Abstract
Nature at the lab level in biology and chemistry can be described by the application of quantum mechanics. In many cases, a reasonable approximation to quantum mechanics is classical mechanics realized through Newton’s equations of motion. Dr. Pedersen began his career using quantum mechanics to describe the properties of small molecular complexes that could serve as models for biochemical systems. To describe large molecular systems required a drop-back to classical means and this led surprisingly to a major improvement in the classical treatment of electrostatics for all molecules, not just biological molecules. Recent work has involved the application of quantum mechanics for the putative active sites of enzymes to gain greater insight into the key steps in enzyme catalysis.
Keywords: Blood coagulation cascade, Classical mechanics, DNA repair enzymes, Particle mesh Ewald, Quantum mechanical/molecular mechanical, Quantum mechanics
INTRODUCTION AND EDUCATIONAL EXPERIENCE
Dr. Lee Pedersen (Figure 1) is a Professor in the Department of Chemistry at the University of North Carolina at Chapel Hill in Chapel Hill, North Carolina. He received his bachelor’s degree in Chemistry from the University of Tulsa (Tulsa, OK) in 1961. His graduate work was done in Physical Chemistry at the University of Arkansas (Fayetteville, AS) where he focused on quantum chemistry and the potential energy surfaces for dynamical calculations (1965). Upon receiving a National Science Foundation Postdoctoral award (1965), he continued his postgraduate training in Theoretical Chemistry with Martin Karplus at Columbia University (New York, NY) and with a National Institutes of Health Postdoctoral award (1966) at Harvard University (Cambridge, MA). Dr. Pedersen has been an invited speaker at international meetings, and is a peer reviewer for many publications and granting agencies. He has won a teaching award at UNC-CH and has been honored by the establishment (UNC-CH) of the Lee G. Pedersen distinguished Professor Chair. In addition to his roles at UNC-CH, he has also been associated for more than 20 years with the Laboratory of Structural Biology at NIEHS in the Research Triangle Park, NC.
Figure 1.
Lee G Pedersen, PhD, Professor, Department of Chemistry, University of North Carolina at Chapel Hill, CB#3290, Chapel Hill, NC 27599, United States.
ACADEMIC STRATEGY AND GOALS
At the beginning of Lee Pedersen’s career in the early 1960s, computers were slow and could solve only relatively limited problems. Today, following Moore’s Law, the size of problems that can be reduced to computation or simulation is much larger. Throughout Dr. Pedersen’s career he has utilized computers in a two-pronged effort involving quantum chemistry and classical mechanics to solve essential problems in chemistry and more recently biochemistry. Currently, his group at UNC-CH is focusing on building three dimensional equilibrium representations of bio-molecular complexes (blood coagulation proteins) in electrically neutral solvents. This work is done in conjunction with experimentalists in the UNC-CH Departments of Biology and Medicine. His group at NIEHS is focused on developing mathematical tools for improving force fields for bio-molecules and on introducing quantum chemical tools into the study of enzyme reactions.
ACADEMIC ACHIEVEMENTS
The following contributions highlight Dr. Pedersen’s activities in the field of computational chemistry and biochemistry.
Barriers to internal rotation
The flexibility of bio-macromolecules is determined to a large extent by the ability of internal rotation to occur about the covalent bonds in the system. Early in his career, Dr. Pedersen, with collaborator Keiji Morokuma, computed quantum chemically the energy required to rotate simple C-C, C-N, C-O, N-N, O-O bonds. This was the first study[1] that systematically examined this problem with ab initio techniques. They found that relatively simple quantum chemical basis sets were sufficient to describe the systems chosen in agreement with known spectroscopic data. They were also able to show the importance of inclusion of polarization functions for certain systems. Similar techniques are used today to parameterize molecular force fields.
Hydrogen bond
Understanding the hydrogen bond is essential to understanding many intra and intermolecular interactions. Early in his career, with collaborator Keiji Morokuma, Dr. Pedersen computed for the first time, quantum chemically the binding energy of the water dimer in a number of possible configurations[2]. This calculation showed the lowest energy form and also showed generally what was possible with ab initio quantum chemistry. The study also discussed the new concept of basis set superposition energy in the context of an important problem. Morokuma has gone on to become one of the world’s leading theoreticians, splitting time in Japan and the USA at Emory University.
Book of problems
With Professor Johnson CS Jr, Dr. Pedersen compiled a large group of problems and detailed solutions, placing them in a pedagogical order and interspersing theoretical introductions to central topics. The resulting book[3] made quantum mechanics much more accessible to many thousands of students all over the world. The book has been revised and is now a Dover classic.
