificity of anti-STAT92E and anti-pSTAT92E. (a) A 3rd instar wing imaginal disc bearing Stat92E−/− cells (marked by lack of GFP) was stained by anti-STAT92E (red). Note that the red signals closely matches STAT92E protein levels, and that no signal is detected in Stat92E−/− cells. (b) S2 cells were transfected with Hop-V5 and/or STAT92E-V5. Total cell lysates were subjected to SDS-PAGE and transferred to nitrocellulose membrane and immunoblotted sequentially with anti-pSTAT92E, anti-V5, and anti-STAT92E, respectively. Note that anti-pSTAT92E does not cross-react with non-phospho-STAT92E (lack of signals in lane 1, 2, left panel), and that anti-STAT92E does not detect other proteins (right panel). Extra bands on the right panel were due to incomplete stripping. Anit-V5 detected non-specific bands (bands across all 3 lanes in middle panel).
Figure S5 HP1 and STAT92E distribution in hopGOF mutant larvae. Salivary glands isolated from hopGOF/+ (hopTum−l/+) larvae were stained with anti-HP1 (red) and anti-STAT92E (green). (a) In glands isolated from hopTum−l/+ larvae raised at 29°C, both HP1 and STAT92E appear diffused, lacking prominent heterochromatin foci. (b) In hopTum−l/+ larvae raised at 25°C, a moderately nonpermissive temperature1, a fraction of the nuclei (32%; n=64/198 nuclei) exhibited nearly normal patterns of HP1 foci (left panel), whereas in nearly all of these nuclei (95%; n=61/64 nuclei), STAT92E appeared dispersed and did not colocalize with HP1 (middle panel). These results suggest that STAT92E disperses in response to HopTum−l phosphorylation, and this process precedes changes in HP1 subnuclear localization.
Figure S6 Effects of H2O2/vanadate and CHX on STAT92E phosphorylation and histone H3 K9 methylation. S2 cells were cultured with or without CHX for 1 h, and then stimulated with H2O2/vanadate for indicated times (a), or incubated in the presence of CHX for indicated times (b). Cell extracts were blotted sequentially with anti-pSTAT92E and anti-STAT92E, or with anti-H3mK9, respectively. Membrane was stripped between blots.
Figure S7 Full-length protein gels. Note that Fig. 2f used a lower percentage gel and longer run, which resulted in a greater separation phospho-STAT92E from STAT92E bands (compared with Fig. 5b).