Abstract
Purpose
Glioma-associated oncogene family zinc finger 1 (GLI1) expression was assessed to determine a potential role of hedgehog (Hh) signaling in head and neck squamous cell carcinoma (HNSCC). Additional proteins known to be modulated by Hh signaling, including beta-catenin (CTNNB1) and epidermal growth factor receptor (EGFR), were also assessed to determine the correlation among these distinct signaling pathways.
Patients and Methods
Nuclear GLI1 and CTNNB1 expression levels were determined in tumors from patients enrolled on Radiation Therapy Oncology Group (RTOG) 9003, a radiation fractionation trial. The results were also correlated with previously determined EGFR expression. The expression levels were evaluated in relation to three end points: time to metastasis (TTM), time to disease progression (TDP), and overall survival (OS).
Results
Among 1,068 eligible patients, data on GLI1, CTNNB1, and EGFR were available in 339, 164, and 300 patients, respectively. Although CTNNB1 expression did not differentiate prognosis, GLI1 was associated with poorer outcomes, adjusted for age, TNM stages, and Karnofsky performance score, and the significant influence persisted in a multivariable analysis (quartile 4 [Q4] v Q1 to Q3: TTM hazard ratio [HR], 2.7; 95% CI, 1.5 to 4.9; TDP HR, 1.6; 95% CI, 1.1 to 2.5; OS HR, 1.9; 95% CI, 1.4 to 2.7). The significance of GLI1 persisted in a multivariable analysis that included EGFR expression levels.
Conclusion
These data suggest that Hh signaling may play an important role in metastasis and that GLI1 could serve as a marker in HNSCC, but the regulatory mechanisms and oncogenic significance need further investigation. Risk classification based on this analysis needs a validation in independent cohorts.
INTRODUCTION
Although clinical outcomes for patients with locally advanced head and neck squamous cell carcinoma (HNSCC) have improved significantly with multimodality treatment approaches, that is not the case for patients with recurrence and/or distant metastasis. Therefore, understanding the biologic processes of treatment resistance and metastasis is critical for successful intervention. Furthermore, identification of novel therapeutic targets for addressing metastasis and development of clinically effective targeted agents to improve survival remains a difficult challenge.
Hedgehog (Hh) signaling is one of the key master regulators of both invertebrate and vertebrate development.1–4 Constitutive Hh signal activation due to mutations or activating deregulation is implicated in numerous neoplastic or hyperplastic conditions. For instance, constitutive activation of the Hh pathway has been shown to play a critical role in tumorigenesis in malignant medulloblastoma, basal cell carcinoma of the skin, and breast, urogenital, GI, pancreatic, and lung cancers.5–14 Although significant expression of proteins involved in Hh signaling has been reported,15 the oncogenic role of protein expression has not been examined in HNSCC.
There are three members of the Hh family of extracellular signaling molecules—sonic hedgehog (SHH), Indian hedgehog, and desert hedgehog—that activate a membrane receptor complex (Appendix Fig A1, online only). Binding of Hh to a transmembrane receptor, patched 1 (PTCH1), releases its inhibition of smoothened (SMO), a distant cousin of the 7-transmembrane G-protein coupled receptor family.16 Activation of SMO by Hh results in activation of glioma-associated oncogene family zinc finger 1 (GLI1) along with GLI2 and GLI3, which are thought to mediate most of the cellular effects.1 In the absence of Hh, GLI transcription factors are phosphorylated, ubiquitinated, and degraded. Although GLI1 and GLI2 generally function as transcriptional activators, the partially degraded GLI3 can function as a transcriptional repressor.17–21
Hh signaling is known to be induced in bronchial epithelial cells exposed to cigarette smoke; thus, it contributes to cell proliferation, anchorage-independent growth, and tumor formation in nude mice.22 In addition, GLI can be activated by noncanonical pathways, including RAS-MEK, AKT, and TGF-beta in the absence of Hh ligands,23–26 and epidermal growth factor (EGF) signaling has been shown to modulate Hh signaling in keratinocytes.27 Furthermore, Hh signaling has been shown to induce epithelial-to-mesenchymal transition (EMT) by inhibition of WNT/beta-catenin (CTNNB1) signaling and by upregulating secreted frizzled-related protein 1 (SFRP1). EMT-related genes were one of the three most significant gene sets that were enriched in high-risk patients with HNSCC in our previous study.28,29 In recent studies, a novel SMO inhibitor, GDC-0449, induced response and disease stabilization in patients with advanced basal cell carcinoma and medulloblastoma who frequently harbor inactivating mutations of PTCH1 or less common activating mutations of SMO.30,31 As expected, overexpression of GLI1 mRNA, indicating activation of the Hh pathway, was associated with clinical benefits. The oncogenic significance and clinical development of Hh pathway inhibitors were also summarized by Low et al32 in a recent comprehensive review.
