Abstract
Purpose
Regional lymph node disease (RLND) is a component of the risk-based treatment stratification in rhabdomyosarcoma (RMS). The purpose of this study was to determine the contribution of RLND to prognosis for patients with RMS.
Patients and Methods
Patient characteristics and survival outcomes for patients enrolled onto Intergroup Rhabdomyosarcoma Study IV (N = 898, 1991 to 1997) were evaluated among the following three patient groups: nonmetastatic patients with clinical or pathologic negative nodes (N0, 696 patients); patients with clinical or pathologic positive nodes (N1, 125 patients); and patients with a single site of metastatic disease (77 patients).
Results
Outcomes for patients with nonmetastatic alveolar N0 RMS were significantly better than for patients with N1 RMS (5-year failure-free survival [FFS], 73% v 43%, respectively; 5-year overall survival [OS], 80% v 46%, respectively; P < .001). Patients with a single site of alveolar metastasis had even worse FFS and OS (23% FFS and OS, P = .01) when compared with patients with N1 RMS; however, the differences was not as large as the differences between patients with N0 RMS and N1 RMS. For embryonal RMS, there was no statistically significant difference in FFS or OS (P = .41 and P = .77, respectively) for patients with N1 versus N0 RMS. Gene array analysis of primary tumor specimens identified that genes associated with the immune system and antigen presentation were significantly increased in N1 versus N0 alveolar RMS.
Conclusion
RLND alters prognosis for alveolar but not embryonal RMS. For patients with N1 disease and alveolar histology, outcomes were more similar to distant metastatic disease rather than local disease. Current data suggest that more aggressive therapy for patients with alveolar N1 RMS may be warranted.
INTRODUCTION
Rhabdomyosarcoma (RMS) is the most common malignant soft tissue tumor of childhood.1 Through clinical trials using multimodality therapy, under the auspices of the Intergroup Rhabdomyosarcoma Group and the Children's Oncology Group (COG), survival has steadily improved over the last three decades in North America.2,3 Approximately 15% of children present with distant metastatic RMS, and their prognosis has not significantly improved.4 However, several groups have noted that the outcomes in children with metastatic disease might not be uniformly poor because some subsets of patients demonstrate improved outcomes.5
Prognostic factors and clinical outcomes have been evaluated in patients with RMS by COG and other international cooperative study groups. The COG evaluated prognostic factors for patients with localized and distant metastatic RMS during the Intergroup Rhabdomyosarcoma Study (IRS) III and IRS-IV therapeutic trials. For patients with localized RMS, the factors most strongly associated with failure-free survival (FFS) included stage, clinical group, alveolar histology, unfavorable primary tumor sites, invasive tumors (T2), tumors larger than 5 cm, and age less than 1 or more than 10 years.6 N1 disease was identified as an independent prognostic factor only in patients with stage III alveolar disease. In addition, for patients with otherwise localized disease, such as an extremity, N1 disease may be associated with an inferior outcome.7,8 For patients with distant metastatic RMS, overall survival (OS) and FFS were significantly influenced by alveolar histology and an increasing number of metastatic sites.5 The purpose of this study was to evaluate the effect of regional lymph node disease (RLND) on prognosis for all patients with RMS. In addition, we analyzed gene expression profiles from a subset of patients with alveolar N0 and N1 RMS to determine whether a particular gene expression signature was associated with RNLD and outcome.
PATIENTS AND METHODS
Patient Population
Patients evaluated were enrolled onto IRS-IV (N = 898 patients, 1991 to 1997). Details of the chemotherapy and radiotherapy treatment have been previously published.4, 7, 9 All treatment arms were combined for our analysis because there was no significant difference in outcome. The data presented in this analysis compare patient characteristics and survival outcome among the following three groups of eligible IRS-IV patients: nonmetastatic patients with clinically or pathologically confirmed N0 disease (n = 696); nonmetastatic patients with clinically or pathologically confirmed N1 disease (n = 125); and patients with metastatic disease at a single site (n = 77). IRS-IV required lymph node biopsy of all clinically positive nodes but did not require biopsy for extremity or paratesticular (> 10 years old) primary tumors, as was required on subsequent COG clinical trials, potentially leading to an underestimation of N1 incidence. Patients with clinically confirmed N1 disease by imaging or physical examination and patients with pathologically confirmed N1 disease were combined. Patient characteristics of 125 patients classified as N1 either clinically or pathologically are presented in Appendix Table A1 and Appendix Figure A1 (online only). Although there were differences in patient and disease characteristics between groups (clinically v pathologically confirmed N1), the similar FFS and OS allowed a combined analysis of the two groups. There was good agreement between clinical and pathologic determination of nodal status (κ = 0.78; 95% CI, 0.69 to 0.86). Clinical/radiographic determination of nodal disease was correct in 84% of patients with N1 RMS and 93% of patients with N0 RMS who had pathologic confirmation (n = 222 patients with both radiographic and pathologic nodal status).
Statistical Methods
FFS is defined as the time from the start of treatment to disease progression or death, reflecting the percentage of participants who remained alive without relapse, progressive disease, or a fatal event from any cause. OS was defined as the time from the start of treatment to death from any cause. The Kaplan-Meier method was used to estimate the FFS and OS distributions.10 Differences between survival curves were analyzed by the log-rank test.11 Cox proportional hazards regression models were used to describe the association between the risk of failure and nodal status while accounting for potential confounding factors.12 The distributions of categorical patient characteristics were compared between the groups using a χ2 test or Fisher's exact test. The κ statistic was used to evaluate agreement between clinical and pathologic determination of nodal status. P < .05 was considered statistically significant, unless otherwise indicated. The site of first recurrence was defined as local if the tumor recurred only at the site of primary disease; as regional if regional lymph nodes were involved, with or without local recurrence; and as distant if any metastatic disease was present.