Modeling a membrane surface with ions interactions
As theoretical techniques for modeling grew in the 1980s, it became possible to think about doing simulations with lipid models of membranes. An early realization was a study performed with Paul Charifson for which an X-ray structure of dilaurolyl-DL-phosphatidyl ethanolamine was used to generate a PS/PS monolayer. Parameters were developed for PS and PC, calcium ions and solvent added, and then subsequent molecular dynamics executed[4]. The general stability of the resulting system telescoped to a bright future with membrane model simulations, a future that is being realized today.
Particle mesh Ewald development for molecular dynamics
In early struggles while attempting to perform long time molecular dynamics at the 1990-level of development on charged proteins, Tom Darden, Darrin York and Lee Pedersen found generally that aberrant behavior occurred-β sheets melted, helices unwound-proteins fell apart. A major part of the problem was discovered to be the manner in which electrostatic forces were computed at that time. Dr. Darden was a mathematician who had an unusual (for a mathematician) bent toward practical problems and his skills were applied to this longstanding problem. His efforts led to the development and application of the particle mesh Ewald method by our groups at NIEHS and UNC-CH. Two of the papers[5,6] resulting from this work have been cited more than 6500 times. This method, and the methods derived from it, was implemented into most molecular dynamics codes with the result that simulations on large systems could be performed accurately and much more rapidly. York is now a Professor of Chemistry and Chemical Biology at Rutgers University and Darden is at Open Eye Research, a think-tank-like company in Santa Fe, NM.
Coagulation proteins
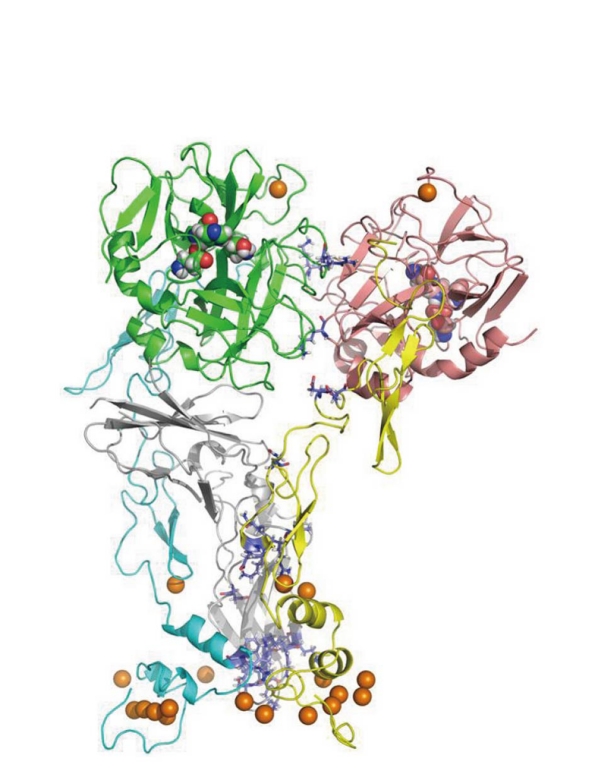
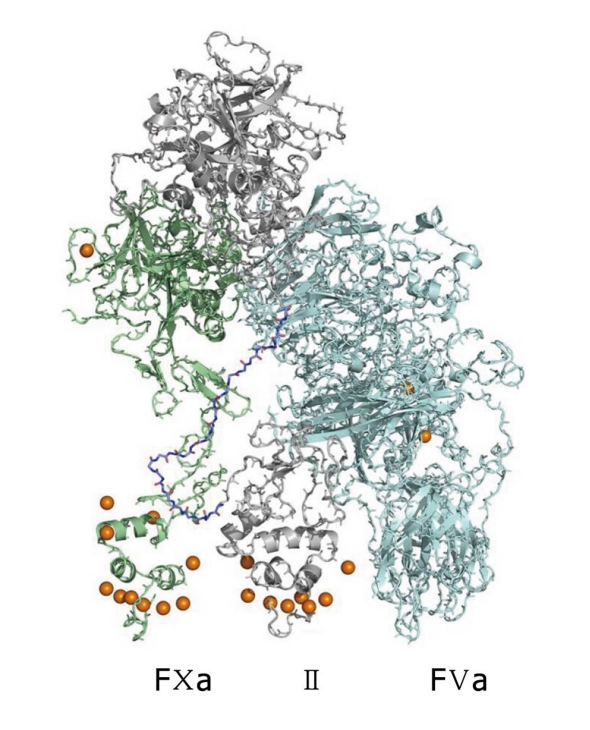
The Blood Coagulation cascade is a set of sequential reactions of enzymes that describes the phenomenon of blood clotting. Our view of what this set of reactions is changes continually. Many of these reactions involve complexes of enzymes and cofactors that react on the surfaces of cellular units such as platelets. Since the early 1990s, X-ray crystal structures of single enzymes have begun to appear, as well as complexes of the enzymes with each other and/or bio-molecular cofactors. These structures are often missing key segments and are not equilibrated in realistic solvent. The Pedersen group at UNC has developed all-atom, solution equilibrated structures for many systems, including tissue factor (TF)/factor VIIa (FVIIa)/factor Xa (FXa) (Figure 2)[7,8] and factor Va (FVa)/FXa/prothrombin(II) (Figure 3)[9] and has recently presented a kinetic/structural model for the activation of prothrombin, the penultimate step in the coagulation cascade. The figures are for all-atom models for which there is as yet no experimental structure determination.
