Abstract
The internalization of essential nutrients, lipids and receptors is a crucial process for all eukaryotic cells. Accordingly, endocytosis is highly conserved across cell types and species. Once internalized, small cargo-containing vesicles fuse with early endosomes (also known as sorting endosomes), where they undergo segregation to distinct membrane regions and are sorted and transported on through the endocytic pathway. Although the mechanisms that regulate this sorting are still poorly understood, some receptors are directed to late endosomes and lysosomes for degradation, whereas other receptors are recycled back to the plasma membrane; either directly or through recycling endosomes. The Rab family of small GTP-binding proteins plays crucial roles in regulating these trafficking pathways. Rabs cycle from inactive GDP-bound cytoplasmic proteins to active GTP-bound membrane-associated proteins, as a consequence of the activity of multiple specific GTPase-activating proteins (GAPs) and GTP exchange factors (GEFs). Once bound to GTP, Rabs interact with a multitude of effector proteins that carry out Rab-specific functions. Recent studies have shown that some of these effectors are also interaction partners for the C-terminal Eps15 homology (EHD) proteins, which are also intimately involved in endocytic regulation. A particularly interesting example of common Rab-EHD interaction partners is the MICAL-like protein, MICAL-L1. MICAL-L1 and its homolog, MICAL-L2, belong to the larger MICAL family of proteins, and both have been directly implicated in regulating endocytic recycling of cell surface receptors and junctional proteins, as well as controlling cytoskeletal rearrangement and neurite outgrowth. In this review, we summarize the functional roles of MICAL and Rab proteins, and focus on the significance of their interactions and the implications for endocytic transport.
Keywords: Rab, MICAL, Eps15 homology, Endosomes, Endocytosis, Trafficking, Cytoskeleton
INTRODUCTION
Internalization of extracellular materials, plasma membrane proteins, and lipids is a highly conserved cellular process. Small molecules, such as ions, amino acids, and sugars can cross the plasma membrane through channels or with the aid of pumps, whereas macromolecules must be carried into the cells through endocytosis, a process in which a small region of the plasma membrane invaginates and pinches off to form an endocytic vesicle.
Upon internalization, cargo-containing vesicles are transported to early endosomes (EE). Depending on the ultimate destination, some cargo, such as the epidermal growth factor receptor (EGFR), is delivered to lysosomes for degradation. These receptors contain sorting signals in their cytosolic tails that facilitate recruitment of sorting machinery that directs the receptor to late endosomal membranes. In turn, the late endosomes (LEs) fuse with lysosomes that contain proteolytic enzymes that are required for the degradation of receptors. In contrast, cargo that does not immediately undergo degradation is often recycled back to the plasma membrane, either directly (fast recycling) or through recycling endosomes (slow recycling).
REGULATION OF ENDOCYTIC TRAFFICKING PATHWAYS BY RAB PROTEINS AND THEIR EFFECTORS
Of the many proteins that are involved in the regulation of the various trafficking pathways, the small Ras-like Rab protein family plays an especially significant role. Rab-GTP binding proteins function as molecular switches that are either in the GTP-bound “on” form or the GDP-bound “off” form[1]. A variety of specific guanine nucleotide exchange factors (GEFs) catalyze the exchange of GDP with GTP. Upon GTP-binding, Rabs can interact with a range of specific molecular effectors that include adaptors, tethering molecules, kinases, phosphatases, and motor proteins. Rab proteins function in vesicle targeting, tethering and fusion. Upon vesicular fusion with the target membrane, Rab-GTPs are then hydrolyzed to Rab-GDP and recycled to the cytoplasm. GTP to GDP hydrolysis is facilitated by GTPase-activating proteins (GAPs)[1]. Although the GDP-bound Rabs are considered inactive, there are a small number of selective Rab effectors that also bind to them.
Rab5: a master regulator of trafficking via EEs
More than 60 Rab proteins regulate distinct trafficking pathways in mammalian cells[2]. One of the best-characterized Rab proteins is Rab5. It localizes to EEs, phagosomes, caveosomes, and the plasma membrane. Accordingly, it is involved in endocytosis, fusion of clathrin-coated vesicles, macropinocytosis, and the maturation of early phagosomes.
Upon activation by its GEF known as Rabex5[3,4], Rab5 can recruit many of its molecular effectors, such as Rabaptin5[5], phosphatidylinositol-3-kinase (PI3K)[6], early endosomal antigen 1 (EEA1)[7], and Rabenosyn-5[8]. Rab5 can interact with Rabaptin5, which in turn binds to Rabex5 and stabilizes GTP-bound Rab5 on EE membranes[1]. Subsequently, Rab5 can recruit other effectors, such as PI3K. Activation of PI3K leads to phosphorylation of phosphatidylinositol (PI) and its conversion to phosphatidylinositol 3-phosphate (PI3P) on EE membranes[9,10]. EEA1 and Rabenosyn-5 are then recruited to the PI3P-enriched membrane through their FYVE (Fab1, YOTB, Vac1, and EEA1) zinc finger domains, regions that specifically bind to PI3P[8,11-13]. EEA1 interacts with both Syntaxin 13[14] and Syntaxin 6[15], two soluble NSF attachment receptor (SNARE) proteins that regulate vesicular fusion, and facilitate the fusion of transport vesicles with recycling endosomes (REs) and the Golgi, respectively. Like EEA1, Rabenosyn-5 mediates transport of EE-derived vesicles to REs and the Golgi[16,17]. The function of Rabenosyn-5 depends upon its interaction with human vacuolar protein sorting 45 (hVps45), a member of the Sec1/Munc18 protein family that is involved in vesicular fusion through its interaction with SNARE proteins[18]. Rabenosyn-5 also interacts with EHD3, a member of the C-terminal Eps15 homology domain (EHD)-containing protein family that facilitates transport from the EEs[16]. Rabenosyn-5 regulates vesicular transport of transferrin and MHC class I from EEs to the REs through its interaction with EHD1, another member of the EHD protein family. Both EHD1 and Rab11 control the recycling of receptors from REs back to the plasma membrane[16,19,20].
Regulation of LEs and lysosomes by Rab proteins
In yeast, Vps8 is a subunit of the Vps39-Vps8-class C intermediate HOPS (homotypic fusion and vacuole protein sorting) complex that is recruited by Rab5[21]. Vps39 indirectly activates Rab7 and recruits it to the early endosomal membrane[22,23]. Subsequently, GTP-bound Rab5 is hydrolyzed to GDP-bound Rab5 and released from the early endosomal membrane. EEs thus mature into LEs, which are then transported to and fuse with lysosomes. An effector of Rab7, Rab7-interacting lysosomal protein (RILP), interacts with Rab7 on LEs and lysosomes[24]. It induces the recruitment of dynein-dynactin motor complexes to Rab7-containing LEs and lysosomes[25]. These organelles are transported to the minus end of microtubules by the motor complexes and are prevented from movement toward the cell periphery.
RILP is not only an effector of Rab7, but also of Rab34 and Rab36[26,27]. Rab34 is a Golgi-associated Rab protein that regulates lysosomal positioning through its interaction with RILP[26]. Overexpression of either the wild-type or the constitutively active mutant of Rab34 results in the redistribution of clusters of lysosomes to the peri-Golgi region[26]. A similar phenotype is observed when Rab36, which shares 56% homology with Rab34, is overexpressed. Through its interaction with RILP, Rab36 regulates the spatial distribution of LEs and lysosomes through a similar mechanism to Rab34[27].
Regulation of parallel trafficking routes by Rab proteins
There are at least two different recycling routes that channel cargo back to the plasma membrane: fast and slow recycling. Transferrin receptor (TfR) is an example of cargo that is recycled back to the plasma membrane. Upon sorting from Rab5-containing EE microdomains into Rab4 microdomains, TfR is recycled directly back to the plasma membrane (fast recycling). In contrast, sorting at the EE to Rab4/Rab11 microdomains leads to delivery of TfR to REs before being returned to the plasma membrane (slow recycling)[28]. Accordingly, Rab4 regulates both fast and slow recycling of TfR[29-32]. An example of a Rab protein that regulates fast recycling is Rab35[33,34]. Rab35 localizes to both endosomes and the plasma membrane and regulates the fast recycling of MHC class I molecules[34]. In addition, Rab35 interacts with Rme-1 (a Caenorhabditis elegans ortholog of mammalian EHD1), a requirement for recycling of the KCa2.3 Ca2+-activated K+ channels[35].
