Abstract
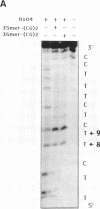
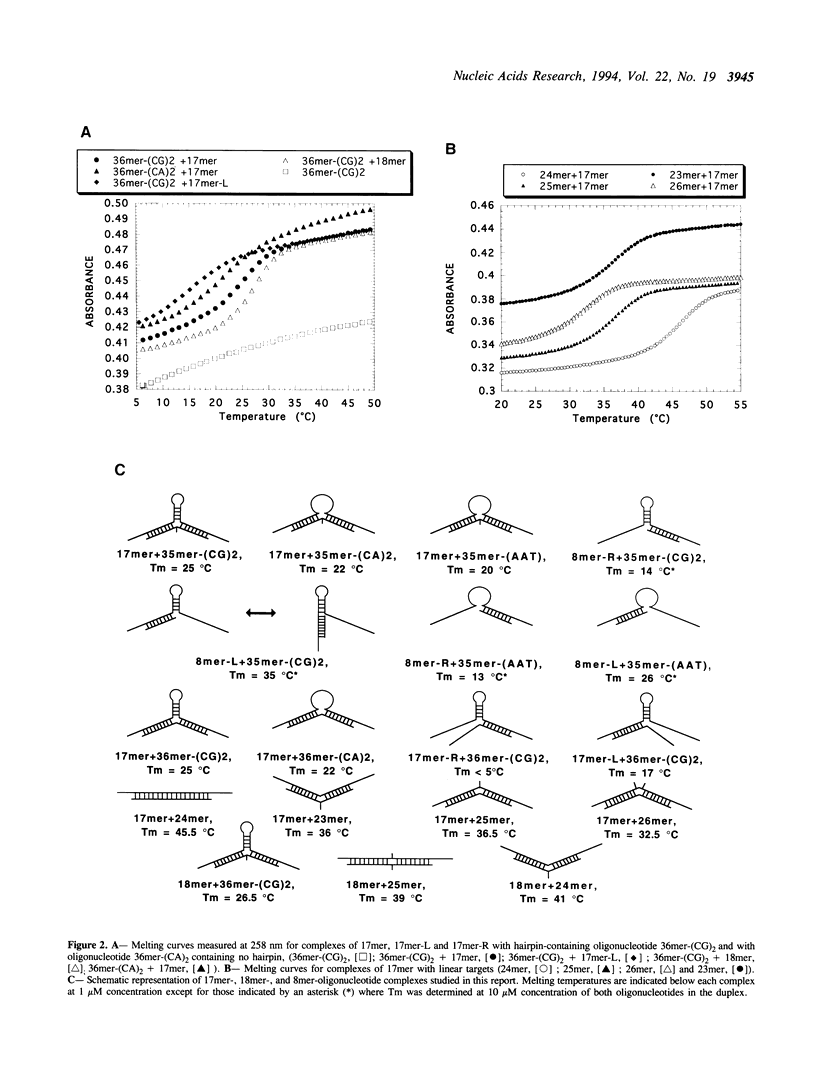
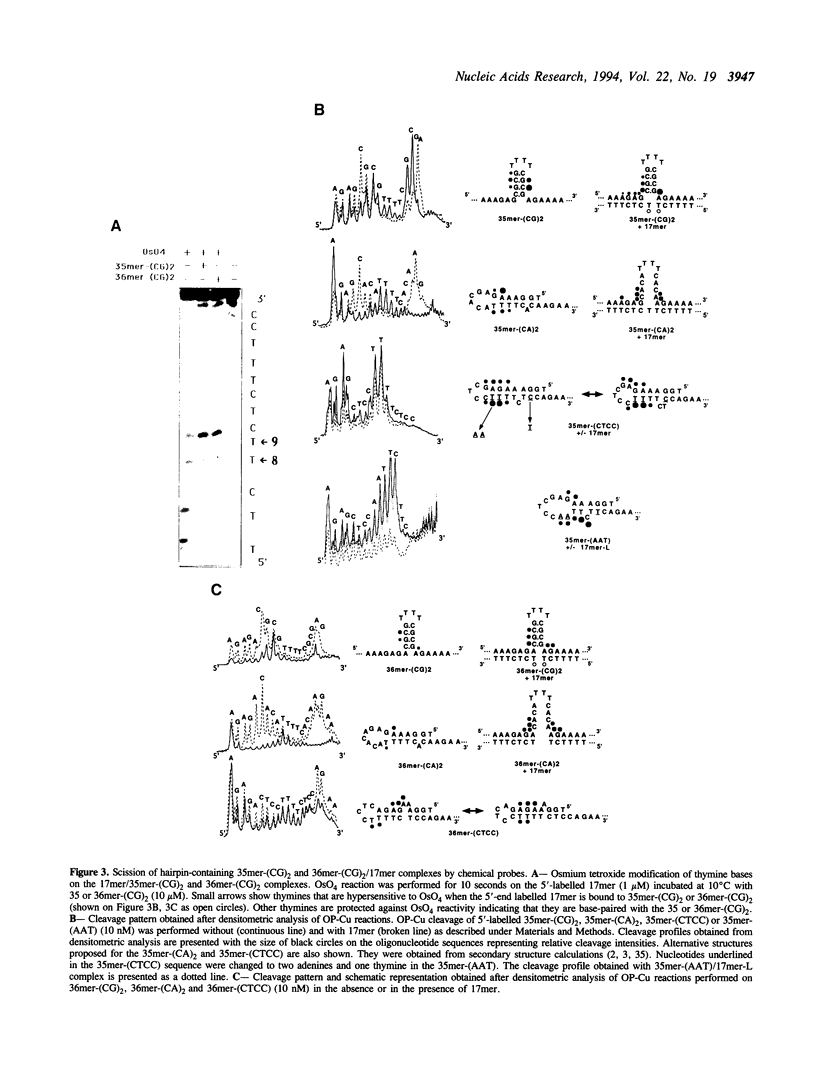
The possibility of designing antisense oligodeoxynucleotides complementary to non-adjacent single-stranded sequences containing hairpin structures was studied using a DNA model system. The structure and stability of complexes formed by a 17mer oligonucleotide with DNA fragments containing hairpin structures was investigated by spectroscopic measurements (melting curves) and chemical reactions (osmium tetroxide reaction, copper-phenanthroline cleavage). A three-way junction was formed when the oligonucleotide was bound to both sides of the hairpin structure. When the complementary sequences of the two parts of the oligonucleotide were separated by a sequence which could not form a hairpin, the oligonucleotide exhibited a slightly weaker binding than to the hairpin-containing target. An oligodeoxynucleotide-phenanthroline conjugate was designed to form Watson-Crick base pairs with two single-stranded regions flanking a hairpin structure in a DNA fragment. In the presence of Cu2+ ions and a reducing agent, two main cleavage sites were observed at the end of the duplex structure formed by the oligonucleotide-phenanthroline conjugate with its target sequence. Competition experiments showed that both parts of the oligonucleotide must be bound in order to observe sequence-specific cleavage. Cleavage was still observed with target sequences which could not form a hairpin, provided the reaction was carried out at lower temperatures. These results show that sequence-specific recognition and modification (cleavage) can be achieved with antisense oligonucleotides which bind to non-adjacent sequences in a single-stranded nucleic acid.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boulard Y., Cognet J. A., Gabarro-Arpa J., Le Bret M., Sowers L. C., Fazakerley G. V. The pH dependent configurations of the C.A mispair in DNA. Nucleic Acids Res. 1992 Apr 25;20(8):1933–1941. doi: 10.1093/nar/20.8.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor C. R., Warshaw M. M., Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9(9):1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- Dessen P., Fondrat C., Valencien C., Mugnier C. BISANCE: a French service for access to biomolecular sequence databases. Comput Appl Biosci. 