Analysis of Gene Expression
Microarray data for this study was from a previously published study13 and can be found at the National Cancer Institute Web site.14 From a total of 27 alveolar histology patients with annotated complete pathologic staging data, a subset of 25 IRS-IV patients with alveolar histology (N0, n = 15; N1, n = 10), with no distant metastases, and with microarray data from the primary tumor site were evaluated for differential gene expression using Affymetrix U133Av2 GeneChips (Affymetrix, Santa Clara, CA). Two samples obtained from regional lymph nodes rather than the primary site were excluded from analysis (complete pathologic data can be found in the Data Supplement). Using routine histologic examination and CD45 immunohistochemistry, all primary tumor samples evaluated for microarray analysis were screened for lymphocyte contamination (> 1%). Gene expression data were available for only three patients with embryonal N1 RMS, precluding a similar analysis. Supervised testing for differentially expressed genes was determined using selection criteria of at least 1.5 mean-fold difference in expression, filtered using a t test for a significance level of P < .05. Functional annotation was performed using the DAVID online tool (http://david .abcc.ncifcrf.gov/ease) for over-representation analysis of differentially expressed genes, and hierarchical clustering for expression matrix was performed using Pearson's correlation coefficient and complete-linkage distance metrics.15–17
Immunohistochemistry
Five-micron sections cut from paraffin blocks were stained using mouse antimyogenin antibody (clone F5D; BD Pharmingen, San Jose, CA) and polyclonal rabbit antihuman immunoglobulin (Ig) A, IgG, IgM, and κ and λ light chains (DakoCytomation, Glostrup, Denmark) after antigen retrieval using Leica Bond Epitope Retrieval Solution 2 (Leica Microsystems, Newcastle upon Tyne, United Kingdom). Primary antibodies were detected using species-specific secondary antibodies tagged with Cy3 donkey antimouse or fluorescein isothiocyanate goat antirabbit (Jackson ImmunoResearch, West Grove, PA). Images were captured using a Leica inverted fluorescence microscope, equipped with an Optronics MagnaFire digital microscope camera (Optronics, Goleta, CA); composite images were prepared using Photoshop CS3 (Adobe Systems, Mountain View, CA).
RESULTS
Patient Characteristics
Table 1 lists patient characteristics by the following three analysis groups: N0, N1, and single site of metastasis. There are significant differences between the analysis groups. Compared with patients with N0 disease, patients with N1 disease were more likely to be older than 10 years, have unfavorable primary sites, have alveolar histology, have a higher stage and group disease, and have large invasive primary tumors. The disease and patient characteristics for patients with N1 disease more closely approximate those of patients with a single site of metastatic disease. By definition, stage and group will be different between patients with nonmetastatic disease, regardless of nodal status, versus metastatic disease.
Table 1.
Demographics and Baseline Disease Characteristics According to Grouping Status
| Demographic or Disease Characteristic | Node Negative (n = 696) |
Node Positive (n = 125) |
Single Site of Metastasis (n = 77) |
P | |||
|---|---|---|---|---|---|---|---|
| No. of Patients | % | No. of Patients | % | No. of Patients | % | ||
| Age, years | .0048 | ||||||
| < 1 | 31 | 4 | 6 | 5 | 3 | 3 | |
| 1-9 | 485 | 70 | 66 | 53 | 50 | 65 | |
| ≥ 10 | 180 | 26 | 53 | 42 | 24 | 31 | |
| Sex | .0972 | ||||||
| Male | 438 | 63 | 66 | 53 | 46 | 60 | |
| Female | 258 | 37 | 59 | 47 | 31 | 40 | |
| Race | .038 | ||||||
| White | 506 | 73 | 76 | 61 | 50 | 65 | |
| Hispanic | 59 | 8 | 17 | 14 | 12 | 16 | |
| Black | 108 | 16 | 23 | 18 | 13 | 17 | |
| Other | 23 | 3 | 9 | 7 | 2 | 3 | |
| Site | < .001 | ||||||
| Favorable | 273 | 39 | 28 | 22 | 10 | 13 | |
| Unfavorable | 423 | 61 | 97 | 78 | 67 | 87 | |
| Histology | < .001 | ||||||
| Alveolar | 118 | 17 | 56 | 45 | 30 | 39 | |
| Embryonal | 505 | 73 | 54 | 43 | 36 | 47 | |
| Undifferentiated | 29 | 4 | 13 | 10 | 2 | 3 | |
| Other | 44 | 6 | 2 | 2 | 9 | 12 | |
| Stage | < .001 | ||||||
| I | 43 | 37 | 30 | 24 | 0 | 0 | |
| II | 35 | 30 | 0 | 0 | 0 | 0 | |
| III | 37 | 32 | 95 | 76 | 0 | 0 | |
| IV | 0 | 0 | 0 | 0 | 77 | 100 | |
| Group | < .001 | ||||||
| I | 196 | 28 | 0 | 0 | 0 | 0 | |
| IIA | 95 | 14 | 0 | 0 | 0 | 0 | |
| IIB | 1 | 0.1 | 12 | 10 | 0 | 0 | |
| IIC | 0 | 0 | 11 | 9 | 0 | 0 | |
| III | 404 | 58 | 102 | 82 | 0 | 0 | |
| IV | 0 | 0 | 0 | 0 | 77 | 100 | |
| Size, cm | < .001 | ||||||
| < 5 | 380 | 55 | 35 | 28 | 13 | 17 | |
| ≥ 5 | 316 | 45 | 90 | 72 | 64 | 83 | |
| Tumor stage | < .001 | ||||||
| T1 | 397 | 57 | 29 | 23 | 8 | 11 | |
| T2 | 299 | 43 | 96 | 77 | 68 | 89 | |
Incidence of RLND
Although the overall frequency of N1 disease was 23%, higher rates were seen at the following selected primary sites: perineum (50%), retroperitoneum (28%), and extremity (23%; Table 2).
Table 2.
