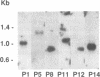
Abstract
Plant sequences that act as transcriptional activation domains in yeast as well as in plants have been isolated by genetic selection in yeast. The selection was based on the reconstitution of a functional GAL4 transcriptional activator. Since the peptides show no homology with reported activation domains, they represent a new class of activating sequences. The sequence P1, which is 10 amino acids long, is the shortest functional activation domain reported. A cDNA that encodes the P14 class (peptides P14-P18) activating sequence have been cloned. The protein exhibits strong homology (higher than 50% amino acid identity) with the BBC1-related sequences, a highly conserved family of basic proteins containing nuclear localization signals. The P14 and P15 peptides are the most effective plant activating sequences. The P14 and P15 peptides are highly hydrophilic, positively charged and mostly unstructured. These properties are at odds with the ones usually found in known activation domains.
Full text
PDF
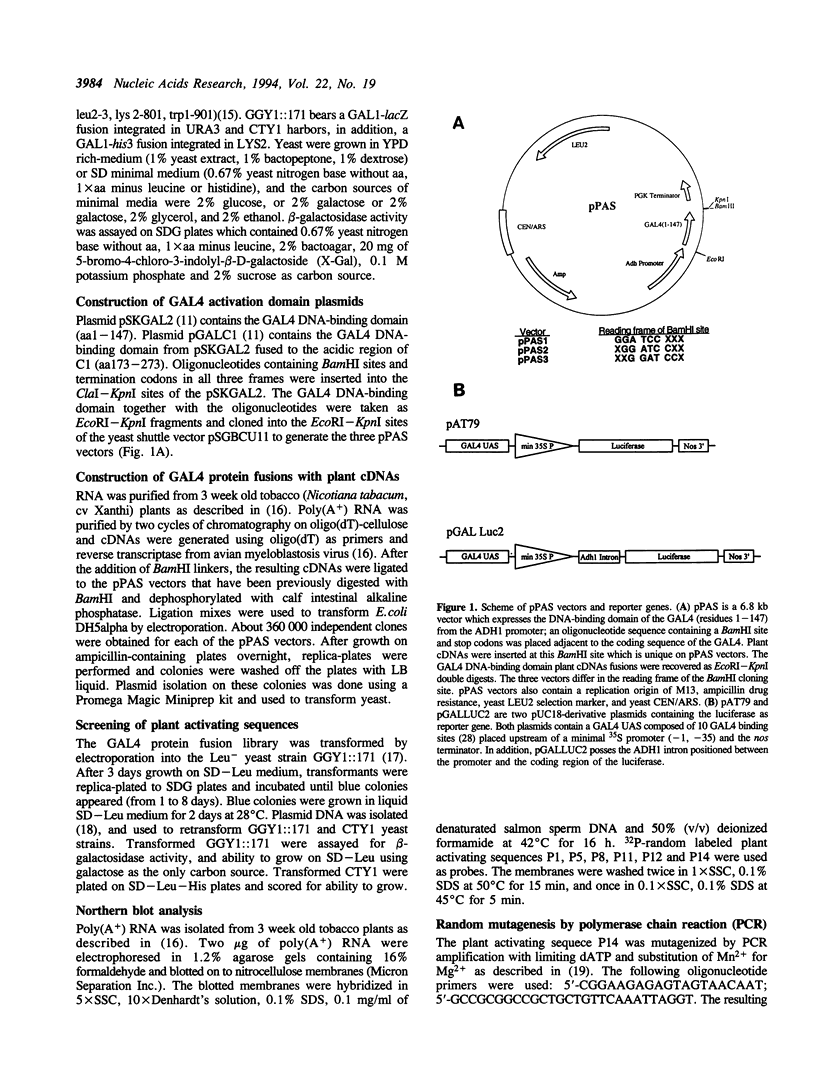





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams S. M., Helps N. R., Sharp M. G., Brammar W. J., Walker R. A., Varley J. M. Isolation and characterization of a novel gene with differential expression in benign and malignant human breast tumours. Hum Mol Genet. 1992 May;1(2):91–96. doi: 10.1093/hmg/1.2.91. [DOI] [PubMed] [Google Scholar]
- Berger S. L., Piña B., Silverman N., Marcus G. A., Agapite J., Regier J. L., Triezenberg S. J., Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992 Jul 24;70(2):251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- Callis J., Fromm M., Walbot V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987 Dec;1(10):1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988 Dec 2;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Cress W. D., Triezenberg S. J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991 Jan 4;251(4989):87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- Estruch J. J., Goff A., Crossland L. Transformation of yeast cells by electroporation in combination with a screening method that avoids catabolic repression. Biotechniques. 1994 Apr;16(4):610–612. [PubMed] [Google Scholar]
- Gill G., Ptashne M. Mutants of GAL4 protein altered in an activation function. Cell. 1987 Oct 9;51(1):121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- Giniger E., Ptashne M. Transcription in yeast activated by a putative amphipathic alpha helix linked to a DNA binding unit. Nature. 1987 Dec 17;330(6149):670–672. doi: 10.1038/330670a0. [DOI] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Goff S. A., Cone K. C., Chandler V. L. Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 1992 May;6(5):864–875. doi: 10.1101/gad.6.5.864. [DOI] [PubMed] [Google Scholar]
- Goff S. A., Cone K. C., Fromm M. E. Identification of functional domains in the maize transcriptional activator C1: comparison of wild-type and dominant inhibitor proteins. Genes Dev. 1991 Feb;5(2):298–309. doi: 10.1101/gad.5.2.298. [DOI] [PubMed] [Google Scholar]