In the course of slow recycling, a number of Rabs are implicated in the transport of vesicles from EEs to REs. For example, Rab15 co-localizes with Rab4, Rab5, and Rab11[36,37]. Overexpression of the wild-type and the constitutively active Rab15 mutant leads to a reduction in the rate of homotypic EE fusion and decreased fluid-phase and receptor-mediated endocytosis, without affecting the rate of recycling from EEs[37]. In contrast, expression of the constitutively inactive Rab15 stimulates EE homotypic fusion that differentially affects the rate of internalization and recycling from EEs[37]. Rab15 also regulates trafficking from apical REs to the basolateral plasma membrane in polarized cells[38]. Rab15 effector protein (REP15) co-localizes with Rab15 and Rab11 on REs, but not with Rab15, Rab4 or EEA1 on EEs[39]. Indeed, REP15 regulates receptor recycling from REs to the plasma membrane[39].
Although REP15 does not interact with Rab11, the two proteins have similar functions; both regulate vesicular trafficking from REs to the plasma membrane. Rab11 has a number of its own effectors, including Rab11 family-interacting protein 5 (Rab11-FIP5), which mediates transport from EEs to REs through binding with Kif3B, a component of the kinesin II motor protein[40]. Rab11-FIP2, another Rab11 effector, facilitates the recycling of receptors through its interaction with both EHD1 and EHD3, which serve as a link between the EHDs and Rab11[41]. Rab11-FIP2 also functions as an adaptor that connects Rab11a-positive endocytic recycling vesicles to myosin Vb[42]. Moreover, myosin Vb interacts directly with both Rab11 and Rab8[43].
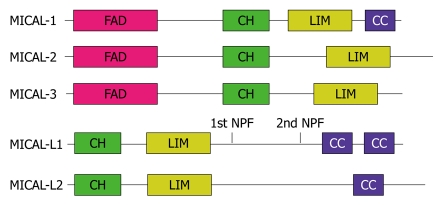
Rab8 mediates secretion from the trans-Golgi network (TGN) to the plasma membrane in polarized Madin-Darby canine kidney (MDCK) cells, the recycling of receptors from REs to the plasma membrane, ciliogenesis (along with Rab17 and Rab23), and Glut4 vesicle translocation to the plasma membrane (along with Rab10 and Rab14)[38,44,45]. One of the Rab8 effectors, MICAL-L1 (molecule interacting with CasL-like1), regulates receptor recycling and recruits Rab8 and EHD1 to tubular RE membranes that emanate from the perinuclear region[46]. MICAL-L1 belongs to the MICAL protein family that consists of four other members: MICAL-1, MICAL-2, MICAL-3, and MICAL-L2 (Figure 1). These proteins regulate vesicular transport and cytoskeleton rearrangement[46-60]. In polarized cells, MICAL-L2, is an effector of both Rab8 and Rab13, and mediates transport of tight and adherens junction proteins from the TGN to the plasma membrane[50,52-57,59,60].
Figure 1.
Schematic diagram of the MICAL protein family domain architecture. FAD: Flavin-adenine dinucleotide binding domain; CH: Calponin homology domain; LIM: LIM domain; CC: Coiled-coil; NPF: Asparagine-proline-phenylalanine motif.
MICAL-1 is also a Rab1 effector that plays a role in vesicular trafficking between the endoplasmic reticulum (ER) and Golgi[47,49,61-63]. However, unlike other Rab1 effectors (such as the Golgi-membrane associated GM130), MICAL-1 generally displays a predominantly cytosolic localization[61].
THE MICAL PROTEIN FAMILY
MICALs are large multi-domain proteins that are expressed in specific neuronal and non-neuronal tissues both during development and in adulthood[51,64]. Much information on the function of MICAL proteins comes from studies of Drosophila melanogaster, which contains a single MICAL ortholog known as D-MICAL. MICAL proteins are also Rab effectors for various Rab proteins[65]. This suggests a complex regulatory role for MICAL proteins in endocytic trafficking. Since MICAL proteins are comprised of multiple functional domains, in this review, we define the various domains of the MICAL family members and describe their functional significance.
DOMAINS OF MICAL FAMILY MEMBERS
MICAL family members all contain calponin homology (CH), LIM and coiled-coil (CC) domains, which share a high level of homology between the family members. These domains are all known for their involvement in protein-protein interactions. CH and LIM domain-containing proteins have been implicated in signal transduction and cytoskeletal organization. MICAL-1, MICAL-2 and MICAL-3 have an FAD (flavin adenine dinucleotide binding mono-oxygenase domain) domain at their N-terminal region, which is absent in the MICAL-like proteins.
CH domains
In MICAL-like proteins, the CH domain is localized near the N terminus, whereas in MICAL proteins, this domain is localized very close to the FAD domain (Figure 1). The CH domain is a structurally conserved 100-110-residue domain that is mainly found in actin-binding proteins such as calponin, Cdc42, and actinin[66]. MICAL family members have been classified as having type-2 CH domains, similar to those found in proteins such as smoothelin and microtubule-associated RP/EBs (end binding proteins)[67,68]. Although type-2 CH domains cannot interact directly with F-actin, they can abet the F-actin binding activity of type-1 CH domains[66]. Type-2 CH domains also possess the ability to interact with phosphatidylinositol-(4,5)-bisphosphate (PIP2)[69] and certain cytoskeletal proteins. Based on their nuclear magnetic resonance structures, the MICAL-1 and MICAL-3 CH domains are highly conserved with other known CH domains[67]. The MICAL-1 CH domain is comprised of six α helices and one 310 helix (a type of helical structure with three residues per turn). Folding dynamics studies have indicated that the MICAL-1 CH domain is comprised of four distinct cooperative unfolding units with a hydrophobic core[70]. The conserved hydrophobic character indicates that the three-dimensional structure of the MICAL-1 CH domain is maintained by hydrophobic interactions.
LIM domains
In MICAL family proteins, CH domains are followed by LIM domains. However, the distance between the CH and LIM domain varies between the family members, with MICAL-2 having the largest gap between the two domains[71]. The LIM domain has been described as a cytoskeletal interaction element that is also capable of interacting with different types of motifs/domains[72]. This domain was identified and named after the three LIM-domain-containing homeo-domain proteins Linl-1, Isl-1 and Mec3. LIM domains contain two zinc finger motifs that are separated by two amino acids. Each zinc finger domain consists of a cluster of eight cysteine and histidine residues at conserved locations. Some LIM-domain-containing proteins are localized in the nucleus and play a role in the regulation of gene expression, whereas others are localized in the cytoplasm and regulate cytoskeletal organization. LIM-domain-containing proteins have been classified into four groups, based on the arrangement of their LIM domains and the overall structure of the proteins. The first group, which generally localizes to the nucleus and serves as transcription factors, consists of the N-terminal tandem LIM-domain-containing proteins. The second group, which localizes both to the nucleus and to the cytoplasm, includes proteins that consist primarily of multiple LIM domains. The third group consists of the C-terminal LIM domain proteins, which also contain various other protein-protein interaction motifs. The MICAL family and the LIM kinase family of proteins, belong to the fourth group, known as the mono-oxygenase or kinase catalytic motif group[72,73].
FAD domains
The FAD domain comprises two subdomains: the FAD-binding subdomain and the mono-oxygenase subdomain. It is a catalytic domain that is localized to the N terminus of MICAL-1, MICAL-2 and MICAL-3. However, this domain is absent in the MICAL-like proteins. The FAD domain contains an FAD permanently bound in the interface between the mono-oxygenase and FAD-binding subdomains, with its catalytic activity dependent on NADPH[74,75]. MICAL-1 FAD domain structure resembles that of a flavo-enzyme, p-hydroxybenzoate hydroxylase[74,75]. In the presence of NADPH and oxygen, the MICAL-1 FAD domain uses the reducing equivalent of NADPH to produce H2O2. Recently, it has been demonstrated that the FAD domain of D-MICAL (D. melanogaster MICAL) binds filamentous actin in vitro and is required for actin filament destabilization in vivo[58]. In addition, the catalytic activity of the FAD domain is required for MICAL proteins to act as neuromuscular junction regulators[64].
Additional MICAL domains
Another protein-protein interaction domain that is localized to the C terminus of some MICAL family proteins is the CC domain, also known as an ezrin/radixin/moesin α-like motif. One of the hallmarks of some MICAL family proteins is that they localize to a striking array of tubular membranes[46,48] that participate in the recycling of surface receptors[46]. Indeed, a recent study has demonstrated that the CC domain is required for the association of MICAL-L1 to tubular membranes[46]. MICAL-L1 recruits both EHD1 and Rab8a to tubular structures and these tubular membrane associations are required for efficient recycling of receptors such as transferrin and β1 integrins[46] (Figures 2 and 3A). Mutations within the CC domain affect its ability to associate with tubular membranes, thus affecting MICAL-L1 function[46]. Based on a recent proteomic study that has demonstrated the interaction of various Rab proteins with MICAL family members, it has been postulated that CC domains are required for the binding of MICAL proteins to Rabs[65]. In contrast, however, Rab13 interacts with the region of MICAL-L2 that is adjacent to its CC domain[76].