Incidence of Regional Lymph Node Disease by Site of Primary Disease
| Site | Node Negative (n = 696) |
Node Positive (n = 125) |
Pathologic Node Positive (n = 59) |
Clinical Node Positive (n = 66) |
||||
|---|---|---|---|---|---|---|---|---|
| No. of Patients | % | No. of Patients | % | No. of Patients | % | No. of Patients | % | |
| Extremity | 79 | 77 | 24 | 23 | 16 | 15 | 8 | 8 |
| Head/neck/orbit | 130 | 91 | 13 | 9 | 7 | 5 | 6 | 4 |
| Bladder/prostate | 81 | 88 | 11 | 12 | 3 | 3 | 8 | 9 |
| GI | 7 | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Parameningeal | 169 | 82 | 36 | 18 | 14 | 7 | 22 | 11 |
| Paratestis | 113 | 89 | 14 | 11 | 9 | 7 | 5 | 4 |
| Perineum | 7 | 50 | 7 | 50 | 6 | 43 | 1 | 7 |
| Retroperineum | 41 | 72 | 16 | 28 | 3 | 5 | 13 | 23 |
| Trunk | 35 | 94 | 2 | 6 | 1 | 3 | 1 | 3 |
| Uterine/vagina | 30 | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
Patient Outcomes
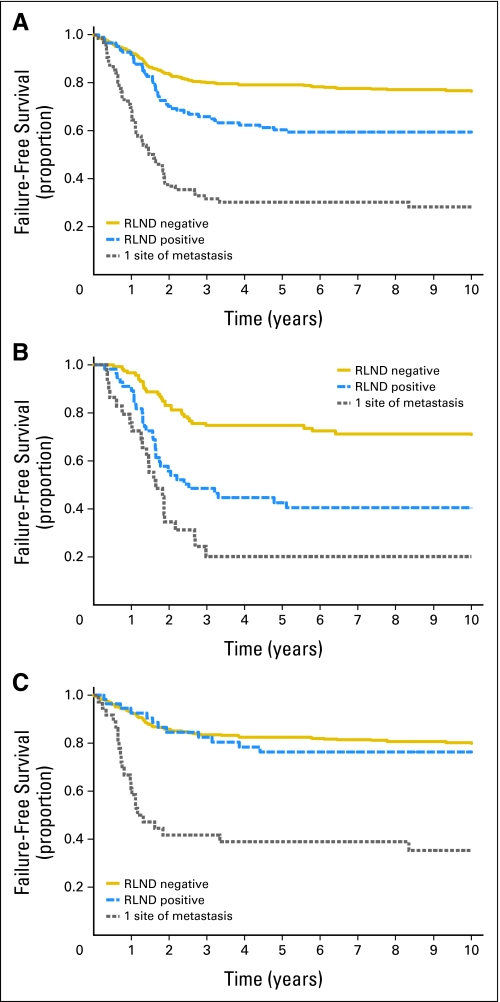
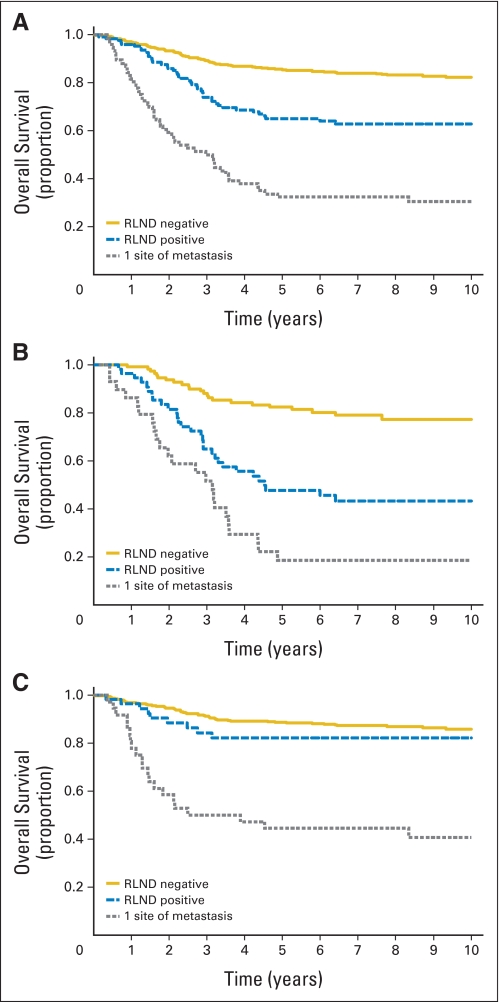
Comparing the different analysis groups, there was a statistically significant difference in FFS (Fig 1A) and OS (Appendix Fig A2A, online only). Patients with N0 disease had the best outcome, and patients with a single metastatic site had the worst outcome. Patients with N1 disease had an intermediate outcome for both FFS and OS (P < .001 for all comparisons).
Fig 1.
Failure-free survival curves for patients who are regional lymph node disease (RLND) negative, patients who are RLND positive, and patients with one site of metastatic disease in (A) both embryonal and alveolar rhabdomyosarcoma (RMS), (B) alveolar RMS only, and (C) embryonal RMS only.
Patient outcomes based on tumor histology (alveolar and embryonal) were also evaluated. Patients with alveolar tumors, either N1 or a single metastatic focus, had similar FFS rates that were significantly worse when compared with N0 alveolar tumors (P < .001; Fig 1B). OS for patients with alveolar histology was similar to FFS (P < .001; Appendix Fig A2B). In contrast, patients with embryonal RMS had similar FFS rates regardless of nodal status; however, patients with a single distant metastatic focus had significantly worse FFS (P < .001; Fig 1C) and OS (P < .001; Appendix Fig A2C).
Predictors of Outcome
To determine whether RLND is an independent predictor of outcome, we used a multivariate regression analysis model. Stepwise, multivariate regression models initially included RLND, group, age, and tumor stage, size, and site and their association with FFS and OS separately for alveolar and embryonal histology. Statistically significant predictors of FFS are displayed in Table 3; identical predictors were identified for OS. Node status was the only statistically significant predictor of outcome for patients with alveolar tumors. For patients with embryonal tumors, the statistically significant predictors of outcome included RLND, patient age, tumor invasion, and site of primary tumors. However, after adjusting for the other predictors in the model, there were no statistically significant differences in outcome for N0 versus N1 disease in embryonal tumors (FFS, P = .41; OS, P = .77). Patients with a single site of metastasis with embryonal histology had significantly poorer outcomes than patients with N1 disease (FFS, P < .001; OS, P < .001) after adjusting for the other statistically significant factors.
