Abstract
The sphingosine kinases, SK1 and SK2, produce the potent signaling lipid sphingosine-1-phosphate (S1P). These enzymes have garnered increasing interest for their roles in tumorigenesis, inflammation, vascular diseases, and immunity, as well as other functions. The sphingosine kinases are considered signaling enzymes by producing S1P, and their activity is acutely regulated by a variety of agonists. However, these enzymes are also key players in the control of sphingolipid metabolism. A variety of sphingolipids, such as sphingosine and the ceramides, are potent signaling molecules in their own right. The role of sphingosine kinases in regulating sphingolipid metabolism is potentially a critical aspect of their signaling function. A central aspect of signaling lipids is that their hydrophobic nature constrains them to membranes. Most enzymes of sphingolipid metabolism, including the enzymes that degrade S1P, are membrane enzymes. Therefore the localization of the sphingosine kinases and S1P is likely to be important in S1P signaling. Sphingosine kinase localization affects sphingolipid signaling in several ways. Translocation of SK1 to the plasma membrane promotes extracellular secretion of S1P. SK1 and SK2 localization to specific sites appears to direct S1P to intracellular protein effectors. SK localization also determines the access of these enzymes to their substrates. This may be an important mechanism for the regulation of ceramide biosynthesis by diverting dihydrosphingosine, a precursor in the ceramide biosynthetic pathway, from the de novo production of ceramide.
Keywords: Sphingolipid, Lipid signaling, Ceramide, Sphingosine-1-phosphate, Sphingosine
INTRODUCTION
Sphingosine-1-phosphate (S1P) has repeatedly lived up to its namesake, the sphinx, as an intriguing and enigmatic molecule. S1P, and the enzymes that produce it, the sphingosine kinases (SKs), have become of relatively recent interest, stemming largely from the seminal work of Olivera et al[1]. The explosion of research into their function has produced fascinating insights into the basic mechanisms of sphingolipid biosynthesis, metabolism, and signaling, and on the clinical importance of sphingosine-1-phoshate and the sphingosine kinases. This review focuses on the role of subcellular compartmentalization and translocation of the sphingosine kinases and S1P as a critical element of their function. Understanding lipid signaling requires embracing the concept that both the substrates and products of the signaling enzymes are lipophilic and therefore are incorporated in the membranes of the cell. This places particular constraints on how the substrates are accessed by the relevant enzymes and how the signaling products encounter their effectors. These effectors can be conventional cell surface receptors, as reviewed in other articles in this compendium, as well as by intracellular effectors that, in the case of S1P, are now just starting to be elucidated. There is another layer of control by sphingosine kinase that is now coming to light. The sphingolipids are metabolically inter-related (Figure 1). Sphingosine kinase and the enzymes that metabolize S1P are now recognized as key elements in the metabolic control of sphingolipid levels. In this review we will address how the localization of sphingosine kinase is as important as its level of activity, and how subcellular localization may affect the role that sphingosine kinase plays in controlling SK signaling and sphingolipid metabolism.
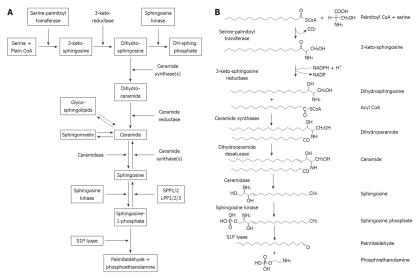
Figure 1.
The sphingolipid metabolic network. A: Schematic representation of the sphingolipid metabolic pathway. Note that the degradative enzymes for sphingosine kinase, the specific sphingosine-1-phosphate phosphatases SPP1/2 and the sphingosine-1-phosphate lyase are intrinsic membrane proteins of the endoplasmic reticulum. Similarly, the enzymes for ceramide biosynthesis, serine palmitoyl transferase, 3-keto sphingosine reductase, and the ceramide synthases, are intrinsic membrane enzymes of the endoplasmic reticulum; B: Structures of lipids in the sphingosine and ceramide biosynthetic pathway. Note that sphingosine is initially produced by the hydrolysis of ceramide by ceramidases. Dihydroceramide is directly produced by the de novo biosynthetic pathway of ceramide biosynthesis. Modified from Siow et al[44], 2010.