Figure 2.
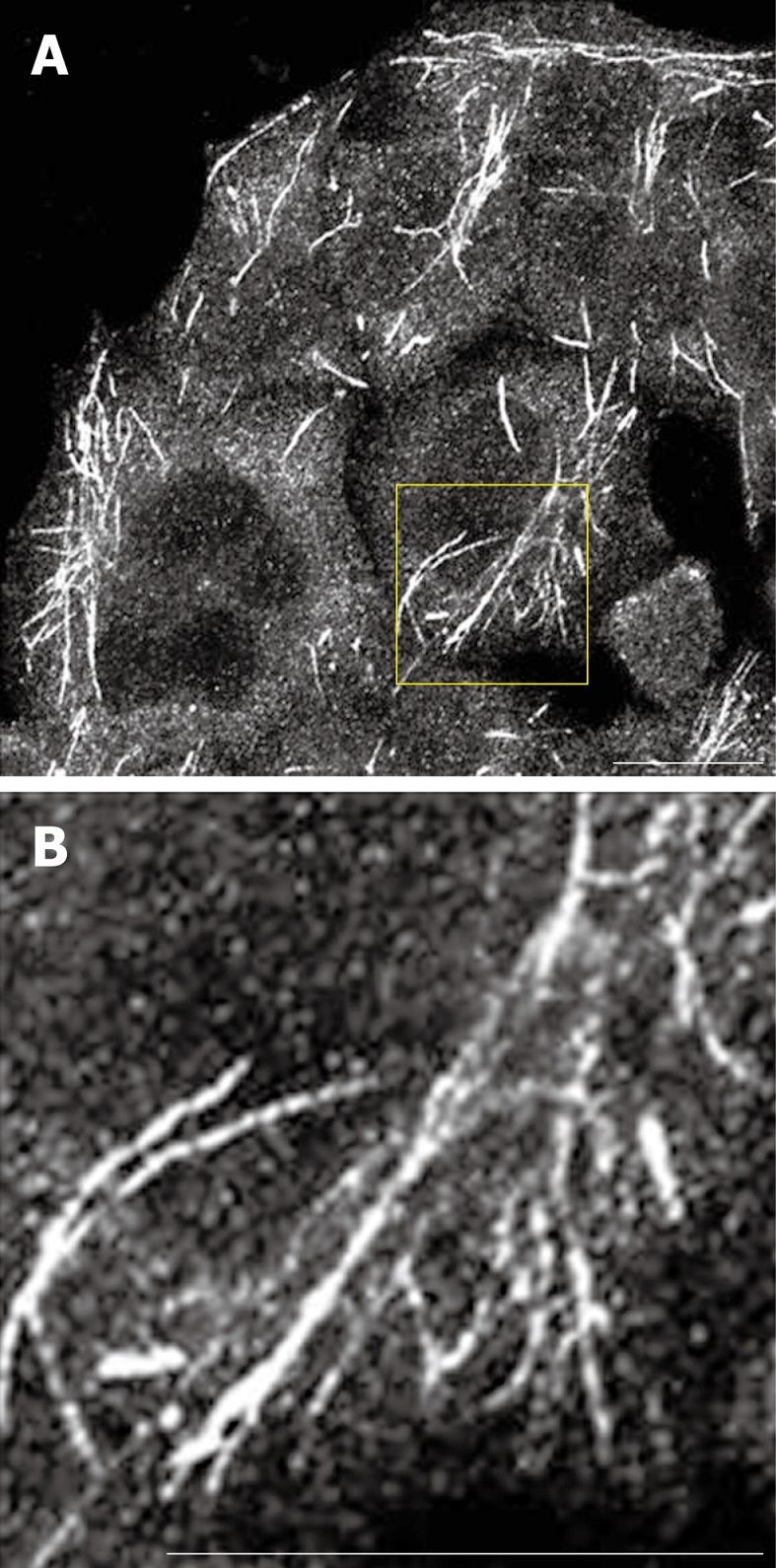
Endogenous MICAL-L1 localized to tubular recycling endosomes. A and B: HeLa cells were grown on coverslips, fixed with 4% paraformaldehyde in PBS, immunostained with anti-MICAL-L1 antibody, followed by 568-conjugated goat anti-mouse antibody. A: MICAL-L1; B: MICAL-L1 (inset). Bar, 10 μm.
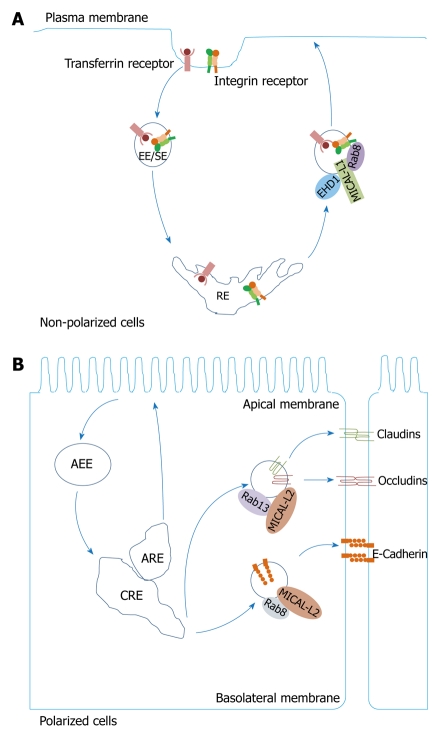
Figure 3.
MICAL-L1 and MICAL-L2 regulate endocytic recycling. A: MICAL-L1 recruits Eps15 homology domain 1 (EHD1) and Rab8A to endocytic membranes and regulates recycling of transferrin and β1 integrin receptors; B: MICAL-L2/Rab13 interactions control transport of claudin-1 and occludin to the plasma membrane, whereas MICAL-L2/Rab8 complexes regulate recycling of E-cadherin. EE/SE: Early endosome/sorting endosome; RE: Recycling endosome; AEE: Apical early endosome; CRE: Common recycling endosome; ARE: Apical recycling endosome.
MICAL-1 has a proline-rich ‘PPKPP’ motif that interacts with the src homology 3 (SH3) domain of CasL [47], a multi-domain docking protein localized at focal adhesions and stress fibers.
FUNCTIONAL ROLES OF MICAL-1, 2 AND 3
Initial studies have identified MICAL-1 as an interaction partner of CasL [a member of the p130cas (Cas) family] and vimentin (an intermediate filament interacting protein)[47]. Hence, the name MICAL is an abbreviation of either molecules interacting with CasL or microtubule associated CH and LIM domain containing proteins. It is of interest that nocodazole-induced loss of microtubules causes a change in the localization of MICAL-1 and MICAL-3 and their absence from tubular membranes[49].
MICAL-1, MICAL-2 and MICAL-3 are expressed both during development and in adulthood in various tissues such as lung, heart, brain and thymus[64]. In particular, however, MICAL proteins have been best characterized in neuronal systems. Their protein expression levels are elevated in oligodendrocytes and in meningeal fibroblasts, at the sites of spinal cord injury, and also in muscular tissues. In mammals, various splice variants exist for each MICAL protein. MICAL-2 isoforms (MICAL-2-PVa and MICAL-2-PVb) are highly expressed in the progression of prostate cancer. Given the high Gleason score for MICAL-2, indicating that cancer cells are more proliferative and become more invasive when expressed, this suggests MICAL-2 involvement and its potential use as a marker and/or target for prostate cancer[77].
MICALs and axonal guidance in Drosophila
It has been demonstrated that the D. melanogaster ortholog, D-MICAL, has a functional role in neuronal development[48]. This involves the interpretation of a myriad of guidance cues present within the neuronal environment by the neuron to reach its target. Semaphorins are one of the largest families of guidance cues known for axon path finding, fasciculation, branching, and neuronal cell migration[78]. Semaphorin signaling is enabled through its interaction with plexin proteins, an event that initiates various cellular changes, including rapid growth cone collapse, which is accompanied by the depolymerization of F-actin and a decreased ability to polymerize. The mechanism of plexin-mediated effects was unclear until the identification of D-MICAL as a plexin-A-interacting protein and a mediator of semaphorin 1-plexin A signaling[48]. The D-MICAL-plexin A interaction and D-MICAL mono-oxygenase activity are required for semaphorin-1-induced motor axon repulsion in neuronal development. Mutations in the D-MICAL mono-oxygenase domain lead to abnormally structured neuromuscular junctions[51]. Recent studies have shown that D-MICAL is a crucial regulator of F-actin instability that is sufficient to reorganize F-actin networks both in vivo and in vitro[58]. D-MICAL is both necessary and sufficient for semaphorin-plexin mediated actin filament dynamics. In other words, D-MICAL acts as an F-actin disassembly factor, which provides spatiotemporal cell morphological changes in response to semaphorins, thereby regulating neuronal growth. It has also been established that D-MICAL acts as a cytoskeletal regulator of myofilament organization and neuromuscular junctions[64]. Mutations within the MICAL proteins show high penetration defects in the patterning and arrangements of synaptic structures. Moreover, Sox14, a transcription factor that is sufficient to induce dendrite severing, functions by regulating the expression level of its target gene, D-MICAL[79].