Table 3.
Multivariate Analysis of FFS and OS
| Predictor | Hazard Ratio | 95% CI | P |
|---|---|---|---|
| Alveolar FFS | |||
| Node group | |||
| Node positive | 1 | — | |
| Node negative | 0.4 | 0.2 to 0.6 | < .001 |
| Single site of metastasis | 1.8 | 1.0 to 3.0 | .04 |
| Alveolar OS | |||
| Node group | |||
| Node positive | 1 | — | |
| Node negative | 0.3 | 0.2 to 0.5 | < .001 |
| Single site of metastasis | 2.0 | 1.1 to 3.4 | .01 |
| Embryonal FFS | |||
| Node group | |||
| Node positive | 1 | — | |
| Node negative | 1.3 | 0.7 to 2.4 | .41 |
| Single site of metastasis | 5.2 | 2.6 to 10.5 | < .001 |
| Age at diagnosis, years | |||
| < 10 | 1 | — | |
| ≥ 10 | 2.2 | 1.5 to 3.2 | < .001 |
| Tumor stage | |||
| T1 | 1 | — | |
| T2 | 2.5 | 1.7 to 3.7 | < .001 |
| Site | |||
| Other | 1 | — | |
| Extremity | 2.5 | 1.3 to 5.1 | .008 |
| Embryonal OS | |||
| Node group | |||
| Node positive | 1 | — | |
| Node negative | 1.1 | 0.5 to 2.3 | .77 |
| Single site of metastasis | 6.0 | 2.7 to 13.3 | < .001 |
| Age at diagnosis, years | |||
| < 10 | 1 | — | |
| ≥ 10 | 2.2 | 1.5 to 3.2 | < .001 |
| Tumor stage | |||
| T1 | 1 | — | |
| T2 | 2.5 | 1.7 to 3.7 | < .001 |
| Site | |||
| Other | 1 | — | |
| Extremity | 2.5 | 1.3 to 5.1 | < .001 |
| Parameningeal | — | — | .002 |
Abbreviations: FFS, failure-free survival; OS, overall survival.
Site of Relapse
Of the total 253 patients who experienced relapse, 151 (22% rate of relapse) had N0 disease, 48 (38%) had N1 disease, and 54 (70%) had metastatic disease. Complete information concerning site of relapse was only available for 225 study participants. The types of relapse are similar between the analysis groups (P = .54, Appendix Table A2, online only).
Gene Array Analysis
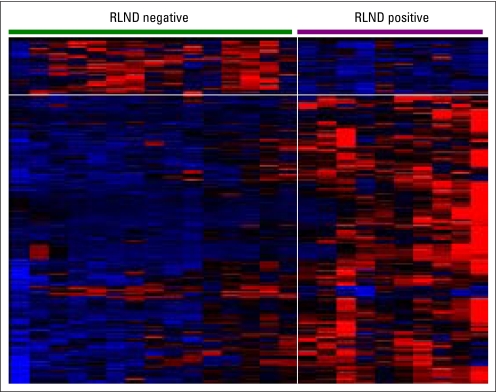
To explore whether the significant differences in FFS and OS observed in alveolar RMS for N1 versus N0 disease were associated with significant differences in tumor biology, we analyzed a subset of tumors of 25 IRS-IV patients using oligonucleotide microarrays to profile gene expression. All of the expression profiles analyzed were from primary tumor material, and none contained more than 1% leukocyte infiltration. We found 198 probe sets that were differentially expressed (160 and 38 probe sets at increased expression in N1 and N0 tumors, respectively; Appendix Fig A3, online only, and Data Supplement). Table 4 lists the most differentially expressed probe sets and shows that most of the genes overexpressed in N1 alveolar tumors are immune markers including β2-microglobulin, complement components, major histocompatibility complex molecules, and Igs. This pattern of overexpressed immune markers was present in 80% of N1 alveolar tumors. Rigorous over-representation analysis of Gene Ontology categories confirms that in N1 alveolar tumors, 40% of the overexpressed genes encode immune response proteins whose most frequent molecular function and cellular compartment were antigen binding and major histocompatibility complex protein complex, respectively (Data Supplement). Particularly prominent were those associated with antigen-presenting cells such as B cells. The immune response signatures detected in this subset of N1 alveolar tumors suggests activation of a gene expression program that may contribute to evasion of immune cell–mediated tumor apoptosis. A similar analysis for embryonal tumors was not possible given the low number of patients (n = 3) with N1 disease.
Table 4.