SPHINGOSINE KINASE-A SIGNALING ENZYME WITH A HIGH LEVEL OF BASAL ACTIVITY
Signaling vs metabolic function of sphingosine kinase
There are two isoforms of SK, generated from separate genes, SK-1 and SK-2[2]. SK-1 is more abundant in most cell types and has been the best studied. SK-1 is activated by a host of agonists, including growth factors and G-protein linked receptor activation[3]. SK-2 may also be activated in response to some agonists[3]. Most signaling enzymes are activated manyfold by agonist stimulation. In contrast, agonist stimulation of sphingosine kinase only elevates activity by 2- or 3-fold at the most[4]. Is this activation significant? Studies using a catalytically inactive form of SK-1 demonstrates that it is[5]. Treatment of cells with tumor necrosis factor (TNF), or the PKC activator PMA, leads to increased SK-1 activity. Overexpression of a catalytically inactive form of SK-1 (due to a single point mutation in the ATP binding site) blocks this activation. Remarkably, although activation is blocked, the basal activity of the activity is unaffected. This indicates that the inactive SK-1 interferes with the agonist-induced activation, but not with the activity, of the endogenous SK. Furthermore, while TNF activation leads to the activation of the MAP kinases ERK1/2 in control cells, this activation is blocked in cells overexpressing the inactive SK-1. This demonstrates that the activation of SK-1, as modest as it is, is critical for downstream signaling events. What, then, are we to make of the basal activity? As will be outlined below, the basal activity of SK may be required for a role entirely separate from that of signaling, as a metabolic regulator of sphingolipids. But how do we know that the basal activity is truly “basal” and not simply a reflection of a low level of stimulation of cells in culture?
Sphingosine kinase activation by phosphorylation
The evidence for a true basal activity of sphingosine kinase is three-pronged. First, the enzyme, as purified from rat or human tissues, has substantial activity[4,6]. Secondly, the enzyme can be produced in an active form in Escherichia coli and so presumably lacks any of the eukaryotic stimulating influences[4]. Finally, the agonist stimulation results in phosphorylation of SK-1 at serine225[7]. Mutation of serine225 to alanine ablates the ability of agonists to stimulate the activity of SK-1 when expressed in cells. However, this mutant retains basal activity. Recombinant SK-1 is activated in vitro by ERK2 by 14-fold when stoichiometrically phosphorylated. This conclusively demonstrates that SK-1 is an enzyme with both a substantial basal activity as well as a phosphorylation-dependent activated state.
PHOSPHORYLATION OF SK MEDIATES AGONIST-DEPENDENT TRANSLOCATION TO THE PLASMA MEMBRANE
Translocation of SK to the cell surface and cellular transformation
SK-1 transforms 3T3 fibroblasts when overexpressed[8]. The ability of SK-1 to promote growth in soft agar and in low serum is consistent with the large body of evidence correlating high levels of SK expression and tumorogenesis. This has been reviewed extensively elsewhere, including in this volume by James Van Brocklyn. The relationship between the transforming ability of SK and phosphorylation at serine225 was revealed in a fascinating series of experiments by Pitson et al[9]. They overexpressed both the wild-type and phosphorylation-deficient (serine225 to alanine mutant) form of SK-1 in 3T3 fibroblasts. The levels of SK enzyme activity in extracts from these two sets of cells were identical and manyfold above that of the endogenous enzyme. Strikingly, while the wild-type enzyme promoted transformation of the transfected 3T3 cells, expression of the phosphorylation-deficient mutant had no effect in promoting the transformed phenotype. Something about phosphorylation of SK, beyond simply enzymatic activity, is required for transformation. Ongoing studies indicated that SK-1 translocates, at least partially, to the cell surface upon agonist stimulation. Pitson et al[9] demonstrated that this translocation was dependent on phosphorylation at serine225. Could a block in translocation to the cell surface underlie the block in the ability of the serine225 mutant to transform 3T3 cells? To test this, a membrane targeting sequence was appended to the amino terminus of SK. This signal, derived from the kinase Lck, leads to constitutive plasma membrane targeting of SK. When tested in 3T3 cells, the phosphorylation-deficient form of SK, if artificially targeted to the plasma membrane, acquires the ability to readily transform 3T3 cells. These experiments illustrate that the activity of SK alone is not sufficient to promote pro-oncogenic signaling. Localization of the enzyme is an over-riding factor in this ability.
Localization and translocation of the sphingosine kinases
Localization of SK-1 and -2 in unstimulated cells: Neither isoform of sphingosine kinase has an obvious membrane anchoring sequence, and by this criterion is considered a cytosolic enzyme. Indeed, when cells are analyzed by subcellular fractionation, the bulk of SK-1 remains in the cytosol fraction[10]. Similarly, the purified protein is completely soluble[4]. However, a small percentage of SK-1 is associated with the membrane fraction, even from resting cells. In mast cells, a portion of SK-1 appears to associate with the actin cytoskeleton[11].