To assess a potential oncogenic role of Hh signaling in HNSCC, we examined the nuclear expression of GLI1 in patients treated with radiation therapy in the Radiation Therapy Oncology Group (RTOG) 9003 clinical trial. We also assessed the correlation of GLI1 expression with CTNNB1 to determine the inhibitory effect of GLI1 on WNT signaling and correlation with EGF receptor (EGFR) expression to examine a potential noncanonical activation of GLI1 through the EGFR pathway. Additionally, these protein expression levels were correlated with clinical outcomes, including time to metastasis (TTM), time to disease progression (TDP), and overall survival (OS), to evaluate their roles as prognostic biomarkers.
PATIENTS AND METHODS
Patient Characteristics
The characteristics of patients enrolled on the RTOG 9003 clinical trial have been reported in detail.33 Briefly, RTOG 9003 was a phase III trial that compared hyperfractionation, split-course accelerated fractionation, and accelerated fractionation with concomitant boost with standard fractionation radiotherapy in American Joint Committee on Cancer (AJCC) stage III and IV but M0 HNSCC. The paraffin blocks from the primary tumor and/or lymph node metastasis were obtained from 430 patients, and tissue microarrays (TMAs) were constructed for laboratory analyses.
AQUA Staining of GLI1 and CTNNB1
Automated quantitative protein expression analysis (AQUA) was performed by using the HistoRx System (HistoRx, New Haven, CT). TMA slides were processed in Dako Target Retrieval Solution (Dako Cytomation, Carpinteria, CA) by using a Biocare Medical Decloaking Chamber (Biocare Medical, Concord, CA) for 6 minutes at 121°C. Slides were blocked in Cell Signaling Technology's antibody dilution/protein blocking solution (Cell Signaling Technology, Danvers, MA). Areas of tumor were labeled by using a 1:100 dilution of either the mouse anticytokeratin (AE1/AE3) antibody or the rabbit antibovine cytokeratin polyclonal antibody (Dako Cytomation) and visualized by using the goat antimouse or goat antirabbit Alexa 555 SFX kit (Invitrogen, Carlsbad, CA). GLI1 was stained by using H-300 rabbit anti-GLI1 polyclonal antibody at a concentration of 1:200 (Santa Cruz Biotechnology, Santa Cruz, CA). CTNNB1 was stained by using anti–beta-catenin-1 mouse monoclonal antibody at a concentration of 1:1,000 (Dako Cytomation). Primary antibodies were visualized with either the mouse or rabbit EnVision Plus DAB Kit (Dako Cytomation) by using the TSA-CY5 tyramide amplification kit (PerkinElmer, Fremont, CA). Slides were mounted by using ProLong Gold Anti-Fade mounting medium containing 4,6-diamidino-2-phenylindole-2-HCI (Invitrogen). In addition to data for these two proteins, data on EGFR staining from two of the four arms of the RTOG 9003 trial were obtained from a previously published study34 and examined for correlation with data for the proteins in this study. Automated image acquisition and analysis using AQUA were performed as previously described.35
Statistical Methods
Three time-to-event end points were evaluated: (1) TTM, in which failure was defined as regional or distant progression (as a first failure site, with local progression, second primary tumor, and death before progression treated as competing events that censor TTM); (2) TDP, in which failure was defined as local, regional, or distant progression event (with second primary cancer and death treated as competing events that censor TDP); and (3) OS, in which failure was defined as death due to any cause. In relation to the TTM end point, we also evaluated time to distant metastasis events alone. All event times were measured from date of random assignment to the date of event occurrence, censored by competing event occurrence or last follow-up.
Patient and disease characteristics and event frequencies among those with and without marker values were compared to establish whether the analysis cohorts were representative of all trial participants. Because the population of patients with the two array–derived biomarker values consisted of overlapping but not identical cohorts, we re-evaluated main results on a single cohort consisting of those who had all array marker values. Descriptive statistics were used to evaluate distributional properties of GLI1, CTNNB1, and EGFR. Graphical methods and correlation statistics were used to assess associations between markers. Associations between markers and patient and disease characteristics were evaluated by comparing marker summary statistics within categories of characteristics and examining cross-classified tables of marker quartiles and characteristics.
For the three end points, average annual failure rates (events per total person-time) were computed within quartiles of each marker. The Cox proportional hazards model was used to evaluate the influence of markers on risk of failure for OS and the event-specific end points of metastasis and progression.36,37 We estimated hazard ratios (HRs) and 95% CIs for each marker by using martingale residual plots to guide choice of the functional form.38 The forms that were considered included the addition of a quadratic term for nonlinearity and a logarithmic transform for skewness and to down-weight outliers. Marker effects on the HR appeared to be either linear or to have a threshold effect (ie, only large values were important), and thus we opted for quartile categories to represent marker effects. Graphical methods were used to assess the appropriateness of the proportional hazards assumption. Subsequently, other potentially prognostic factors were included to control for confounding effects on each marker. Finally, all markers were considered together to assess the joint prognostic effect. Cumulative incidence functions39 for TTM and TDP and Kaplan-Meier survival functions40 for OS were estimated in strata by marker value category. Curves generated by using nonparametric methods and the models (averaged over other covariates41) were similar, and we presented the former in this study.