The 40 Most Significant Probe Sets (ranked by P value) From the List of Differentially Expressed Probe Sets Between Patients With Alveolar RMS RLND Positive and Negative Disease
| Gene Name | Gene Symbol | Mean FD | P (ttest) |
|---|---|---|---|
| Apolipoprotein E | APOE | 1.7 | < .001 |
| Apolipoprotein E | APOE | 1.6 | < .001 |
| β2-microglobulin | B2M | 2.2 | < .001 |
| Complement component 1, q subcomponent, A chain | C1QA | 1.8 | < .001 |
| Complement component 1, r subcomponent | C1R | 1.6 | < .001 |
| Complement component 1, s subcomponent | C1S | 1.8 | < .001 |
| Complement component 7 | C7 | 2.1 | < .001 |
| Calcium channel, voltage-dependent, P/Q type, α1A subunit | CACNA1A | −1.6 | < .001 |
| CD74 molecule, major histocompatibility complex, class II invariant chain | CD74 | 2.3 | < .001 |
| Carboxypeptidase E | CPE | 1.8 | < .001 |
| GTP binding protein overexpressed in skeletal muscle | GEM | 1.7 | < .001 |
| Major histocompatibility complex, class I, A | HLA-A | 1.9 | < .001 |
| Major histocompatibility complex, class I, A | HLA-A | 2.4 | < .001 |
| Major histocompatibility complex, class I, B | HLA-B | 2.6 | < .001 |
| Major histocompatibility complex, class I, B | HLA-B | 2.4 | < .001 |
| Major histocompatibility complex, class I, C | HLA-C | 2.6 | < .001 |
| Major histocompatibility complex, class I, C | HLA-C | 2.4 | < .001 |
| Major histocompatibility complex, class I, C | HLA-C | 2.3 | < .001 |
| Major histocompatibility complex, class II, DM α | HLA-DMA | 1.5 | < .001 |
| Major histocompatibility complex, class II, DP α1 | HLA-DPA1 | 2.1 | < .001 |
| Major histocompatibility complex, class II, DP α1 | HLA-DPA1 | 2.5 | < .001 |
| Major histocompatibility complex, class II, DP β1 | HLA-DPB1 | 2.0 | < .001 |
| Major histocompatibility complex, class II, DQ β1 | HLA-DQB1 | 1.6 | < .001 |
| Major histocompatibility complex, class II, DR α | HLA-DRA | 2.2 | < .001 |
| Major histocompatibility complex, class II, DR α | HLA-DRA | 2.2 | < .001 |
| Major histocompatibility complex, class II, DR β1 | HLA-DRB1 | 2.2 | < .001 |
| Major histocompatibility complex, class II, DR β1 | HLA-DRB1 | 2.2 | < .001 |
| Major histocompatibility complex, class II, DR β1 | HLA-DRB1 | 2.0 | < .001 |
| Major histocompatibility complex, class II, DR β1 | HLA-DRB1 | 2.5 | < .001 |
| Major histocompatibility complex, class I, F | HLA-F | 1.8 | < .001 |
| Insulin-like growth factor binding protein 7 | IGFBP7 | 2.3 | < .001 |
| Immunoglobulin heavy locus | IGH@ | 6.0 | < .001 |
| Immunoglobulin κ locus | IGK@ | 2.4 | < .001 |
| Immunoglobulin κ constant | IGKC | 2.0 | < .001 |
| Immunoglobulin κ constant | IGKC | 3.7 | < .001 |
| Immunoglobulin κ constant | IGKC | 4.0 | < .001 |
| Immunoglobulin κ variable 1D-13 | IGKV1D-13 | 1.5 | < .001 |
| Immunoglobulin λ locus | IGL@ | 4.8 | < .001 |
| Immunoglobulin λ-like polypeptide 3 | IGLL3 | 1.7 | < .001 |
| Interleukin-6 signal transducer (gp130, oncostatin M receptor) | IL6ST | 1.8 | < .001 |
Abbreviations: RMS, rhabdomyosarcoma; RLND, regional lymph node disease; FD, fold difference.
Relative to patients who are RLND positive.
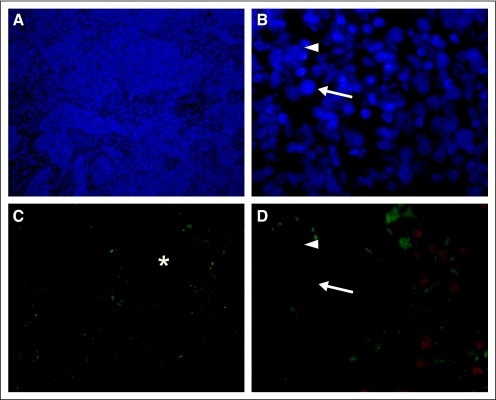
To support the gene array findings, we performed immunohistochemistry on a subset of primary tumors from four patients identified by the gene array to highly express Ig κ constant (Fig 2). Manual counting of 10 myogenin-positive cells in five random fields for each patient revealed that 45% (standard deviation, 28%) of tumor cells coexpressed Ig chains. These findings show that both B-cell–specific genes and proteins are expressed in N1 alveolar RMS tumor cells.
Fig 2.
Representative photomicrograph of rhabdomyosarcoma tissue after dual immunofluorescence staining for myogenin (MGN, red) and immunoglobulin (Ig, green). (A, C) Low magnification (×100) shows densely cellular tumor (4,6-diamidino-2-phenylindole) in which regions containing MGN- and Ig-expressing cells generally overlap (*). (B, D) Higher magnification (×630) shows that some MGN-expressing nuclei are in cells expressing Ig on the cell surface (arrow), whereas others are not (arrowhead). Staining here is representative of four separate samples.
DISCUSSION
This study demonstrates several important clinical findings and biologic associations for patients with N1 RMS. Patients with N1 disease, compared with patients with N0 disease, more commonly have disease characteristics associated with poor prognosis. In addition, N1 disease was present in 23% of all patients with RMS, with sites such as perineum, retroperitoneum, extremity, bladder/prostate, parameningeal, and paratesticular having a greater than 10% incidence of N1. N1 disease alters both FFS and OS for alveolar but not embryonal RMS. For patients with alveolar histology and N1 disease, outcomes were more similar to patients with a single site of metastatic disease than patients with N0 disease. For alveolar RMS, multivariate analysis confirmed the importance of N1 as an independent prognostic factor. However, these findings did not extend to embryonal RMS. Finally, gene array analysis of tumors shows that different categories of genes are upregulated in patients with N1 disease depending on tumor histology. Specifically, there are associations between the expression of immune response genes and patients with N1 disease and alveolar histology.
The unfavorable clinical features in patients with N1 disease mirror those traditionally seen in patients with distant metastatic disease, who are more likely to be older and have alveolar histology, larger invasive tumors, and unfavorable primary tumor sites.4,5,18 The European Intergroup studies (MMT4-89 and MMT4-91) additionally found a 55% incidence of N1 disease.18
The outcomes analysis for the current study identified significant differences between analysis groups for both FFS and OS. Patients with N0 disease had the best outcomes, and patients with a single metastatic site had the worst outcomes. Patients who had N1 disease had an intermediate prognosis. Outcomes analysis based on tumor histology showed that patients with N1 alveolar tumors or a single metastatic focus had similar outcomes that were significantly worse when compared with patients with N0 disease. However, patients with embryonal histology with either N0 or N1 disease had similar outcomes, whereas patients with a single metastatic focus still faired poorly.