In the absence of stimulation, therefore, SK-1 is a predominantly cytosolic enzyme. One group has proposed that this cytoplasmic localization is the steady state result of shuttling between the nucleus and the cytoplasm, due to active import and export signals in SK-1[12]. There have not been reports, however, of conditions which lead to a nuclear localization of SK-1. This contrasts with reports of a nuclear localization of SK-2. SK-2 is found both in the cytoplasm and the nucleus[13,14]. SK-2 also shuttles between the cytoplasm and the nucleus, but the steady-state localization is tipped such that nuclear localization is evident. This suggests that the rate of nuclear export is somewhat lower for SK-2 than SK-1. Indeed, as outlined below, regulation of the rate of SK-2 export from the nucleus shifts its distribution.
Even though SK-1 and, in part, SK-2, are cytosolic enzymes, there is evidence that they demonstrate an organelle-specific interaction with the endoplasmic reticulum (ER)[15]. It should be remembered that substrate for these enzymes, sphingosine, is associated with membranes, and SK encounters its substrate in the context of a membrane. The ER is an extensive network of membranes and even without any specific targeting to the ER, simply by mass action, this is the membrane that would be most often encountered by soluble SKs. The behavior of SK-1 with respect to utilization of substrate mirrors that of endoplasmic-reticulum anchored SK-1[15]. This would also be expected of the cytosolic fraction of SK-2.
Extracellular SKs: Two reports suggest that SKs may have a role outside of cells. Ancellin et al[16] have detected a minor amount of SK release from cells. They propose that this may promote the formation of extracellular S1P. SK-2 has been reported to be released from apoptotic cells resulting from a caspase-1 induced cleavage at the amino-terminus[17]. In the latter studies, it is difficult to determine if this release is simply due to disruption of the cell membrane, but it is interesting that the release depends on truncation of the amino-terminus.
Translocation of SK-1: SK-1 translocates to the plasma membrane upon activation by a number of agonists[3]. This translocation is the result of serine225 phosphorylation by Erk1/2 or a related kinase[9]. The precise mechanism of translocation is not clear. In vitro studies demonstrates that SK prefers to associate with the anionic phospholipids characteristic of the inner leaflet of the plasma membrane[18]. Changes in the composition of that leaflet may influence the translocation of SK. Those studies also demonstrate that serine225 phosphorylation increases the affinity of SK-1 with anionic lipids, consistent with the role of that phosphorylation in the promoting translocation. Recently the role of calcium and integrin binding protein-1 (CIB1) in SK-1 translocation has been established[19]. siRNA knockdown of CIB1 inhibits SK-1 translocation. The details of how CIB1 mediates this movement remain to be elucidated, and the relationship to the lipid-binding studies of Stahelin and colleagues remains to be understood. Not all SK-1 translocation depends on serine225 phosphorylation, however. SK-1 robustly translocates to the plasma membrane in response to carbochol stimulation of the M3 muscarinic receptor[20]. This translocation is independent of serine 225 phosphorylation. SK also translocates to the membranes of forming phagosomes in macrophages in response to uptake of killed mycobacterium or latex beads[21]. This translocation also does not require the phosphorylation of serine225 as does plasma membrane translocation. Clearly, the translocation mechanism of SK is incompletely understood.
Translocation of SK-2: SK-2 is also phosphorylated in response to agonist treatment. In this case, however, the translocation is from a nuclear localization into the cytoplasm. Phosphorylation, which is mediated by protein kinase D, occurs in a nuclear export region[22]. As mentioned, SK-2 appears to constantly shuttle into and out of the nucleus. Disruption of the nuclear export signal by protein kinase D-mediated phosphorylation therefore leads to a predominantly cytoplasmic localization.
THE ROLE OF SUBCELLULAR COMPARTMENTALIZATION AND TRANSLOCATION IN SK SIGNALING
In principle, the SK localization could determine the function of SK in signaling in several ways. First, localization to the plasma membrane could enhance the secretion of S1P. Secondly, localization to the endoplasmic reticulum could affect the downstream metabolism of S1P. Thirdly, localization to specific intracellular sites could promote association with intracellular targets. Finally, SK localization could enhance the utilization of specific pools of its substrate, sphingosine. Below, we will discuss each of these potential roles of SK localization and translocation.