1990 Oct;6(4):355–356. doi: 10.1093/bioinformatics/6.4.355. [DOI] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Duckett D. R., Lilley D. M. The three-way DNA junction is a Y-shaped molecule in which there is no helix-helix stacking. EMBO J. 1990 May;9(5):1659–1664. doi: 10.1002/j.1460-2075.1990.tb08286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Lilley D. M. The role of metal ions in the conformation of the four-way DNA junction. EMBO J. 1990 Feb;9(2):583–590. doi: 10.1002/j.1460-2075.1990.tb08146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova O. S., Podust L. M., Maksakova G. A., Gorn V. V., Knorre D. G. The influence of the target structure on the efficiency of alkylation of single-stranded DNA with the reactive derivatives of antisense oligonucleotides. FEBS Lett. 1992 May 4;302(1):47–50. doi: 10.1016/0014-5793(92)80281-k. [DOI] [PubMed] [Google Scholar]
- François J. C., Saison-Behmoaras T., Barbier C., Chassignol M., Thuong N. T., Hélène C. Sequence-specific recognition and cleavage of duplex DNA via triple-helix formation by oligonucleotides covalently linked to a phenanthroline-copper chelate. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9702–9706. doi: 10.1073/pnas.86.24.9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q., Lu M., Churchill M. E., Tullius T. D., Kallenbach N. R. Asymmetric structure of a three-arm DNA junction. Biochemistry. 1990 Dec 11;29(49):10927–10934. doi: 10.1021/bi00501a010. [DOI] [PubMed] [Google Scholar]
- Hunter W. N., Brown T., Anand N. N., Kennard O. Structure of an adenine-cytosine base pair in DNA and its implications for mismatch repair. Nature. 1986 Apr 10;320(6062):552–555. doi: 10.1038/320552a0. [DOI] [PubMed] [Google Scholar]
- Hunter W. N., Brown T., Kennard O. Structural features and hydration of a dodecamer duplex containing two C.A mispairs. Nucleic Acids Res. 1987 Aug 25;15(16):6589–6606. doi: 10.1093/nar/15.16.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc D. A., Morden K. M. Thermodynamic characterization of deoxyribooligonucleotide duplexes containing bulges. Biochemistry. 1991 Apr 23;30(16):4042–4047. doi: 10.1021/bi00230a031. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Palecek E. The supercoil-stabilised cruciform of ColE1 is hyper-reactive to osmium tetroxide. EMBO J. 1984 May;3(5):1187–1192. doi: 10.1002/j.1460-2075.1984.tb01949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima W. F., Monia B. P., Ecker D. J., Freier S. M. Implication of RNA structure on antisense oligonucleotide hybridization kinetics. Biochemistry. 1992 Dec 8;31(48):12055–12061. doi: 10.1021/bi00163a013. [DOI] [PubMed] [Google Scholar]