Figure 2.
A solvent-equilibrated model of human factor VIIa/tissue factor/factor Xa. Left: Factor VIIa (FVIIa)-the SP domain is green, while the EGF2-EGF1-GLA domains are blue. Right: FXa-the SP domain is maroon, while the EGF2-EGF1-GLA domains are yellow. Center: Tissue factor-the two domains are shown in dark gray. The gold colored spheres are Ca2+ ions. The three residues that define the active sites of the serine protease domains are shown by space filling atoms. There is no experimental structure to date for this complex.
Figure 3.
A solvent-equilibrated model of human factor Va/factor Xa/II. factor Xa (FXa) is on the left in green, FVa is on the right in blue and II (prothrombin) is in the center in gray. There is no experimental structure to date for this complex
Analysis of gene expression data
Analysis programs for noisy gene expression microarray data taken from tumors were developed at NIEHS with collaborator Dr. Leping Li, Dr. Clarice Weinberg and Dr. Tom Darden[10,11]. These programs were based on a genetic algorithm/k-nearest neighbor (GA/KNN) approach that allowed differentiation between multiple subtypes of the same cancer. Part of the motivation for the approach was derived from our earlier successful application of genetic algorithms to noisy NMR data[12]. The gene analysis programs have been avidly downloaded and used worldwide.
DNA repair models
As X-ray crystal structures of binary and ternary complexes of DNA polymerases have become available, the opportunity to understand the chemistry of the repair process through modeling has arisen. The Pedersen group, working with crystallographers and kineticists at NIEHS, has been able, via quantum mechanical/molecular mechanical (QM/MM) calculations, to perform all-atom model scenarios of bond forming/bond breaking that mimic the insertion step of DNA repair. Such calculations have been carried out when the inserting NTP is correctly or incorrectly matched to the template strand of DNA. The model calculations, reported in the Proceedings of the National Academy[13,14], show the cost of energy for incorrect insertion and also show the intricate details of proton transfer that characterizes the reaction.
Electrostatic potentials
The molecular electrostatic potential (MEP) at the nucleus of an atom in a molecule is a fundamental quantum mechanical property. In collaboration with Dr. Shubin Liu at UNC-CH, Dr. Pedersen has recently been able to uncover a fundamental correlation of the MEP of electrostatic atoms in a molecule with acidity (i.e. the pKa) and to derive relationships that provide an understanding of why this correlation holds over many orders of magnitude[15,16].
CONCLUSION
The dominant variable in the career of Dr. Pedersen has been the exponential increase in the power of high performance computing over the past 50 years. It was apparent even in the 1950s that the theoretical framework provided by quantum mechanics would be sufficient to describe most chemical and biological systems and ultimately be predictive. The contributions on rotational barriers and the hydrogen bond were useful as they provided a taste of the future-the impact of quantum chemistry on biochemical problems. The ability to now examine the core of enzyme reactions with QM/MM simulations is most fulfilling. Of special importance has been the development of the particle mesh Ewald method for performing accurate and rapid computations of the electrostatic forces in large biological systems. These methods allow us to predict the equilibrium structures of solvated bio-molecular systems, including blood coagulation complexes.
Acknowledgments
I am grateful for the support of my wife Barbara and my children Lars and Kurt. I am also grateful for research support from a number of agencies: mostly NSF, NIH and the state of North Carolina. Both UNC-CH and NIEHS have provided me with nurturing environments for learning, teaching and research. The students and collaborators with whom I have worked have taught me new concepts and have provided me every now and then with the thrill that comes with original discovery.