TRANSLOCATION OF SK TO THE SURFACE PROMOTES S1P SECRETION
The best characterized role of S1P as a signaling molecule is the engagement of the 5 G-protein-linked receptors of the S1P family as reviewed in other contributions to this volume. The SKs (with the possible exception of the extracellular forms, mentioned above) are intracellular enzymes. How, then, does S1P get out of the cell? The mechanism is still incompletely understood. However, members of the ABC transporter family[23-25], as well as a novel protein of the Drosophila spinster-like family of proteins, have been implicated as ATP-drive exporters of S1P[26]. These are all cell surface transporters and so it is reasonable to expect that translocation of SK to the plasma membrane would enhance S1P secretion. This is, in fact, what is found. For example, the PKC activator PMA enhances secretion of S1P by approximately 4-fold over the stimulation of total S1P production[7,19]. One role of the stimulated translocation of SK to the surface is clearly, therefore, to enhance secretion.
LOCALIZATION OF SK DOES NOT AFFECT THE DOWNSTREAM METABOLISM OF S1P
There are two sets of enzymes that degrade S1P. There are two S1P-specific phosphatases, the SPPs[27]. Dephosphorylation of S1P results in the generation of sphingosine, which can either be incorporated into ceramide by a salvage pathway, or re-utilized by SK to regenerate S1P. Irreversible degradation of S1P is mediated by an S1P-specific lyase, which generates phosphoethanolamine and palmitaldehyde[28]. This is a critical enzyme because it mediates the only pathway in the cell for the elimination of the sphingosine backbone. Genetic ablation of the S1P lyase in mice has dramatic effects, leading to shortened lifespan and profound changes in lipid metabolism. Prominent among these is an increase in ceramides, sphingosine, and, as expected, S1P[29]. The SPPs[30,31] and the S1P lyase[32,33] are membrane enzymes, localized to the ER. It might be expected that localization of SK to the endoplasmic reticulum would enhance degradation of S1P. Conversely it would be predicted that localization of SK to the plasma membrane would reduce degradation of S1P, by segregating the S1P produced at that site from the degradative enzymes in the ER. Surprisingly, however, the localization of SK has no noticeable effect on the subsequent fate of S1P[15]. The most straightforward explanation for this observation is that the rate of S1P transport through the cell must be rapid in relation to the rate of S1P downstream metabolism.
DIRECTING S1P TO INTRACELLULAR EFFECTORS BY SITE-SPECIFIC TARGETING OF SK
Despite the conclusion that S1P can rapidly move throughout the cell from its site of synthesis, there is evidence that localizing SK to specific sites can promote the association of S1P with specific intracellular effectors. This is best illustrated for the nuclear localization of SK2 and its effect on the histone deacetylases HDAC-1 and -2. Hait et al[34] have recently shown that S1P is an inhibitor of these histone deacetylases. The S1P that affects HDAC activity is generated by SK2, but not SK-1. Remarkably, they found that the localization of SK2 to the nucleus is required for HDAC inhibition. They showed directly that, when SK2 is directed to the cytosol (by blocking SK2 nuclear localization), S1P levels in the nucleus drop. This indicates that while S1P may move rapidly between cytosolic membranes, its movement into and out of the nucleus is restricted. Similarly, Alvarez et al[35] has also reported that S1P is an effector of Traf2. Traf2 is a component of the TNF receptor signaling complex. Data from this group suggests that S1P is a required co-factor to stimulate Traf2’s activity as an ubiquitin ligase. Their data further suggests that specific targeting of SK1 to the TNF receptor complex promotes the binding of S1P to Traf2. These observations again suggest that site-specific targeting of SK can, in some situations, promote the formation of high local concentrations of S1P required to interact with intracellular effector proteins.