The significance of N1 disease on prognosis for either localized or metastatic tumors has varied in prior large clinical trials. For patients with metastatic RMS, OS at 3 years was significantly influenced by tumor histology and increasing numbers of metastatic sites, whereas the prognostic significance of RLND was marginal by univariate analysis.4,5 In multivariate analysis, the only factor that correlated significantly with improved FFS and OS was the presence of two or fewer metastatic sites (P = .007 and P = .006, respectively). The European Intergroup studies (MMT4-89 and MMT4-91) similarly showed that age between 1 and 10 years, the absence of bone or bone marrow metastases, and primary tumor in head and neck or genitourinary sites (nonbladder/prostate) were found to be independently favorable factors for both OS and FFS.18 Meza et al6 demonstrated in their multivariate analysis of North American patients with nonmetastatic RMS that only stage and group were significantly associated with FFS for most patients with alveolar RMS. However, for patients with stage III, group III alveolar RMS, N1 was associated with poorer FFS and OS. In contrast, in patients with embryonal RMS stage and group, large tumor size (> 5 cm), unfavorable primary site, age less than 1 or more than 10 years, and invasive tumors were associated with inferior FFS.9 Evaluating a pooled cohort of 951 international patients with nonmetastatic RMS, important prognostic factors determined by univariate analysis included tumor invasiveness, tumor size, primary site, and N1 disease.19 However, by multivariate Cox regression, only tumor invasiveness and primary site remained significant predictors of survival. These observations have resulted in current European Sarcoma Study Group treatment protocols assigning patients with alveolar N1 RMS, but not embryonal N1 RMS, to the very high–risk treatment protocol.20 Thus, N1 status impacts treatment intensity in alveolar but not embryonal RMS. The lack of significance for N1 disease on outcome in the majority of prior COG and European studies most likely reflects that the majority of RMS disease is embryonal; therefore, the preponderance of these patients in the data analysis may have masked the effect of N1 disease on outcome for alveolar RMS.
Genomic analysis of human tumor specimens has redefined tumor classes based on molecular features and identified new subclasses previously unrecognized by conventional histology or cytogenetics. Genetic analysis is currently being evaluated as a clinical tool to determine patient prognosis and identify patients at risk for development of nodal and metastatic disease. For example, Rickman et al21 analyzed head and neck squamous cell carcinoma. They concluded that metastatic and poorly differentiated tumors were characterized a pattern of gene expression that was distinct from localized and well-differentiated tumors. More specifically regarding nodal disease, genetic expression profiling was performed by Kashiwazaki et al22 on N0 and N1 oral squamous cell carcinoma. Genes expressed at higher levels in N1 disease included angiogenesis-related molecules, cell adhesion molecules, and proteolytic enzymes. Similarly, Tamoto et al23 showed that N1 esophageal carcinomas overexpressed genes involved in both cell adhesion and cell membrane receptors; whereas those with N0 disease overexpressed genes involved in cell cycle regulation and intracellular signaling. These studies suggest that tumors likely to metastasize to regional lymph nodes may be identified by gene expression profiling.
Davicioni et al15 recently have shown that RMS variants (alveolar, embryonal, and undifferentiated) are associated with distinct gene expression profiles, reflecting their inherent biologic and clinicopathologic differences. In this more focused study, we find additional heterogeneity within the alveolar tumors not previously recognized. The genetic profile of patients with N1 alveolar RMS showed an overexpression of genes encoding proteins involved in immune response functions, such as antigen processing, presentation, and binding, in the vast majority of patients. We observed that most of the N1 alveolar RMS tumors expressed B-cell markers20 and expressed many markers normally associated with lymphoid tissue (CD20, CD19, and CD24) in addition to hallmark alveolar RMS molecular features (eg, PAX/FKHR translocation, MyoD1, actin, myoglobin, desmin expression). The increased expression of this immune response signature may provide an explanation for the distinct biologic behavior and poorer prognosis in patients with N1 alveolar RMS and thus could potentially lead to novel therapeutic strategies targeted to these patients. It should be noted that the numbers of patients in this genetic analysis are too small to provide any definitive conclusions. However, an analysis of adult patients with sarcoma supports our findings, having shown the expression of B-cell genes and proteins in tumor cells.24
The clinical implications of our findings, concerning the prognostic impact of N1 disease in alveolar RMS, are currently under discussion within the COG. We believe that N1 status is important for all patients with RMS, including embryonal RMS. We postulate that given current therapy, which is increased in N1 disease with radiotherapy to the positive nodal basin, it is possible that the negative effect of N1 disease in embryonal RMS is abrogated. Therefore, if we stopped treating N1 embryonal RMS with additional radiotherapy, it is possible that these patients' outcomes would worsen. It is our conclusion that we are not overtreating patients with N1 embryonal RMS but, more likely, that we are undertreating patients with N1 alveolar RMS and that, for these patients, intensification of therapy may improve their outcomes.
Supplementary Material
Acknowledgment
We thank Karen Bachmeyer and Raphael Wilson for performing the immunohistochemistry.
Appendix
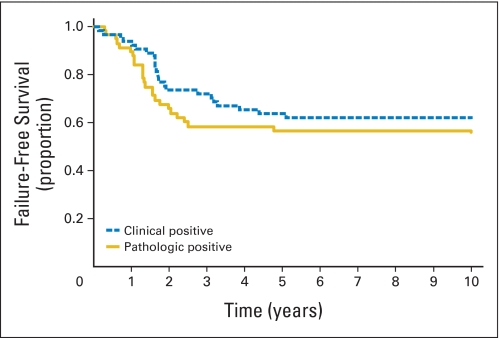
Fig A1.
Failure-free survival curve for patients with rhabdomyosarcoma with clinically positive or pathologically confirmed regional lymph node disease. No statistical difference between patients with clinically positive or pathologically confirmed disease (P = .35).
Fig A2.