The ability of D-MICAL to destabilize both individual and bundled actin filaments is dependent on its FAD and CH domains, which are highly homologous to the mammalian orthologs[58]. Like D-MICAL, both MICAL-1 and MICAL-2 bind to the cytoplasmic region of plexin A via their C terminus that contains the CC domain[48].
Collapsin response mediators and MICALs
Another family of proteins implicated in plexin-mediated signaling is the collapsin response mediator protein (CRMP) family. CRMP1 and MICAL-1 physically associate, which is induced upon semaphorin signaling. In addition to its role as an upstream MICAL activator, CRMP functions downstream of MICAL, which inhibits the activity of the MICAL FAD domain[80]. Plexin/CRMP/MICAL interactions transduce semaphorin signaling into axon guidance. The expression pattern of MICAL-1, MICAL-2 and MICAL-3 in embryonic, postnatal and adult neurons[64] reinforces the notion that these proteins play significant roles in neuronal development and plasticity.
THE ROLE OF MICAL-L1 IN ENDOCYTIC RECYCLING
MICAL-L1 contains a single CH domain at its N terminus, followed by a LIM domain, and two asparagine-proline-phenylalanine (NPF) motifs at residues 425-427 and 633-635[46]. The first of these two NPF motifs, which is followed by a cluster of acidic residues[81,82], is responsible for its interaction with the EH domain of EHD1[46,81]. It has been demonstrated that EHD1 plays a key role in receptor recycling and localizes to vesicular and tubular membranes that emanate from RE[16,43,46,83-95]. MICAL-L1 and the endocytic regulatory protein EHD1 are highly co-localized on these tubular membranes in HeLa cells and directly interact[46]. Moreover, upon MICAL-L1 depletion, there is both a significant delay in receptor recycling, and the loss of EHD1 localized to these tubular membranes[46]. This suggests that the interaction between the two proteins is required for sustained localization of EHD1 to membrane tubules, and consequently, the regulation of receptor recycling.
MICAL-L1 as a linker between EHD1 and Rab8a
Although EHD1 and Rab8a co-localize to the same tubular membranes[43], they do not interact directly. Therefore, the interaction between MICAL-L1 and EHD1 provides an explanation for how Rab8a, which also plays a key role in the recycling of internalized receptors, associates with EHD1. The interaction of MICAL-L1 with GTP-bound Rab8a facilitates bridging of an indirect interaction between Rab8a and EHD1[46]. MICAL-L1 is considered an effector of Rab8a, therefore, it is surprising that Rab8a is dispensable for MICAL-L1 localization to the tubular membranes[46]. Moreover, depletion of MICAL-L1 causes dissociation of Rab8a from the tubular membranes, which suggests the function of MICAL-L1 as an unusual effector of Rab8a, because it recruits Rab8a to EHD1-containing tubular REs[46].
Recruitment of MICAL-L1 to tubular membranes
How is MICAL-L1 recruited to the tubular membranes? MICAL-L1 might be capable of interacting directly with phosphoinositides, which is common to some Rab effectors, such as Rabenosyn-5 and EEA1. Both of these Rab5 effectors are able to bind directly to PI3P through their FYVE domains[8,11-13]. Two sites within the CC regions of MICAL-L1 are necessary for association with membranes[46]. The first site, which contains a hydrophobic stretch of amino acids, can allow membrane binding and/or insertion, whereas the second site, which contains positively charged residues, mighty allow its interaction with phosphoinositides.
THE ROLE OF MICAL-L2 IN ASSEMBLY AND RECYCLING OF TIGHT JUNCTIONS AND ADHERENS JUNCTIONS
Regulation of occludin
MICAL-L2, which shares only 30% of homology with MICAL-L1, also regulates endocytic recycling. A recent study has shown that it controls the recycling of occludin[50,53-57,59], an integral plasma membrane protein that is located specifically at tight junctions (Figure 3B). Unlike MICAL-L1, however, MICAL-L2 does not mediate recycling of TfR[50]. MICAL-L2, an effector of Rab13, interacts with Rab13 through its C-terminal region[50,56,59]. Rab13 regulates the recruitment of junctional complexes that include tight junctions and adherens junctions from a cytosolic pool after cell-cell contact formation[59]. This Rab protein localizes to perinuclear membrane structures, vesicular structures in the cytosol, and the plasma membrane in addition to tight junctions[59]. Overexpression of a constitutively active Rab13 mutant as well as the N-terminal region of MICAL-L2, which contains both the CH and LIM domains, inhibits recycling of occludin, but not its internalization from the plasma membrane[59]. Rab13 is not essential for MICAL-L2 localization to tight junctions, because even upon deletion of the C-terminal region that binds to Rab13, MICAL-L2 still localizes to the plasma membrane[59].
Regulation of claudin and E-cadherin
MICAL-L2 is also required for recycling of claudin-1[59], a member of the claudin protein family that serves as an important component of tight junctions. Moreover, MICAL-L2 regulates the recycling of E-cadherin[59], a type-1 transmembrane protein involved in cell adhesion. The expression of the C-terminal region of MICAL-L2 inhibits the transport of claudin-1, occludin, and E-cadherin to the plasma membrane[59]. However, Rab13 only regulates the transport of claudin-1 and occludin to the plasma membrane, but not E-cadherin (Figure 3B). Indeed, Rab8a has been identified as a MICAL-L2 interaction partner that is responsible for transport of E-cadherin to the plasma membrane (Figure 3B).
Interactions of MICAL-L2 with Rab8 and Rab13
One question is how the interaction between MICAL-L2 and both Rab8 and Rab13 is regulated? Although the subcellular distributions of Rab8, Rab13 and MICAL-L2 overlap at the perinuclear region and the plasma membrane, the Rab8/MICAL-L2 interaction is primarily detected at the perinuclear region, whereas the Rab13/MICAL-L2 interaction is preferentially observed at the plasma membrane[59]. Furthermore, the interaction between Rab8 and MICAL-L2 is crucial for E-cadherin recycling[59]. In contrast, the Rab13/MICAL-L2 complex is required for recycling of occludin[59]. Therefore, Rab8/MICAL-L2 and Rab13/MICAL-L2 complexes are regulated by their interactions with the corresponding junctional proteins, and possibly by other proteins that regulate their primary localization[59].
ROLE OF MICAL-L2 IN THE REGULATION OF ACTIN REMODELING AND EPITHELIAL CELL SCATTERING
Both Rab8 and Rab13 compete for their interactions with MICAL-L2, therefore a crucial question is what controls how these complexes are formed or stabilized? One possibility is related to actinin-4. Actinin-4 is a member of the actinin protein family that contains two CH domains that are needed for binding to F-actin, four spectrin-like repeats, and two EF-hand (EFh) motifs. Actinin-4 functions as an actin crosslinking protein and anchors integral membrane proteins or scaffolding proteins to the actin cytoskeleton. In MTD-1A cells, not only do actinin-4 and MICAL-L2 co-localize at cell-cell junctions, but they also exist in a complex[54]. Their direct interaction is mediated through an uncharacterized domain of MICAL-L2 that is located between the LIM domain and the CC domain, and the C-terminal domain of actinin-4 that contains two EFh motifs[54]. Actinin-4 links MICAL-L2 to the actin cytoskeleton through its interaction with F-actin[54]. Actinin-4 recruits MICAL-L2 and consequently, occludin to cell-cell junctions, and its depletion leads to impaired recruitment of occludin to tight junctions[54]. Actinin-4 also recruits MICAL-L2 from the cytosol to cell-cell junctions during epithelial cell polarization[54].
The maintenance of adherens junctions is required for epithelial cell-cell adhesion. Therefore, endocytosis of integral adherens junction proteins, such as E-cadherin, coupled with actin cytoskeleton remodeling is needed to decrease cell-cell adhesion. Consequently, cell motility is regulated by actin cytoskeleton rearrangement and endocytic recycling of plasma membrane proteins, such as integrins and integral adherens junction proteins, which leads to membrane protrusion and adhesion to the extracellular matrix. Rab13 is transiently activated during the scattering of polarized MDCK cells in response to12-O -tetradecanoylphorbol-13-acetate[60]. This activation leads to redistribution of Rab13 and MICAL-L2, along with remodeling of the actin cytoskeleton from cell-cell contacts to the lamellipodia at the leading edge[60]. Although the mechanisms for the involvement of Rab13 and MICAL-L2 in epithelial cell scattering are still unknown, it is possible that these proteins regulate the remodeling of the actin cytoskeleton, which in turn induces epithelial cell scattering[60]. Another possibility is that Rab13 and MICAL-L2 regulate endocytic recycling of as-yet-unidentified molecules that are needed for epithelial cell scattering[60].