SK LOCALIZATION DETERMINES SUBSTRATE UTILIZATION
An underappreciated principle in the control of S1P production is that the substrate for SK, sphingosine, is subsaturating[36]. This means that increasing access of SK to sphingosine, either by increasing overall levels of sphingosine or by moving SK to a site where sphingosine levels are high, will increase S1P output without changing SK activity per se. For example, adding sphingosine to cells increases S1P production[36]. More physiologically, stimulation of sphingosine production by increasing ceramidase levels (to cleave ceramide to sphingosine) raises S1P levels[37]. Does SK localization affect the access that it has to substrate? The subcellular localization of sphingosine has not been conclusively determined, although it is presumed that a proportion resides in lysosomes[38]. However it would not be unexpected that sphingosine might be compartmentalized. Direct evidence that localization of SK affects the pools of sphingosine to which it has access comes from a series of studies from several laboratories which indicate an unexpected role for SK in controlling levels of the pro-apoptotic lipid, ceramide[15,39,40]. High levels of SK-1 expression lead to moderate increases in S1P levels. However, a much more dramatic increase in dihydrosphingosine-1-phosphate levels was observed. What is the implication of SK utilization of dihydrosphingosine as a substrate? Dihydrosphingosine is a precursor of (dihydro)ceramide in the ceramide biosynthetic pathway (Figure 1). Sphingosine is only produced by hydrolysis of ceramide because the 2, 3-double bond is only introduced when dihydrosphingosine is incorporated into dihydroceramide. These data demonstrate that SK can divert dihydrosphingosine from the ceramide biosynthetic pathway. Moreover, localization of SK has a strong influence on this effect. When SK is targeted to the plasma membrane, it no longer utilizes significant quantities of dihydrosphingosine[15]. This result is not entirely surprising. All of the enzymes involved in ceramide biosynthesis; the rate limiting and initiating enzyme serine palmitoyltransferases, 3-keto reductase (which produces dihydrosphingosine from 3-ketosphingosine), and the ceramide synthases, are localized to the endoplasmic reticulum[41]. Both an ER-targeted form of SK1 and the native, untargeted SK1 avidly use dihydrosphingosine. This indicates that even without obvious ER binding, SK1 encounters the ER constantly. Therefore, localizing SK to the plasma membrane would segregate it from the site of dihydrosphingosine production.
Two important points arise from these series of experiments. First, SK appears to control ceramide biosynthesis by utilizing precursors in this pathway. A number of chemotherapeutic agents induce cancer cell apoptosis by stimulating de novo synthesis of ceramide[42,43]. The ability of SK to interfere with ceramide biosynthesis may be an important component in its action to promote the growth, survival, and chemoresistance of cancer cells. Secondly, these studies indicate that the localization of SK is a key regulator of the effect of SK on sphingolipid metabolism.
CONCLUSION AND FUTURE DIRECTIONS
The localization and translocation of both SK1 and SK2 has clear consequences for the ability of these enzymes to effect cell life and death. The mechanisms that control the subcellular localization of the SKs, and the function of these enzymes at these sites, remain a fertile area for future exploration. The dual roles of the SKs, like many lipid signaling enzymes, as both producers of lipid signaling molecules and modulators of lipid metabolic pathways, poses a challenge to understanding the biological roles of these enzymes. The increasing use of overexpression and genetic knockout of lipid signaling enzymes in both cells and animals has expanded our understanding of the role of these enzymes in a number of physiological and pathophysiological settings. However, the interpretation of these studies is complicated by an increasingly sophisticated view of the multiple roles these signaling enzymes play. For example, overexpression or knockdown of the SK’s can no longer be interpreted by the assumption that the effects are solely due to changes in S1P levels without also considering the effects on the metabolism of other sphingolipids. The ability to make accurate and sensitive measurements of lipid levels by mass spectroscopy is an essential tool to complete the picture. These analyses will have to be combined with artful manipulations of sphingolipid metabolic pathways to complete the picture of signaling mediated by these enigmatic lipids and the enzymes that control them.