Overall survival curves for patients who are regional lymph node disease (RLND) negative, patients who are RLND positive, and patients with one site of metastatic disease in (A) both embryonal and alveolar rhabdomyosarcoma (RMS), (B) alveolar RMS only, and (C) embryonal RMS only.
Fig A3.
Expression matrix derived from hierarchical clustering of 198 probe sets representing genes differentially expressed between regional lymph node disease (RLND) –negative and RLND-positive alveolar rhabdomyosarcoma. Colored heat map depicts expression above (red) and below (blue) the median (black) expression level for each probe set (rows) across the samples (columns).
Table A1.
Demographics and Baseline Disease Characteristics of Patients Classified As N1 Either Clinically or Pathologically
| Demographic or Clinical Characteristic | Pathologically Positive (n = 59) |
Clinically Positive (n = 66) |
P | ||
|---|---|---|---|---|---|
| No. of Patients | % | No. of Patients | % | ||
| Age, years | .04 | ||||
| < 1 | 2 | 3 | 4 | 6 | |
| 1-10 | 25 | 42 | 41 | 62 | |
| > 10 | 32 | 54 | 21 | 32 | |
| Sex | .86 | ||||
| Male | 32 | 54 | 34 | 52 | |
| Female | 27 | 46 | 32 | 48 | |
| Race | .49 | ||||
| White | 40 | 68 | 36 | 55 | |
| Hispanic | 6 | 10 | 11 | 17 | |
| Black | 9 | 15 | 14 | 21 | |
| Other | 4 | 7 | 5 | 8 | |
| Site | .0079 | ||||
| Extremity | 16 | 27 | 8 | 12 | |
| Head/neck/orbit | 7 | 12 | 6 | 9 | |
| Bladder/prostate | 3 | 5 | 8 | 12 | |
| Parameningeal | 14 | 24 | 22 | 33 | |
| Paratestis | 9 | 15 | 5 | 8 | |
| Perineum | 6 | 10 | 1 | 2 | |
| Retroperitoneum | 3 | 5 | 13 | 20 | |
| Trunk | 1 | 2 | 1 | 2 | |
| Other | 0 | 2 | 3 | ||
| Histology | <.001 | ||||
| Alveolar | 41 | 69 | 15 | 23 | |
| Embryonal | 17 | 24 | 37 | 56 | |
| Other | 1 | 2 | 14 | 21 | |
| Stage | .14 | ||||
| I | 18 | 31 | 12 | 18 | |
| II | 0 | 0 | 0 | 0 | |
| III | 41 | 69 | 54 | 82 | |
| IV | 0 | 0 | 0 | 0 | |
| Group | < .001 | ||||
| I | 0 | 0 | 0 | 0 | |
| IIA | 0 | 0 | 0 | 0 | |
| IIB | 11 | 19 | 1 | 2 | |
| IIC | 10 | 17 | 1 | 2 | |
| III | 38 | 64 | 64 | 97 | |
| Size, cm | .11 | ||||
| 5 | 21 | 36 | 14 | 21 | |
| > 5 | 38 | 64 | 52 | 79 | |
| T stage | .4 | ||||
| T1 | 16 | 27 | 13 | 20 | |
| T2 | 43 | 73 | 53 | 80 | |
Table A2.
Type of Relapse for Each Analysis Group
| Type of Relapse | Node Negative |
Node Positive |
Single Site of Metastasis |
All Patients (No.) | |||
|---|---|---|---|---|---|---|---|
| No. of Patients | % | No. of Patients | % | No. of Patients | % | ||
| Local* | 80 | 59 | 22 | 51 | 23 | 50 | 125 |
| Regional* | 17 | 12 | 3 | 7 | 5 | 9 | 25 |
| Distant | 34 | 25 | 14 | 33 | 15 | 33 | 63 |
| CNS extension | 5 | 4 | 4 | 9 | 3 | 7 | 12 |
| Total | 136 | 43 | 46 | 225 | |||
Includes local only plus local as a component of local plus regional relapse and local plus distant relapse.
Includes regional only plus regional as a component of regional and distant relapse.
Footnotes
Supported by Grants No. U10 CA24507 (Intergroup Rhabdomyosarcoma Study Group) and U10 CA98543 (Children's Oncology Group Chair's Grant) from the National Cancer Institute, National Institutes of Health, Bethesda, MD.