THE ROLE OF MICAL-L2 IN NEURITE OUTGROWTH
In neuronal cells, such as PC12, the Rab13/MICAL-L2 interaction is required for neurite outgrowth[76]. Neurite outgrowth is a process in which neuronal cells become polarized and synaptic contacts are generated through elongation of the axon and dendrites during development, and it is crucial for neuronal differentiation and regeneration. Overexpression of Rab13 induces neurite outgrowth, whereas knock-down of Rab13 exhibits a significant reduction in neurite outgrowth in PC12 cells[76]. In addition, overexpression of MICAL-L2 inhibits neurite outgrowth, but this effect can be rescued by overexpression of the constitutively active Rab13 mutant[76]. This phenomenon can be explained by the “open” and “closed” conformational changes of MICAL-L2 that are induced by its interaction with Rab13[76]. Binding of Rab13 to the C-terminal end of MICAL-L2 is required for the removal of MICAL-L2 auto-inhibition by an intramolecular interaction between the MICAL-L2 CH and LIM domains and its CC region[76]. Since the mechanism of neurite outgrowth involves cytoskeletal rearrangement, the interaction between MICAL-L2 and actinin-4 (enhanced by the expression of the GTP-bound Rab13) provides an explanation of the molecular cascade that leads to the accumulation of molecules involved in actin dynamics at the tips of neurites: the complex of MICAL-L2/actinin-4 is induced and transported by Rab13 to the neurite tips, which leads to the formation of fingerlike filopodia that are important for neurite outgrowth[76].
FUTURE DIRECTIONS
Interactions between Rabs and MICAL proteins
Through a large-scale yeast two-hybrid screening of MICAL-1, MICAL-L1, MICAL-L2, and the C-terminal region of MICAL-3, it has been demonstrated that these MICAL proteins are capable of binding to Rab8a, Rab8b, Rab10, Rab13, and Rab15[65]. In addition, MICAL-1 and MICAL-L1 interact with Rab35 and Rab36, whereas the C-terminal region of MICAL-3 binds to Rab35. However, unlike the MICAL-like proteins, MICAL-1 and the C-terminal region of MICAL-3 interact with Rab1a and Rab1b via the MICAL protein CC domains. Furthermore, as discussed earlier, the C-terminal region of MICAL-L2 is required for its interaction with Rab13. Indeed, the CC regions of MICAL-1, MICAL-3 and the MICAL-like proteins are highly conserved. Therefore, it is likely that the interactions between MICAL proteins and the Rab protein family are mediated through their CC domains.
The CC region of MICAL-L2 is required for its interaction with Rab8a or Rab13; the latter being crucial for the recycling of E-cadherin or junctional proteins, respectively, in polarized cells[50,53-57,59,60]. However, the functional significance of MICAL-L2 in non-polarized cells remains unknown. On the other hand, MICAL-L1 is important in controlling recycling of cell surface receptors, such as TfR and β1 integrin receptors in non-polarized cells[46]. However, a crucial line of research will be to determine the role of MICAL-L1 in polarized cells. In addition, it has been shown that MICAL-L1 interacts with Rab13 through the yeast two-hybrid assays, but it is still unknown whether the two proteins interact in vivo, and what the significance of their interaction might be.
Similarly, potential MICAL interactions with Rab10 and Rab15 in vivo have yet to be explored. Rab10, which is involved in secretory transport from the Golgi to the plasma membrane and Glut4 translocation to the plasma membrane along with Rab8, might require MICAL interactions for its activity. The interaction between Rab15 and the MICAL protein family might mediate Rab15 function in regulating transport of vesicles between EEs and REs.
MICAL-L1 as a putative Rab35 effector protein
MICAL-L1 is a strong candidate effector for Rab35. Both proteins facilitate the transport of receptors from REs to the plasma membrane. Like MICAL-L1, Rab35 is localized to intracellular endosomal structures and localizes to the plasma membrane and cytoplasm[96]. The putative interaction between MICAL-L1 and Rab35 might link the latter to Rab8 and Rab11, which are required for endocytic recycling. If MICAL-L1 also serves as an effector of Rab35, it would be important to determine whether Rab35 recruits MICAL-L1 to intracellular REs, or whether MICAL-L1 is needed for Rab35 localization to these structures.
MICALs as scaffolds for concentrating cytoskeletal elements at neurite tips
As stated earlier, Rab13 is required to recruit MICAL-L2 and actinin-4 to the neurite tips in neuronal cells[76]. Although it remains unknown whether MICAL-L1 can interact with actin or microtubule binding proteins, interactions between MICAL-L1 and Rab proteins might be required for cytoskeletal rearrangement at the neurite tips and thus, neurite outgrowth. Through a different mechanism, mammalian MICAL-1, MICAL-2 and MICAL-3, which contain FAD and CH domains, might also be involved in actin rearrangement and subsequently control neurite outgrowth. Both FAD and CH domains of D-MICAL are required for redox activity that alters actin filament (F-actin) dynamics, as the homozygous mutant adult flies (Mical-/-) have abnormally shaped bristle cells[58]. Therefore, it is hypothesized that mammalian MICAL-1, MICAL-2 or MICAL-3 carries out redox activity that leads to F-actin rearrangement and neurite outgrowth. Overall, the interactions between MICAL and Rab proteins may be crucial for F-actin rearrangement to occur at the tip of the neuronal cells.
Phosphorylation of MICAL proteins
Through several mass spectrometry experiments, it has been discovered that MICAL proteins are phosphorylated at multiple sites. MICAL-1 undergoes phosphorylation on Y483 (located between the FAD and CH domains) and S818 (before the CC domain) (www.phosphosite.org), whereas MICAL-2 is phosphorylated on Y653 (between the LIM and CH domains) in some non-small lung cancer cell lines and tumors[97]. Phosphorylation of MICAL-3 on residues S649 and T684 is linked to mitosis[98].
MICAL phosphorylation and potential regulation of the cell cycle
MICAL-L1 phosphorylation: Human MICAL-L1 is phosphorylated on many of its serine and threonine residues localized between the LIM and the CC domains. Phosphorylation of these residues is required for differentiation of human embryonic stem cells, chemotaxis, and mitosis[98-104]. For example, phosphorylated residues S295, S391, T547, and S578 of MICAL-L1 specifically interact with the Polo-box domain (PBD) of Polo-like kinase 1 (Plk1) during mitosis[102]. PBD is a specialized phosphoserine-threonine binding domain that is postulated to target Plk1 to its substrate during mitosis and cytokinesis. MICAL-L1 contains several S[ST]P and [ED]X[ST][FLIYWVM] motifs that are required for recognition and phosphorylation by Plk1, respectively. In addition, residues T469, S470, and S619 of MICAL-L1 are phosphorylated by cyclin-dependent kinase 2, which regulates cell cycle progression[104].
MICAL-L2 phosphorylation: Human MICAL-L2 is also phosphorylated at many of its serine and tyrosine residues. For example, amino acid Y59 of MICAL-L2, which is located within the CH domain, is phosphorylated by an unidentified tyrosine kinase that might be implicated in the development of lung cancer[97]. Residue S143, which is located between the CH and LIM domains of MICAL-L2, is involved in the early differentiation of human embryonic stem cells[99]. Phosphorylated serine residues 494, 504, 649, 658 and 660 of MICAL-L2, which are located between the LIM domain and CC region, play key roles in mitosis[98]. Similar to MICAL-L1, MICAL-L2 also contains motifs that are required for recognition and phosphorylation by Plk1[102].
Many of their phosphorylated residues are required for mitosis, therefore, the MICAL family of proteins might play key roles in cell division. However, many questions still need to be answered, such as which kinases are involved in the phosphorylation? How do they target MICAL proteins? What are the consequences of these phosphorylation events? At which steps of mitosis are the MICAL proteins involved? At present, it is possible only to speculate that phosphorylation of MICAL-L1 is required for its interaction with Rab35, and that this interaction regulates trafficking of proteins that are crucial for mitosis and subsequently, cytokinesis. Support for this notion comes from studies that have demonstrated that Rab35 regulates cytokinesis, possibly by controlling trafficking of phosphatidylinositol-4-phosphate 5 kinase to the cleavage furrow to generate PIP2 on the plasma membrane of daughter cells[96,105].