Footnotes
Supported by The James Graham Brown Cancer Center and NIH, No. CA111987
Peer reviewers: Jiandi Zhang, PhD, 2108 Clarice Lane, Burlingame, CA 94010, United States; Dr. Rui Wu, Department of Chemistry, Boston University, 590 Commonwealth Ave. Rm299, Boston, MA 02215, United States
S- Editor Cheng JX L- Editor Negro F E- Editor Zheng XM
References
- 1.Olivera A, Kohama T, Edsall L, Nava V, Cuvillier O, Poulton S, Spiegel S. Sphingosine kinase expression increases intracellular sphingosine-1-phosphate and promotes cell growth and survival. J Cell Biol. 1999;147:545–558. doi: 10.1083/jcb.147.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu H, Sugiura M, Nava VE, Edsall LC, Kono K, Poulton S, Milstien S, Kohama T, Spiegel S. Molecular cloning and functional characterization of a novel mammalian sphingosine kinase type 2 isoform. J Biol Chem. 2000;275:19513–19520. doi: 10.1074/jbc.M002759200. [DOI] [PubMed] [Google Scholar]
- 3.Wattenberg BW, Pitson SM, Raben DM. The sphingosine and diacylglycerol kinase superfamily of signaling kinases: localization as a key to signaling function. J Lipid Res. 2006;47:1128–1139. doi: 10.1194/jlr.R600003-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Pitson SM, D'andrea RJ, Vandeleur L, Moretti PA, Xia P, Gamble JR, Vadas MA, Wattenberg BW. Human sphingosine kinase: purification, molecular cloning and characterization of the native and recombinant enzymes. Biochem J. 2000;350 Pt 2:429–441. [PMC free article] [PubMed] [Google Scholar]
- 5.Pitson SM, Moretti PA, Zebol JR, Xia P, Gamble JR, Vadas MA, D'Andrea RJ, Wattenberg BW. Expression of a catalytically inactive sphingosine kinase mutant blocks agonist-induced sphingosine kinase activation. A dominant-negative sphingosine kinase. J Biol Chem. 2000;275:33945–33950. doi: 10.1074/jbc.M006176200. [DOI] [PubMed] [Google Scholar]
- 6.Olivera A, Kohama T, Tu Z, Milstien S, Spiegel S. Purification and characterization of rat kidney sphingosine kinase. J Biol Chem. 1998;273:12576–12583. doi: 10.1074/jbc.273.20.12576. [DOI] [PubMed] [Google Scholar]
- 7.Pitson SM, Moretti PA, Zebol JR, Lynn HE, Xia P, Vadas MA, Wattenberg BW. Activation of sphingosine kinase 1 by ERK1/2-mediated phosphorylation. EMBO J. 2003;22:5491–5500. doi: 10.1093/emboj/cdg540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia P, Gamble JR, Wang L, Pitson SM, Moretti PA, Wattenberg BW, D'Andrea RJ, Vadas MA. An oncogenic role of sphingosine kinase. Curr Biol. 2000;10:1527–1530. doi: 10.1016/s0960-9822(00)00834-4. [DOI] [PubMed] [Google Scholar]
- 9.Pitson SM, Xia P, Leclercq TM, Moretti PA, Zebol JR, Lynn HE, Wattenberg BW, Vadas MA. Phosphorylation-dependent translocation of sphingosine kinase to the plasma membrane drives its oncogenic signalling. J Exp Med. 2005;201:49–54. doi: 10.1084/jem.20040559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohama T, Olivera A, Edsall L, Nagiec MM, Dickson R, Spiegel S. Molecular cloning and functional characterization of murine sphingosine kinase. J Biol Chem. 1998;273:23722–23728. doi: 10.1074/jbc.273.37.23722. [DOI] [PubMed] [Google Scholar]
- 11.Kusner DJ, Thompson CR, Melrose NA, Pitson SM, Obeid LM, Iyer SS. The localization and activity of sphingosine kinase 1 are coordinately regulated with actin cytoskeletal dynamics in macrophages. J Biol Chem. 2007;282:23147–23162. doi: 10.1074/jbc.M700193200. [DOI] [PubMed] [Google Scholar]
- 12.Inagaki Y, Li PY, Wada A, Mitsutake S, Igarashi Y. Identification of functional nuclear export sequences in human sphingosine kinase 1. Biochem Biophys Res Commun. 2003;311:168–173. doi: 10.1016/j.bbrc.2003.09.194. [DOI] [PubMed] [Google Scholar]
- 13.Igarashi N, Okada T, Hayashi S, Fujita T, Jahangeer S, Nakamura S. Sphingosine kinase 2 is a nuclear protein and inhibits DNA synthesis. J Biol Chem. 2003;278:46832–46839. doi: 10.1074/jbc.M306577200. [DOI] [PubMed] [Google Scholar]
- 14.Sankala HM, Hait NC, Paugh SW, Shida D, Lépine S, Elmore LW, Dent P, Milstien S, Spiegel S. Involvement of sphingosine kinase 2 in p53-independent induction of p21 by the chemotherapeutic drug doxorubicin. Cancer Res. 2007;67:10466–10474. doi: 10.1158/0008-5472.CAN-07-2090. [DOI] [PubMed] [Google Scholar]