- Martinez H. M. An RNA secondary structure workbench. Nucleic Acids Res. 1988 Mar 11;16(5):1789–1798. doi: 10.1093/nar/16.5.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Sigman D. S. Chemical nucleases. Biochemistry. 1990 Oct 2;29(39):9097–9105. doi: 10.1021/bi00491a001. [DOI] [PubMed] [Google Scholar]
- Sigman D. S., Graham D. R., D'Aurora V., Stern A. M. Oxygen-dependent cleavage of DNA by the 1,10-phenanthroline . cuprous complex. Inhibition of Escherichia coli DNA polymerase I. J Biol Chem. 1979 Dec 25;254(24):12269–12272. [PubMed] [Google Scholar]
- Sowers L. C., Fazakerley G. V., Kim H., Dalton L., Goodman M. F. Variation of nonexchangeable proton resonance chemical shifts as a probe of aberrant base pair formation in DNA. Biochemistry. 1986 Jul 15;25(14):3983–3988. doi: 10.1021/bi00362a002. [DOI] [PubMed] [Google Scholar]
- Stull R. A., Taylor L. A., Szoka F. C., Jr Predicting antisense oligonucleotide inhibitory efficacy: a computational approach using histograms and thermodynamic indices. Nucleic Acids Res. 1992 Jul 11;20(13):3501–3508. doi: 10.1093/nar/20.13.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers T., Baker B. F., Cook P. D., Zounes M., Buckheit R. W., Jr, Germany J., Ecker D. J. Inhibition of HIV-LTR gene expression by oligonucleotides targeted to the TAR element. Nucleic Acids Res. 1991 Jun 25;19(12):3359–3368. doi: 10.1093/nar/19.12.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J. B., Duckett D. R., Lilley D. M. Structures of bulged three-way DNA junctions. Nucleic Acids Res. 1993 Sep 25;21(19):4548–4555. doi: 10.1093/nar/21.19.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werntges H., Steger G., Riesner D., Fritz H. J. Mismatches in DNA double strands: thermodynamic parameters and their correlation to repair efficiencies. Nucleic Acids Res. 1986 May 12;14(9):3773–3790. doi: 10.1093/nar/14.9.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M., Rashes M. S., Kallenbach N. R. Effect of T-T base mismatches on three-arm DNA junctions. Biochemistry. 1993 Jul 13;32(27):6898–6907. doi: 10.1021/bi00078a013. [DOI] [PubMed] [Google Scholar]
- Zhong M., Rashes M. S., Leontis N. B., Kallenbach N. R. Effects of unpaired bases on the conformation and stability of three-arm DNA junctions. Biochemistry. 1994 Mar 29;33(12):3660–3667. doi: 10.1021/bi00178a024. [DOI] [PubMed] [Google Scholar]
- Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]