In conclusion, despite recent advances, the MICAL family of proteins remains poorly understood. Some of its members function as molecular effectors of the Rab proteins that regulate vesicular targeting and fusion. Therefore, the MICAL protein family plays important roles in vesicular trafficking. The interactions between MICAL-L1, EHD1 and Rab8 are required for the recycling of TfR and β1 integrin receptors. In contrast, the C-terminal domain of MICAL-L2 is critical for its interaction with either Rab8 or Rab13, an association that is necessary for regulating the recycling of junctional proteins. In addition, MICAL-L2/actinin-4 complexes are transported by Rab13 to the neurite tips to regulate actin dynamics and subsequently, neurite outgrowth. Although the C-terminal regions of MICAL-1 and MICAL-3 (which are required for their interactions with certain Rab proteins) share a high degree of homology with the MICAL-like proteins, the putative roles for MICAL-1 and MICAL-3 in vesicular transport remain unknown. However, MICAL-1, MICAL-2 and MICAL-3 might control actin dynamics via a mechanism that differs from that of MICAL-L2; through redox reactions by their FAD and CH domains, as demonstrated for D-MICAL.
Footnotes
Supported by The National Institutes of Health grants R01GM074876 (Caplan S and Naslavsky N), R01GM087455 (Caplan S), the Nebraska Dept. of Health (Naslavsky N), as well as P20 RR018759 from the National Center
Peer reviewers: Victor Faundez, MD, PhD, Associate Professor, Department Cell Biology Emory University, 615 Michael Street., Whitehead 446, Atlanta, GA 30322, United States; Annie Angers, PhD, Department of Biological Sciences, University of Montreal, PO Box 6128, Station Centre-Ville, Montreal, Quebec, H3C 3J7, Canada
S- Editor Cheng JX L- Editor Kerr C E- Editor Zheng XM
References
- 1.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schultz J, Doerks T, Ponting CP, Copley RR, Bork P. More than 1,000 putative new human signalling proteins revealed by EST data mining. Nat Genet. 2000;25:201–204. doi: 10.1038/76069. [DOI] [PubMed] [Google Scholar]
- 3.Horiuchi H, Lippé R, McBride HM, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M, et al. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–59. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- 4.Rybin V, Ullrich O, Rubino M, Alexandrov K, Simon I, Seabra MC, Goody R, Zerial M. GTPase activity of Rab5 acts as a timer for endocytic membrane fusion. Nature. 1996;383:266–269. doi: 10.1038/383266a0. [DOI] [PubMed] [Google Scholar]
- 5.Stenmark H, Vitale G, Ullrich O, Zerial M. Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell. 1995;83:423–432. doi: 10.1016/0092-8674(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 6.Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 7.Merithew E, Stone C, Eathiraj S, Lambright DG. Determinants of Rab5 interaction with the N terminus of early endosome antigen 1. J Biol Chem. 2003;278:8494–8500. doi: 10.1074/jbc.M211514200. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen E, Christoforidis S, Uttenweiler-Joseph S, Miaczynska M, Dewitte F, Wilm M, Hoflack B, Zerial M. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J Cell Biol. 2000;151:601–612. doi: 10.1083/jcb.151.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siddhanta U, McIlroy J, Shah A, Zhang Y, Backer JM. Distinct roles for the p110alpha and hVPS34 phosphatidylinositol 3'-kinases in vesicular trafficking, regulation of the actin cytoskeleton, and mitogenesis. J Cell Biol. 1998;143:1647–1659. doi: 10.1083/jcb.143.6.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 12.Stenmark H, Aasland R. FYVE-finger proteins--effectors of an inositol lipid. J Cell Sci. 1999;112(Pt 23):4175–4183. doi: 10.1242/jcs.112.23.4175. [DOI] [PubMed] [Google Scholar]
- 13.Lawe DC, Patki V, Heller-Harrison R, Lambright D, Corvera S. The FYVE domain of early endosome antigen 1 is required for both phosphatidylinositol 3-phosphate and Rab5 binding. Critical role of this dual interaction for endosomal localization. J Biol Chem. 2000;275:3699–3705. doi: 10.1074/jbc.275.5.3699. [DOI] [PubMed] [Google Scholar]
- 14.McBride HM, Rybin V, Murphy C, Giner A, Teasdale R, Zerial M. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98:377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- 15.Simonsen A, Gaullier JM, D'Arrigo A, Stenmark H. The Rab5 effector EEA1 interacts directly with syntaxin-6. J Biol Chem. 1999;274:28857–28860. doi: 10.1074/jbc.274.41.28857. [DOI] [PubMed] [Google Scholar]
- 16.Naslavsky N, Boehm M, Backlund PS Jr, Caplan S. Rabenosyn-5 and EHD1 interact and sequentially regulate protein recycling to the plasma membrane. Mol Biol Cell. 2004;15:2410–2422. doi: 10.1091/mbc.E03-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naslavsky N, McKenzie J, Altan-Bonnet N, Sheff D, Caplan S. EHD3 regulates early-endosome-to-Golgi transport and preserves Golgi morphology. J Cell Sci. 2009;122:389–400. doi: 10.1242/jcs.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahajeng J, Caplan S, Naslavsky N. Common and distinct roles for the binding partners Rabenosyn-5 and Vps45 in the regulation of endocytic trafficking in mammalian cells. Exp Cell Res. 2010;316:859–874. doi: 10.1016/j.yexcr.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren M, Xu G, Zeng J, De Lemos-Chiarandini C, Adesnik M, Sabatini DD. Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc Natl Acad Sci USA. 1998;95:6187–6192. doi: 10.1073/pnas.95.11.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ullrich O, Reinsch S, Urbé S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peplowska K, Markgraf DF, Ostrowicz CW, Bange G, Ungermann C. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev Cell. 2007;12:739–750. doi: 10.1016/j.devcel.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 22.Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peralta ER, Martin BC, Edinger AL. Differential effects of TBC1D15 and mammalian Vps39 on Rab7 activation state, lysosomal morphology, and growth factor dependence. J Biol Chem. 2010;285:16814–16821. doi: 10.1074/jbc.M110.111633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, Janssen H, Wubbolts R, Neefjes J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- 26.Wang T, Hong W. Interorganellar regulation of lysosome positioning by the Golgi apparatus through Rab34 interaction with Rab-interacting lysosomal protein. Mol Biol Cell. 2002;13:4317–4332. doi: 10.1091/mbc.E02-05-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Hu J, Yun Y, Wang T. Rab36 regulates the spatial distribution of late endosomes and lysosomes through a similar mechanism to Rab34. Mol Membr Biol. 2010;27:24–31. doi: 10.3109/09687680903417470. [DOI] [PubMed] [Google Scholar]
- 28.Sönnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149:901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deneka M, van der Sluijs P. 'Rab'ing up endosomal membrane transport. Nat Cell Biol. 2002;4:E33–E35. doi: 10.1038/ncb0202-e33. [DOI] [PubMed] [Google Scholar]
- 30.van der Sluijs P, Hull M, Webster P, Mâle P, Goud B, Mellman I. The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell. 1992;70:729–740. doi: 10.1016/0092-8674(92)90307-x. [DOI] [PubMed] [Google Scholar]
- 31.Sheff DR, Daro EA, Hull M, Mellman I. The receptor recycling pathway contains two distinct populations of early endosomes with different sorting functions. J Cell Biol. 1999;145:123–139. doi: 10.1083/jcb.145.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Wit H, Lichtenstein Y, Kelly RB, Geuze HJ, Klumperman J, van der Sluijs P. Rab4 regulates formation of synaptic-like microvesicles from early endosomes in PC12 cells. Mol Biol Cell. 2001;12:3703–3715. doi: 10.1091/mbc.12.11.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato M, Sato K, Liou W, Pant S, Harada A, Grant BD. Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 2008;27:1183–1196. doi: 10.1038/emboj.2008.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allaire PD, Marat AL, Dall'Armi C, Di Paolo G, McPherson PS, Ritter B. The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol Cell. 2010;37:370–382. doi: 10.1016/j.molcel.2009.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Y, Balut CM, Bailey MA, Patino-Lopez G, Shaw S, Devor DC. Recycling of the Ca2+-activated K+ channel, KCa2.3, is dependent upon RME-1, Rab35/EPI64C, and an N-terminal domain. J Biol Chem. 2010;285:17938–17953. doi: 10.1074/jbc.M109.086553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuk PA, Elferink LA. Rab15 mediates an early endocytic event in Chinese hamster ovary cells. J Biol Chem. 1999;274:22303–22312. doi: 10.1074/jbc.274.32.22303. [DOI] [PubMed] [Google Scholar]