- 15.Siow DL, Anderson CD, Berdyshev EV, Skobeleva A, Pitson SM, Wattenberg BW. Intracellular localization of sphingosine kinase 1 alters access to substrate pools but does not affect the degradative fate of sphingosine-1-phosphate. J Lipid Res. 2010;51:2546–2559. doi: 10.1194/jlr.M004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ancellin N, Colmont C, Su J, Li Q, Mittereder N, Chae SS, Stefansson S, Liau G, Hla T. Extracellular export of sphingosine kinase-1 enzyme. Sphingosine 1-phosphate generation and the induction of angiogenic vascular maturation. J Biol Chem. 2002;277:6667–6675. doi: 10.1074/jbc.M102841200. [DOI] [PubMed] [Google Scholar]
- 17.Weigert A, Cremer S, Schmidt MV, von Knethen A, Angioni C, Geisslinger G, Brüne B. Cleavage of sphingosine kinase 2 by caspase-1 provokes its release from apoptotic cells. Blood. 2010;115:3531–3540. doi: 10.1182/blood-2009-10-243444. [DOI] [PubMed] [Google Scholar]
- 18.Stahelin RV, Hwang JH, Kim JH, Park ZY, Johnson KR, Obeid LM, Cho W. The mechanism of membrane targeting of human sphingosine kinase 1. J Biol Chem. 2005;280:43030–43038. doi: 10.1074/jbc.M507574200. [DOI] [PubMed] [Google Scholar]
- 19.Jarman KE, Moretti PA, Zebol JR, Pitson SM. Translocation of sphingosine kinase 1 to the plasma membrane is mediated by calcium- and integrin-binding protein 1. J Biol Chem. 2010;285:483–492. doi: 10.1074/jbc.M109.068395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ter Braak M, Danneberg K, Lichte K, Liphardt K, Ktistakis NT, Pitson SM, Hla T, Jakobs KH, Meyer zu Heringdorf D. Galpha(q)-mediated plasma membrane translocation of sphingosine kinase-1 and cross-activation of S1P receptors. Biochim Biophys Acta. 2009;1791:357–370. doi: 10.1016/j.bbalip.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 21.Thompson CR, Iyer SS, Melrose N, VanOosten R, Johnson K, Pitson SM, Obeid LM, Kusner DJ. Sphingosine kinase 1 (SK1) is recruited to nascent phagosomes in human macrophages: inhibition of SK1 translocation by Mycobacterium tuberculosis. J Immunol. 2005;174:3551–3561. doi: 10.4049/jimmunol.174.6.3551. [DOI] [PubMed] [Google Scholar]
- 22.Ding G, Sonoda H, Yu H, Kajimoto T, Goparaju SK, Jahangeer S, Okada T, Nakamura S. Protein kinase D-mediated phosphorylation and nuclear export of sphingosine kinase 2. J Biol Chem. 2007;282:27493–27502. doi: 10.1074/jbc.M701641200. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi N, Kobayashi N, Yamaguchi A, Nishi T. Characterization of the ATP-dependent sphingosine 1-phosphate transporter in rat erythrocytes. J Biol Chem. 2009;284:21192–21200. doi: 10.1074/jbc.M109.006163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitra P, Oskeritzian CA, Payne SG, Beaven MA, Milstien S, Spiegel S. Role of ABCC1 in export of sphingosine-1-phosphate from mast cells. Proc Natl Acad Sci USA. 2006;103:16394–16399. doi: 10.1073/pnas.0603734103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nieuwenhuis B, Lüth A, Chun J, Huwiler A, Pfeilschifter J, Schäfer-Korting M, Kleuser B. Involvement of the ABC-transporter ABCC1 and the sphingosine 1-phosphate receptor subtype S1P(3) in the cytoprotection of human fibroblasts by the glucocorticoid dexamethasone. J Mol Med. 2009;87:645–657. doi: 10.1007/s00109-009-0468-x. [DOI] [PubMed] [Google Scholar]
- 26.Osborne N, Brand-Arzamendi K, Ober EA, Jin SW, Verkade H, Holtzman NG, Yelon D, Stainier DY. The spinster homolog, two of hearts, is required for sphingosine 1-phosphate signaling in zebrafish. Curr Biol. 2008;18:1882–1888. doi: 10.1016/j.cub.2008.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pyne S, Long JS, Ktistakis NT, Pyne NJ. Lipid phosphate phosphatases and lipid phosphate signalling. Biochem Soc Trans. 2005;33:1370–1374. doi: 10.1042/BST0331370. [DOI] [PubMed] [Google Scholar]