Footnotes
Supported by The National Institutes of Health (HL-006350) and National Science Foundation (FRG DMR 0804549)
Peer reviewer: Eugene S Kryachko, PhD, Professor, Senior Scientist, Bogolyubov Institute for Theoretical Physics, National Academy of Sciences of Ukraine, Kiev-143, 03680, Ukraine
S- Editor Cheng JX L- Editor Webster JR E- Editor Zheng XM
References
- 1.Morokuma K, Pedersen L. Molecular - orbital studies of hydrogen bonds. An ab initio calculation for dimeric H2O. J Chem Phys. 1968;48:3275–3282. [Google Scholar]
- 2.Pedersen L, Morokuma K. Ab Initio Calculations of the Barriers to Internal Rotation of CH3CH3, CH3NH2, CH3OH, N2H4, H2O2, and NH2OH. J Chem Phys. 1967;46:3941–3947. [Google Scholar]
- 3.Johnson CS, Pedersen LG. Problems and solutions in quantum chemistry and physics. New York: Dover Publications; 1986. pp. 1–429. [Google Scholar]
- 4.Charifson PS, Hiskey RG, Pedersen LG. Construction and molecular modeling of phospholipid surfaces. J Comp Chem. 1990;11:1181–1186. [Google Scholar]
- 5.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 6.Darden T, York D, Pedersen L. Particle mesh Ewald: An N-log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 7.Venkateswarlu D, Duke RE, Perera L, Darden TA, Pedersen LG. An all-atom solution-equilibrated model for human extrinsic blood coagulation complex (sTF-VIIa-Xa): a protein-protein docking and molecular dynamics refinement study. J Thromb Haemost. 2003;1:2577–2588. doi: 10.1111/j.1538-7836.2003.00421.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee CJ, Chandrasekaran V, Wu S, Duke RE, Pedersen LG. Recent estimates of the structure of the factor VIIa (FVIIa)/tissue factor (TF) and factor Xa (FXa) ternary complex. Thromb Res. 2010;125 Suppl 1:S7–S10. doi: 10.1016/j.thromres.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee CJ, Lin P, Chandrasekaran V, Duke RE, Everse SJ, Perera L, Pedersen LG. Proposed structural models of human factor Va and prothrombinase. J Thromb Haemost. 2008;6:83–89. doi: 10.1111/j.1538-7836.2007.02821.x. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Weinberg CR, Darden TA, Pedersen LG. Gene selection for sample classification based on gene expression data: study of sensitivity to choice of parameters of the GA/KNN method. Bioinformatics. 2001;17:1131–1142. doi: 10.1093/bioinformatics/17.12.1131. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Darden TA, Weinberg CR, Levine AJ, Pedersen LG. Gene assessment and sample classification for gene expression data using a genetic algorithm/k-nearest neighbor method. Comb Chem High Throughput Screen. 2001;4:727–739. doi: 10.2174/1386207013330733. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Darden TA, Freedman SJ, Furie BC, Furie B, Baleja JD, Smith H, Hiskey RG, Pedersen LG. Refinement of the NMR solution structure of the gamma-carboxyglutamic acid domain of coagulation factor IX using molecular dynamics simulation with initial Ca2+ positions determined by a genetic algorithm. Biochemistry. 1997;36:2132–2138. doi: 10.1021/bi962250r. [DOI] [PubMed] [Google Scholar]
- 13.Lin P, Pedersen LC, Batra VK, Beard WA, Wilson SH, Pedersen LG. Energy analysis of chemistry for correct insertion by DNA polymerase beta. Proc Natl Acad Sci USA. 2006;103:13294–13299. doi: 10.1073/pnas.0606006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin P, Batra VK, Pedersen LC, Beard WA, Wilson SH, Pedersen LG. Incorrect nucleotide insertion at the active site of a G:A mismatch catalyzed by DNA polymerase beta. Proc Natl Acad Sci USA. 2008;105:5670–5674. doi: 10.1073/pnas.0801257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S, Pedersen LG. Estimation of molecular acidity via electrostatic potential at the nucleus and valence natural atomic orbitals. J Phys Chem A. 2009;113:3648–3655. doi: 10.1021/jp811250r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Schauer CK, Pedersen LG. Molecular acidity: A quantitative conceptual density functional theory description. J Chem Phys. 2009;131:164107. doi: 10.1063/1.3251124. [DOI] [PMC free article] [PubMed] [Google Scholar]