- Kaiser P., Auer B. Rapid shuttle plasmid preparation from yeast cells by transfer to E. coli. Biotechniques. 1993 Apr;14(4):552–552. [PubMed] [Google Scholar]
- Katagiri F., Chua N. H. Plant transcription factors: present knowledge and future challenges. Trends Genet. 1992 Jan;8(1):22–27. doi: 10.1016/0168-9525(92)90020-5. [DOI] [PubMed] [Google Scholar]
- Leuther K. K., Salmeron J. M., Johnston S. A. Genetic evidence that an activation domain of GAL4 does not require acidity and may form a beta sheet. Cell. 1993 Feb 26;72(4):575–585. doi: 10.1016/0092-8674(93)90076-3. [DOI] [PubMed] [Google Scholar]
- Li J. J., Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993 Dec 17;262(5141):1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Ha I., Maldonado E., Reinberg D., Green M. R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991 Oct 10;353(6344):569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. A new class of yeast transcriptional activators. Cell. 1987 Oct 9;51(1):113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- Martin K. J., Lillie J. W., Green M. R. Evidence for interaction of different eukaryotic transcriptional activators with distinct cellular targets. Nature. 1990 Jul 12;346(6280):147–152. doi: 10.1038/346147a0. [DOI] [PubMed] [Google Scholar]
- Mermod N., O'Neill E. A., Kelly T. J., Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989 Aug 25;58(4):741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- Paz-Ares J., Ghosal D., Wienand U., Peterson P. A., Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987 Dec 1;6(12):3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Rothstein S. J., Lahners K. N., Lotstein R. J., Carozzi N. B., Jayne S. M., Rice D. A. Promoter cassettes, antibiotic-resistance genes, and vectors for plant transformation. Gene. 1987;53(2-3):153–161. doi: 10.1016/0378-1119(87)90003-5. [DOI] [PubMed] [Google Scholar]
- Santoro C., Mermod N., Andrews P. C., Tjian R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988 Jul 21;334(6179):218–224. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- Sigler P. B. Transcriptional activation. Acid blobs and negative noodles. Nature. 1988 May 19;333(6170):210–212. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- Struhl K. Molecular mechanisms of transcriptional regulation in yeast. Annu Rev Biochem. 1989;58:1051–1077. doi: 10.1146/annurev.bi.58.070189.005155. [DOI] [PubMed] [Google Scholar]
- Sáez-Vásquez J., Raynal M., Meza-Basso L., Delseny M. Two related, low-temperature-induced genes from Brassica napus are homologous to the human tumour bbc1 (breast basic conserved) gene. Plant Mol Biol. 1993 Dec;23(6):1211–1221. doi: 10.1007/BF00042354. [DOI] [PubMed] [Google Scholar]
- Van Hoy M., Leuther K. K., Kodadek T., Johnston S. A. The acidic activation domains of the GCN4 and GAL4 proteins are not alpha helical but form beta sheets. Cell. 1993 Feb 26;72(4):587–594. doi: 10.1016/0092-8674(93)90077-4. [DOI] [PubMed] [Google Scholar]
- Wang M. M., Reed R. R. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature. 1993 Jul 8;364(6433):121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]