- 28.Fyrst H, Saba JD. Sphingosine-1-phosphate lyase in development and disease: sphingolipid metabolism takes flight. Biochim Biophys Acta. 2008;1781:448–458. doi: 10.1016/j.bbalip.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bektas M, Allende ML, Lee BG, Chen W, Amar MJ, Remaley AT, Saba JD, Proia RL. Sphingosine 1-phosphate lyase deficiency disrupts lipid homeostasis in liver. J Biol Chem. 2010;285:10880–10889. doi: 10.1074/jbc.M109.081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Stunff H, Galve-Roperh I, Peterson C, Milstien S, Spiegel S. Sphingosine-1-phosphate phosphohydrolase in regulation of sphingolipid metabolism and apoptosis. J Cell Biol. 2002;158:1039–1049. doi: 10.1083/jcb.200203123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogawa C, Kihara A, Gokoh M, Igarashi Y. Identification and characterization of a novel human sphingosine-1-phosphate phosphohydrolase, hSPP2. J Biol Chem. 2003;278:1268–1272. doi: 10.1074/jbc.M209514200. [DOI] [PubMed] [Google Scholar]
- 32.Ikeda M, Kihara A, Igarashi Y. Sphingosine-1-phosphate lyase SPL is an endoplasmic reticulum-resident, integral membrane protein with the pyridoxal 5'-phosphate binding domain exposed to the cytosol. Biochem Biophys Res Commun. 2004;325:338–343. doi: 10.1016/j.bbrc.2004.10.036. [DOI] [PubMed] [Google Scholar]
- 33.Reiss U, Oskouian B, Zhou J, Gupta V, Sooriyakumaran P, Kelly S, Wang E, Merrill AH Jr, Saba JD. Sphingosine-phosphate lyase enhances stress-induced ceramide generation and apoptosis. J Biol Chem. 2004;279:1281–1290. doi: 10.1074/jbc.M309646200. [DOI] [PubMed] [Google Scholar]
- 34.Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 2009;325:1254–1257. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, Maceyka M, Jiang H, Luo C, Kordula T, et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465:1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olivera A, Spiegel S. Sphingosine-1-phosphate as second messenger in cell proliferation induced by PDGF and FCS mitogens. Nature. 1993;365:557–560. doi: 10.1038/365557a0. [DOI] [PubMed] [Google Scholar]
- 37.Sun W, Jin J, Xu R, Hu W, Szulc ZM, Bielawski J, Obeid LM, Mao C. Substrate specificity, membrane topology, and activity regulation of human alkaline ceramidase 2 (ACER2) J Biol Chem. 2010;285:8995–9007. doi: 10.1074/jbc.M109.069203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kågedal K, Zhao M, Svensson I, Brunk UT. Sphingosine-induced apoptosis is dependent on lysosomal proteases. Biochem J. 2001;359:335–343. doi: 10.1042/0264-6021:3590335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berdyshev EV, Gorshkova IA, Usatyuk P, Zhao Y, Saatian B, Hubbard W, Natarajan V. De novo biosynthesis of dihydrosphingosine-1-phosphate by sphingosine kinase 1 in mammalian cells. Cell Signal. 2006;18:1779–1792. doi: 10.1016/j.cellsig.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 40.Taha TA, Kitatani K, El-Alwani M, Bielawski J, Hannun YA, Obeid LM. Loss of sphingosine kinase-1 activates the intrinsic pathway of programmed cell death: modulation of sphingolipid levels and the induction of apoptosis. FASEB J. 2006;20:482–484. doi: 10.1096/fj.05-4412fje. [DOI] [PubMed] [Google Scholar]
- 41.Mandon EC, Ehses I, Rother J, van Echten G, Sandhoff K. Subcellular localization and membrane topology of serine palmitoyltransferase, 3-dehydrosphinganine reductase, and sphinganine N-acyltransferase in mouse liver. J Biol Chem. 1992;267:11144–11148. [PubMed] [Google Scholar]
- 42.Perry DK. The role of de novo ceramide synthesis in chemotherapy-induced apoptosis. Ann N Y Acad Sci. 2000;905:91–96. doi: 10.1111/j.1749-6632.2000.tb06541.x. [DOI] [PubMed] [Google Scholar]
- 43.Perry DK, Carton J, Shah AK, Meredith F, Uhlinger DJ, Hannun YA. Serine palmitoyltransferase regulates de novo ceramide generation during etoposide-induced apoptosis. J Biol Chem. 2000;275:9078–9084. doi: 10.1074/jbc.275.12.9078. [DOI] [PubMed] [Google Scholar]
- 44.Siow DL, Anderson CD, Berdyshev EV, Skobeleva A, Natarajan V, Pitson SM, Wattenberg BW. Sphingosine kinase localization in the control of sphingolipid metabolism. Adv Enzyme Regul. 2010:Epub ahead of print. doi: 10.1016/j.advenzreg.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]