Although the sample size for this analysis is limited by the number of patients with marker values, power is sufficient to detect clinically meaningful relative hazards (in the range of 1.50 and above) associated with the markers.
RESULTS
Patient Cohort Comparison for GLI1, CTNNB1, and EGFR Expression
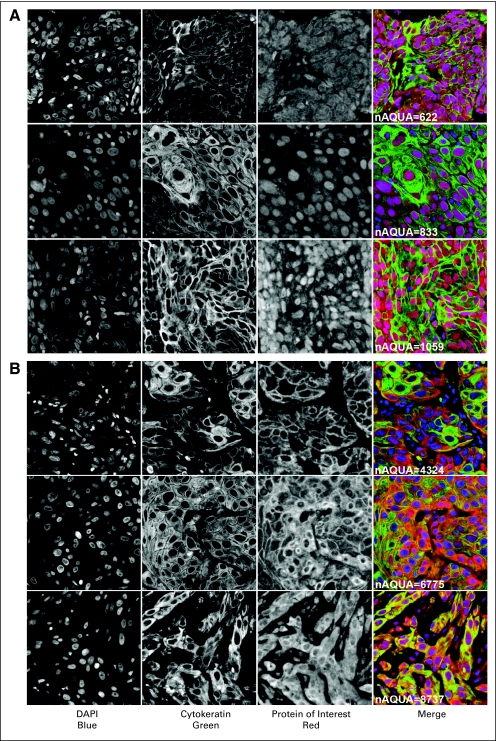
Detailed patient characteristics with protein expression data are summarized in Table 1. Representative tumors with AQUA staining using GLI1 and CTNNB1 are shown in Figure 1. Of 1,068 eligible patients, 339 had GLI1 data, 164 had CTNNB1 data, and 300 had EGFR data. The difference in the number of data points or patient tumors per protein staining is due to progressive depletion of cores in each TMA as the TMA blocks were cut deeper. The EGFR data were obtained from patients in two (standard and accelerated radiation) of the four treatment arms of the trial, as previously published.34 The subcohorts with different protein staining were similar to the entire cohort with respect to patient and disease characteristics. Comparisons of patients with and without data points for each protein did not reveal major differences in characteristics (Appendix Table A1, online only). Finally, the proportion of patients with failures for the end points evaluated in this report did not differ among those with and without protein staining values (data not shown).
Table 1.
Characteristics of RTOG 9003 Cohort and Subcohorts With Protein Expression Data for GLI1, CTNNB1, and EGFR
| Characteristic | RTOG 9003 Cohort (N = 1,068) |
GLI1 (n = 339) |
CTNNB1 (n = 164) |
EGFR (n = 300) |
||||
|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | |
| Treatments | ||||||||
| Standard | 266 | 24.9 | 78 | 23.0 | 32 | 19.0 | 154 | 51.3 |
| Hypofractionation | 261 | 24.4 | 100 | 29.5 | 49 | 29.2 | — | — |
| Split-course accelerated fractionation | 274 | 25.7 | 88 | 26.0 | 54 | 32.1 | — | — |
| Accelerated fractionation with concomitant boost | 267 | 25.0 | 73 | 21.5 | 29 | 17.3 | 146 | 48.7 |
| Sex | ||||||||
| Male | 851 | 79.7 | 263 | 77.6 | 132 | 78.6 | 226 | 75.3 |
| Female | 217 | 20.3 | 76 | 22.4 | 32 | 19.0 | 74 | 24.7 |
| KPS | ||||||||
| 60 | 48 | 4.5 | 16 | 4.7 | 9 | 5.4 | 14 | 4.7 |
| 70 | 121 | 11.3 | 48 | 14.2 | 27 | 16.1 | 39 | 13.0 |
| 80 | 243 | 22.8 | 75 | 22.1 | 43 | 25.6 | 69 | 23.0 |
| 90 | 494 | 46.3 | 153 | 45.1 | 68 | 40.5 | 132 | 44.0 |
| 100 | 162 | 15.2 | 47 | 13.9 | 17 | 10.1 | 46 | 15.3 |
| Age at diagnosis, years | ||||||||
| ≤ 55 | 336 | 31.5 | 109 | 32.2 | 56 | 33.3 | 89 | 29.7 |
| 56-65 | 395 | 37.0 | 119 | 35.1 | 60 | 35.7 | 120 | 40.0 |
| ≥ 66 | 337 | 31.6 | 111 | 32.7 | 48 | 28.6 | 91 | 30.3 |
| Primary site | ||||||||
| Oral cavity | 110 | 10.3 | 33 | 9.7 | 15 | 8.9 | 35 | 11.7 |
| Oropharynx | 646 | 60.5 | 191 | 56.3 | 100 | 59.5 | 166 | 55.3 |
| Hypopharynx | 140 | 13.1 | 43 | 12.7 | 18 | 10.7 | 41 | 13.7 |
| Larynx | 172 | 16.1 | 72 | 21.2 | 31 | 18.5 | 58 | 19.3 |
| T stage | ||||||||
| T1 | 62 | 5.8 | 10 | 2.9 | 6 | 3.6 | 23 | 7.7 |
| T2 | 287 | 26.9 | 83 | 24.5 | 37 | 22.0 | 79 | 26.3 |
| T3 | 405 | 37.9 | 148 | 43.7 | 76 | 45.2 | 109 | 36.3 |
| T4 | 313 | 29.3 | 98 | 28.9 | 45 | 26.8 | 88 | 29.3 |