- 37.Zuk PA, Elferink LA. Rab15 differentially regulates early endocytic trafficking. J Biol Chem. 2000;275:26754–26764. doi: 10.1074/jbc.M000344200. [DOI] [PubMed] [Google Scholar]
- 38.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 39.Strick DJ, Elferink LA. Rab15 effector protein: a novel protein for receptor recycling from the endocytic recycling compartment. Mol Biol Cell. 2005;16:5699–5709. doi: 10.1091/mbc.E05-03-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schonteich E, Wilson GM, Burden J, Hopkins CR, Anderson K, Goldenring JR, Prekeris R. The Rip11/Rab11-FIP5 and kinesin II complex regulates endocytic protein recycling. J Cell Sci. 2008;121:3824–3833. doi: 10.1242/jcs.032441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naslavsky N, Rahajeng J, Sharma M, Jovic M, Caplan S. Interactions between EHD proteins and Rab11-FIP2: a role for EHD3 in early endosomal transport. Mol Biol Cell. 2006;17:163–177. doi: 10.1091/mbc.E05-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hales CM, Vaerman JP, Goldenring JR. Rab11 family interacting protein 2 associates with Myosin Vb and regulates plasma membrane recycling. J Biol Chem. 2002;277:50415–50421. doi: 10.1074/jbc.M209270200. [DOI] [PubMed] [Google Scholar]
- 43.Roland JT, Kenworthy AK, Peranen J, Caplan S, Goldenring JR. Myosin Vb interacts with Rab8a on a tubular network containing EHD1 and EHD3. Mol Biol Cell. 2007;18:2828–2837. doi: 10.1091/mbc.E07-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sano H, Roach WG, Peck GR, Fukuda M, Lienhard GE. Rab10 in insulin-stimulated GLUT4 translocation. Biochem J. 2008;411:89–95. doi: 10.1042/BJ20071318. [DOI] [PubMed] [Google Scholar]
- 45.Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma M, Giridharan SS, Rahajeng J, Naslavsky N, Caplan S. MICAL-L1 links EHD1 to tubular recycling endosomes and regulates receptor recycling. Mol Biol Cell. 2009;20:5181–5194. doi: 10.1091/mbc.E09-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki T, Nakamoto T, Ogawa S, Seo S, Matsumura T, Tachibana K, Morimoto C, Hirai H. MICAL, a novel CasL interacting molecule, associates with vimentin. J Biol Chem. 2002;277:14933–14941. doi: 10.1074/jbc.M111842200. [DOI] [PubMed] [Google Scholar]
- 48.Terman JR, Mao T, Pasterkamp RJ, Yu HH, Kolodkin AL. MICALs, a family of conserved flavoprotein oxidoreductases, function in plexin-mediated axonal repulsion. Cell. 2002;109:887–900. doi: 10.1016/s0092-8674(02)00794-8. [DOI] [PubMed] [Google Scholar]
- 49.Fischer J, Weide T, Barnekow A. The MICAL proteins and rab1: a possible link to the cytoskeleton? Biochem Biophys Res Commun. 2005;328:415–423. doi: 10.1016/j.bbrc.2004.12.182. [DOI] [PubMed] [Google Scholar]
- 50.Terai T, Nishimura N, Kanda I, Yasui N, Sasaki T. JRAB/MICAL-L2 is a junctional Rab13-binding protein mediating the endocytic recycling of occludin. Mol Biol Cell. 2006;17:2465–2475. doi: 10.1091/mbc.E05-09-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beuchle D, Schwarz H, Langegger M, Koch I, Aberle H. Drosophila MICAL regulates myofilament organization and synaptic structure. Mech Dev. 2007;124:390–406. doi: 10.1016/j.mod.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 52.Kolk SM, Pasterkamp RJ. MICAL flavoprotein monooxygenases: structure, function and role in semaphorin signaling. Adv Exp Med Biol. 2007;600:38–51. doi: 10.1007/978-0-387-70956-7_4. [DOI] [PubMed] [Google Scholar]
- 53.Nishimura N, Sasaki T. Regulation of epithelial cell adhesion and repulsion: role of endocytic recycling. J Med Invest. 2008;55:9–16. doi: 10.2152/jmi.55.9. [DOI] [PubMed] [Google Scholar]
- 54.Nakatsuji H, Nishimura N, Yamamura R, Kanayama HO, Sasaki T. Involvement of actinin-4 in the recruitment of JRAB/MICAL-L2 to cell-cell junctions and the formation of functional tight junctions. Mol Cell Biol. 2008;28:3324–3335. doi: 10.1128/MCB.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishimura N, Sasaki T. Cell-surface biotinylation to study endocytosis and recycling of occludin. Methods Mol Biol. 2008;440:89–96. doi: 10.1007/978-1-59745-178-9_7. [DOI] [PubMed] [Google Scholar]
- 56.Nishimura N, Sasaki T. Identification and characterization of JRAB/MICAL-L2, a junctional Rab13-binding protein. Methods Enzymol. 2008;438:141–153. doi: 10.1016/S0076-6879(07)38010-5. [DOI] [PubMed] [Google Scholar]
- 57.Nishimura N, Sasaki T. Rab family small G proteins in regulation of epithelial apical junctions. Front Biosci. 2009;14:2115–2129. doi: 10.2741/3366. [DOI] [PubMed] [Google Scholar]
- 58.Hung RJ, Yazdani U, Yoon J, Wu H, Yang T, Gupta N, Huang Z, van Berkel WJ, Terman JR. Mical links semaphorins to F-actin disassembly. Nature. 2010;463:823–827. doi: 10.1038/nature08724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamamura R, Nishimura N, Nakatsuji H, Arase S, Sasaki T. The interaction of JRAB/MICAL-L2 with Rab8 and Rab13 coordinates the assembly of tight junctions and adherens junctions. Mol Biol Cell. 2008;19:971–983. doi: 10.1091/mbc.E07-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanda I, Nishimura N, Nakatsuji H, Yamamura R, Nakanishi H, Sasaki T. Involvement of Rab13 and JRAB/MICAL-L2 in epithelial cell scattering. Oncogene. 2008;27:1687–1695. doi: 10.1038/sj.onc.1210812. [DOI] [PubMed] [Google Scholar]
- 61.Weide T, Teuber J, Bayer M, Barnekow A. MICAL-1 isoforms, novel rab1 interacting proteins. Biochem Biophys Res Commun. 2003;306:79–86. doi: 10.1016/s0006-291x(03)00918-5. [DOI] [PubMed] [Google Scholar]
- 62.Nuoffer C, Davidson HW, Matteson J, Meinkoth J, Balch WE. A GDP-bound of rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J Cell Biol. 1994;125:225–237. doi: 10.1083/jcb.125.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pind SN, Nuoffer C, McCaffery JM, Plutner H, Davidson HW, Farquhar MG, Balch WE. Rab1 and Ca2+ are required for the fusion of carrier vesicles mediating endoplasmic reticulum to Golgi transport. J Cell Biol. 1994;125:239–252. doi: 10.1083/jcb.125.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pasterkamp RJ, Dai HN, Terman JR, Wahlin KJ, Kim B, Bregman BS, Popovich PG, Kolodkin AL. MICAL flavoprotein monooxygenases: expression during neural development and following spinal cord injuries in the rat. Mol Cell Neurosci. 2006;31:52–69. doi: 10.1016/j.mcn.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 65.Fukuda M, Kanno E, Ishibashi K, Itoh T. Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics. 2008;7:1031–1042. doi: 10.1074/mcp.M700569-MCP200. [DOI] [PubMed] [Google Scholar]
- 66.Gimona M, Djinovic-Carugo K, Kranewitter WJ, Winder SJ. Functional plasticity of CH domains. FEBS Lett. 2002;513:98–106. doi: 10.1016/s0014-5793(01)03240-9. [DOI] [PubMed] [Google Scholar]
- 67.Sun H, Dai H, Zhang J, Jin X, Xiong S, Xu J, Wu J, Shi Y. Solution structure of calponin homology domain of Human MICAL-1. J Biomol NMR. 2006;36:295–300. doi: 10.1007/s10858-006-9062-5. [DOI] [PubMed] [Google Scholar]
- 68.Korenbaum E, Rivero F. Calponin homology domains at a glance. J Cell Sci. 2002;115:3543–3545. doi: 10.1242/jcs.00003. [DOI] [PubMed] [Google Scholar]
- 69.Fraley TS, Pereira CB, Tran TC, Singleton C, Greenwood JA. Phosphoinositide binding regulates alpha-actinin dynamics: mechanism for modulating cytoskeletal remodeling. J Biol Chem. 2005;280:15479–15482. doi: 10.1074/jbc.M500631200. [DOI] [PubMed] [Google Scholar]
- 70.Jin X, Zhang J, Dai H, Sun H, Wang D, Wu J, Shi Y. Investigation of the four cooperative unfolding units existing in the MICAL-1 CH domain. Biophys Chem. 2007;129:269–278. doi: 10.1016/j.bpc.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 71.Friedberg F. Alternative splicing for members of human mosaic domain superfamilies. I. The CH and LIM domains containing group of proteins. Mol Biol Rep. 2009;36:1059–1081. doi: 10.1007/s11033-008-9281-9. [DOI] [PubMed] [Google Scholar]