Presented in part at the 2007 American Academy of Pediatrics National Conference and Exhibition, San Francisco, CA, October 26-30, 2007.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: David A. Rodeberg, David M. Parham, Andrea A. Hayes-Jordan, Timothy J. Triche, Douglas S. Hawkins
Administrative support: William H. Meyer
Provision of study materials or patients: David A. Rodeberg, Elai Davicioni, Sarah S. Donaldson, Timothy J. Triche, William H. Meyer
Collection and assembly of data: David A. Rodeberg, Elizabeth R. Lyden, Elai Davicioni, David M. Parham, Stephen X. Skapek, Andrea A. Hayes-Jordan, Timothy J. Triche
Data analysis and interpretation: David A. Rodeberg, Norbert Garcia-Henriquez, Elizabeth R. Lyden, Elai Davicioni, David M. Parham, Stephen X. Skapek, Kenneth L. Brown, Timothy J. Triche, Douglas S. Hawkins
Manuscript writing: David A. Rodeberg, Norbert Garcia-Henriquez, Elizabeth R. Lyden, Elai Davicioni, David M. Parham, Stephen X. Skapek, Andrea A. Hayes-Jordan, Sarah S. Donaldson, Kenneth L. Brown, Timothy J. Triche, William H. Meyer, Douglas S. Hawkins
Final approval of manuscript: David A. Rodeberg, Norbert Garcia-Henriquez, Elizabeth R. Lyden, Elai Davicioni, David M. Parham, Stephen X. Skapek, Andrea A. Hayes-Jordan, Sarah S. Donaldson, Kenneth L. Brown, Timothy J. Triche, William H. Meyer, Douglas S. Hawkins
REFERENCES
- 1.Gurney JG, Young JL, Roffers SD, et al. Bethesda, MD: National Cancer Institute; 1999. Soft tissue sarcomas, in: SEER Pediatric Monograph. [Google Scholar]
- 2.Raney RB, Anderson JR, Barr FG, et al. Rhabdomyosarcoma and undifferentiated sarcoma in the first two decades of life: A selective review of Intergroup Rhabdomyosarcoma Study Group experience and rationale for Intergroup Rhabdomyosarcoma Study V. J Pediatr Hematol Oncol. 2001;23:215–220. doi: 10.1097/00043426-200105000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Pappo AS, Shapiro DN, Crist WM, et al. Biology and therapy of pediatric rhabdomyosarcoma. J Clin Oncol. 1995;13:2123–2139. doi: 10.1200/JCO.1995.13.8.2123. [DOI] [PubMed] [Google Scholar]
- 4.Breneman JC, Lyden E, Pappo AS, et al. Prognostic factors and clinical outcomes in children and adolescents with metastatic rhabdomyosarcoma: A report from the Intergroup Rhabdomyosarcoma Study IV. J Clin Oncol. 2003;21:78–84. doi: 10.1200/JCO.2003.06.129. [DOI] [PubMed] [Google Scholar]
- 5.Oberlin O, Rey A, Lyden E, et al. Prognostic factors in metastatic rhabdomyosarcomas: Result of a pooled analysis from United States and European cooperative groups. J Clin Oncol. 2008;26:2384–2389. doi: 10.1200/JCO.2007.14.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meza JL, Anderson J, Pappo AS, et al. Analysis of prognostic factors in patients with non-metastatic rhabdomyosarcoma treated on Intergroup Rhabdomyosarcoma studies III and IV: The Children's Oncology Group. J Clin Oncol. 2006;24:3844–3851. doi: 10.1200/JCO.2005.05.3801. [DOI] [PubMed] [Google Scholar]
- 7.Donaldson SS, Meza J, Breneman JC, et al. Results from the IRS-IV randomized trial of hyperfractionated radiotherapy in children with rhabdomyosarcoma: A report from the IRSG. Int J Radiat Oncol Biol Phys. 2001;51:718–728. doi: 10.1016/s0360-3016(01)01709-6. [DOI] [PubMed] [Google Scholar]
- 8.Neville HL, Andrassy RJ, Lobe TE, et al. Preoperative staging, prognostic factors, and outcome for extremity rhabdomyosarcoma: A preliminary report from the Intergroup Rhabdomyosarcoma Study IV (1991-1997) J Pediatr Surg. 2000;35:317–321. doi: 10.1016/s0022-3468(00)90031-9. [DOI] [PubMed] [Google Scholar]
- 9.Crist WM, Anderson JR, Meza JL, et al. Intergroup Rhabdomyosarcoma Study-IV: Results for patients with nonmetastatic disease. J Clin Oncol. 2001;19:3091–3102. doi: 10.1200/JCO.2001.19.12.3091. [DOI] [PubMed] [Google Scholar]
- 10.Kaplan GL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 11.Peto R, Pike MC, Armitage P, et al. Design and analysis of randomized clinical trials requiring prolonged observation of each patient: II. Analysis and examples. Br J Cancer. 1977;35:1–39. doi: 10.1038/bjc.1977.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox DR. Regression models and life tables. J R Stat Soc B. 1972;34:187–220. [Google Scholar]
- 13.Davicioni E, Anderson MJ, Finckenstein FG, et al. Molecular classification of rhabdomyosarcoma: Genotypic and phenotypic determinants of diagnosis—A report from the Children's Oncology Group. Am J Pathol. 2009;174:550–564. doi: 10.2353/ajpath.2009.080631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Cancer Institute: caArray. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. https://array.nci.nih.gov/caarray/project/trich-00099. [DOI] [PubMed]
- 15.Davicioni E, Finckenstein FG, Shahbazian V, et al. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66:6936–6946. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 16.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 18.Carli M, Colombatti R, Oberlin O, et al. European intergroup studies (MMT4-89 and MMT4-91) on childhood metastatic rhabdomyosarcoma: Final results and analysis of prognostic factors. J Clin Oncol. 2004;22:4787–4794. doi: 10.1200/JCO.2004.04.083. [DOI] [PubMed] [Google Scholar]
- 19.Rodary C, Gehan EA, Flamant F, et al. Prognostic factors in 951 nonmetastatic rhabdomyosarcoma in children: A report from the International Rhabdomyosarcoma Workshop. Med Pediatr Oncol. 1991;19:89–95. doi: 10.1002/mpo.2950190204. [DOI] [PubMed] [Google Scholar]
- 20.Bergeron C, Thiesse P, Rey A, et al. Revisiting the role of doxorubicin in the treatment of rhabdomyosarcoma: An up-front window study in newly diagnosed children with high-risk metastatic disease. Eur J Cancer. 2008;44:427–431. doi: 10.1016/j.ejca.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Rickman DS, Millon R, De Reynies A, et al. Prediction of future metastasis and molecular characterization of head and neck squamous-cell carcinoma based on transcriptome and genome analysis by microarrays. Oncogene. 2008;27:6607–6622. doi: 10.1038/onc.2008.251. [DOI] [PubMed] [Google Scholar]
- 22.Kashiwazaki H, Hassan NM, Hamada J, et al. Gene expression profile changes correlated with lymph node metastasis in oral squamous cell carcinoma. Odontology. 2008;96:38–43. doi: 10.1007/s10266-008-0084-1. [DOI] [PubMed] [Google Scholar]
- 23.Tamoto E, Tada M, Murakawa K, et al. Gene-expression profile changes correlated with tumor progression and lymph node metastasis in esophageal cancer. Clin Cancer Res. 2004;10:3629–3638. doi: 10.1158/1078-0432.CCR-04-0048. [DOI] [PubMed] [Google Scholar]
- 24.Chen Z, Huang X, Ye J, et al. Immunoglobulin G is present in a wide variety of soft tissue tumors and correlates well with proliferation markers and tumor grades. Cancer. 2010;116:1953–1963. doi: 10.1002/cncr.24892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.