- 72.Zheng Q, Zhao Y. The diverse biofunctions of LIM domain proteins: determined by subcellular localization and protein-protein interaction. Biol Cell. 2007;99:489–502. doi: 10.1042/BC20060126. [DOI] [PubMed] [Google Scholar]
- 73.Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 74.Siebold C, Berrow N, Walter TS, Harlos K, Owens RJ, Stuart DI, Terman JR, Kolodkin AL, Pasterkamp RJ, Jones EY. High-resolution structure of the catalytic region of MICAL (molecule interacting with CasL), a multidomain flavoenzyme-signaling molecule. Proc Natl Acad Sci USA. 2005;102:16836–16841. doi: 10.1073/pnas.0504997102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nadella M, Bianchet MA, Gabelli SB, Barrila J, Amzel LM. Structure and activity of the axon guidance protein MICAL. Proc Natl Acad Sci USA. 2005;102:16830–16835. doi: 10.1073/pnas.0504838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sakane A, Honda K, Sasaki T. Rab13 regulates neurite outgrowth in PC12 cells through its effector protein, JRAB/MICAL-L2. Mol Cell Biol. 2010;30:1077–1087. doi: 10.1128/MCB.01067-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ashida S, Furihata M, Katagiri T, Tamura K, Anazawa Y, Yoshioka H, Miki T, Fujioka T, Shuin T, Nakamura Y, et al. Expression of novel molecules, MICAL2-PV (MICAL2 prostate cancer variants), increases with high Gleason score and prostate cancer progression. Clin Cancer Res. 2006;12:2767–2773. doi: 10.1158/1078-0432.CCR-05-1995. [DOI] [PubMed] [Google Scholar]
- 78.Pasterkamp RJ, Verhaagen J. Semaphorins in axon regeneration: developmental guidance molecules gone wrong? Philos. Trans R Soc Lond B Biol Sci. 2006;361:1499–1511. doi: 10.1098/rstb.2006.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kirilly D, Gu Y, Huang Y, Wu Z, Bashirullah A, Low BC, Kolodkin AL, Wang H, Yu F. A genetic pathway composed of Sox14 and Mical governs severing of dendrites during pruning. Nat Neurosci. 2009;12:1497–1505. doi: 10.1038/nn.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schmidt EF, Shim SO, Strittmatter SM. Release of MICAL autoinhibition by semaphorin-plexin signaling promotes interaction with collapsin response mediator protein. J Neurosci. 2008;28:2287–2297. doi: 10.1523/JNEUROSCI.5646-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kieken F, Sharma M, Jovic M, Giridharan SS, Naslavsky N, Caplan S, Sorgen PL. Mechanism for the selective interaction of C-terminal Eps15 homology domain proteins with specific Asn-Pro-Phe-containing partners. J Biol Chem. 2010;285:8687–8694. doi: 10.1074/jbc.M109.045666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Henry GD, Corrigan DJ, Dineen JV, Baleja JD. Charge effects in the selection of NPF motifs by the EH domain of EHD1. Biochemistry. 2010;49:3381–3392. doi: 10.1021/bi100065r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rotem-Yehudar R, Galperin E, Horowitz M. Association of insulin-like growth factor 1 receptor with EHD1 and SNAP29. J Biol Chem. 2001;276:33054–33060. doi: 10.1074/jbc.M009913200. [DOI] [PubMed] [Google Scholar]
- 84.Caplan S, Naslavsky N, Hartnell LM, Lodge R, Polishchuk RS, Donaldson JG, Bonifacino JS. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 2002;21:2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Braun A, Pinyol R, Dahlhaus R, Koch D, Fonarev P, Grant BD, Kessels MM, Qualmann B. EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol Biol Cell. 2005;16:3642–3658. doi: 10.1091/mbc.E05-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Naslavsky N, Caplan S. C-terminal EH-domain-containing proteins: consensus for a role in endocytic trafficking, EH? J Cell Sci. 2005;118:4093–4101. doi: 10.1242/jcs.02595. [DOI] [PubMed] [Google Scholar]
- 87.Rapaport D, Auerbach W, Naslavsky N, Pasmanik-Chor M, Galperin E, Fein A, Caplan S, Joyner AL, Horowitz M. Recycling to the plasma membrane is delayed in EHD1 knockout mice. Traffic. 2006;7:52–60. doi: 10.1111/j.1600-0854.2005.00359.x. [DOI] [PubMed] [Google Scholar]
- 88.Jović M, Naslavsky N, Rapaport D, Horowitz M, Caplan S. EHD1 regulates beta1 integrin endosomal transport: effects on focal adhesions, cell spreading and migration. J Cell Sci. 2007;120:802–814. doi: 10.1242/jcs.03383. [DOI] [PubMed] [Google Scholar]
- 89.Naslavsky N, Rahajeng J, Rapaport D, Horowitz M, Caplan S. EHD1 regulates cholesterol homeostasis and lipid droplet storage. Biochem Biophys Res Commun. 2007;357:792–799. doi: 10.1016/j.bbrc.2007.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shi A, Pant S, Balklava Z, Chen CC, Figueroa V, Grant BD. A novel requirement for C. elegans Alix/ALX-1 in RME-1-mediated membrane transport. Curr Biol. 2007;17:1913–1924. doi: 10.1016/j.cub.2007.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grant BD, Caplan S. Mechanisms of EHD/RME-1 protein function in endocytic transport. Traffic. 2008;9:2043–2052. doi: 10.1111/j.1600-0854.2008.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jović M, Kieken F, Naslavsky N, Sorgen PL, Caplan S. Eps15 homology domain 1-associated tubules contain phosphatidylinositol-4-phosphate and phosphatidylinositol-(4,5)-bisphosphate and are required for efficient recycling. Mol Biol Cell. 2009;20:2731–2743. doi: 10.1091/mbc.E08-11-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pant S, Sharma M, Patel K, Caplan S, Carr CM, Grant BD. AMPH-1/Amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat Cell Biol. 2009;11:1399–1410. doi: 10.1038/ncb1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rainey MA, George M, Ying G, Akakura R, Burgess DJ, Siefker E, Bargar T, Doglio L, Crawford SE, Todd GL, et al. The endocytic recycling regulator EHD1 is essential for spermatogenesis and male fertility in mice. BMC Dev Biol. 2010;10:37. doi: 10.1186/1471-213X-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yap CC, Lasiecka ZM, Caplan S, Winckler B. Alterations of EHD1/EHD4 protein levels interfere with L1/NgCAM endocytosis in neurons and disrupt axonal targeting. J Neurosci. 2010;30:6646–6657. doi: 10.1523/JNEUROSCI.5428-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kouranti I, Sachse M, Arouche N, Goud B, Echard A. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr Biol. 2006;16:1719–1725. doi: 10.1016/j.cub.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 97.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 98.Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Van Hoof D, Muñoz J, Braam SR, Pinkse MW, Linding R, Heck AJ, Mummery CL, Krijgsveld J. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 2009;5:214–226. doi: 10.1016/j.stem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 100.Brill LM, Xiong W, Lee KB, Ficarro SB, Crain A, Xu Y, Terskikh A, Snyder EY, Ding S. Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell. 2009;5:204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen RQ, Yang QK, Lu BW, Yi W, Cantin G, Chen YL, Fearns C, Yates JR 3rd, Lee JD. CDC25B mediates rapamycin-induced oncogenic responses in cancer cells. Cancer Res. 2009;69:2663–2668. doi: 10.1158/0008-5472.CAN-08-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lowery DM, Clauser KR, Hjerrild M, Lim D, Alexander J, Kishi K, Ong SE, Gammeltoft S, Carr SA, Yaffe MB. Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J. 2007;26:2262–2273. doi: 10.1038/sj.emboj.7601683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Y, Ding SJ, Wang W, Jacobs JM, Qian WJ, Moore RJ, Yang F, Camp DG 2nd, Smith RD, Klemke RL. Profiling signaling polarity in chemotactic cells. Proc Natl Acad Sci USA. 2007;104:8328–8333. doi: 10.1073/pnas.0701103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chi Y, Welcker M, Hizli AA, Posakony JJ, Aebersold R, Clurman BE. Identification of CDK2 substrates in human cell lysates. Genome Biol. 2008;9:R149. doi: 10.1186/gb-2008-9-10-r149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Echard A. Membrane traffic and polarization of lipid domains during cytokinesis. Biochem Soc Trans. 2008;36:395–399. doi: 10.1042/BST0360395. [DOI] [PubMed] [Google Scholar]