| Tx | 1 | 0.1 | — | — | — | — | 1 | 0.3 |
| N stage | ||||||||
| N0 | 238 | 22.3 | 79 | 23.3 | 40 | 23.8 | 57 | 19.0 |
| N1 | 212 | 19.9 | 78 | 23.0 | 33 | 19.6 | 59 | 19.7 |
| N2a | 103 | 9.6 | 26 | 7.7 | 11 | 6.5 | 31 | 10.3 |
| N2b | 200 | 18.7 | 63 | 18.6 | 33 | 19.6 | 58 | 19.3 |
| N2c | 189 | 17.7 | 60 | 17.7 | 31 | 18.5 | 59 | 19.7 |
| N3 | 126 | 11.8 | 33 | 9.7 | 16 | 9.5 | 36 | 12.0 |
Abbreviations: RTOG, Radiation Therapy Oncology Group; GLI1, glioma-associated oncogene family zinc finger 1; CTNNB1, beta-catenin; EGFR, epidermal growth factor receptor; KPS, Karnofsky performance score.
Fig 1.
Representative images of automated quantitative protein expression analysis (AQUA) of head and neck squamous cell cancer and the control tissues; (A) glioma-associated oncogene family zinc finger 1, and (B) beta-catenin. DAPI, 4,6-diamidino-2-phenylindole-2-HCI.
Summary Statistics for GLI1, CTNNB1, and EGFR Expression
Descriptive statistics on the protein expression levels are provided in Table 2. The CTNNB1 detection range was wider than that for GLI1 because the AQUA score was obtained by using the newer software. Spearman rank correlation statistics for protein pairs indicate statistically significant negative correlation between CTNNB1 and EGFR expression (r = −0.30; P = 0.02 [P for Ho:correlation = 0]; N [patients with values for both proteins] = 60); however, there was no correlation between GLI1 and CTNNB1 (r = −0.07; P = .40; n = 147) and between GLI1 and EGFR (r = 0.02; P = .81; n = 149). In general, there was no strong association between patient or disease characteristics such as TNM stages and protein expression values (Appendix Tables A2 to A4, online only). EGFR expression tended to be higher for patients with higher tumor stage. Nodal stage and primary site did not show an association with any of the protein expression variables. Patient characteristics (age, sex, performance score) also did not show any association with any of the protein expression variables.
Table 2.
Summary Statistics for Protein Staining Intensity Values of GLI1, CTNNB1, and EGFR
| Variable | No. | Mean | Minimum | Maximum | Lower Quartile | Median | Upper Quartile |
|---|---|---|---|---|---|---|---|
| GLI1 | 339 | 453.82 | 1.96 | 877.58 | 410.86 | 462.06 | 508.09 |
| CTNNB1 | 164 | 6,379.99 | 1,138.07 | 9,981.88 | 5,489.25 | 6,520.72 | 7,484.41 |
| EGFR | 300 | 75.81 | 5.96 | 99.98 | 59.15 | 87.39 | 97.58 |
Abbreviations: GLI1, glioma-associated oncogene family zinc finger 1; CTNNB1, beta-catenin; EGFR, epidermal growth factor receptor.
Univariate Analyses
Proportions and rates of failure were similar in patients in the trial as a whole and in the subcohorts defined by availability of protein expression data (data not shown). Each protein expression was examined individually for a prognostic influence on the three outcomes: TTM, TDP, and OS. Each protein was evaluated on a continuous scale with model diagnostics used to evaluate how changes in the covariate might be related to hazard of failure; each protein was also evaluated by using a simple partition of the marker distribution into quartiles and by contrasting hazard of failure in the second through fourth quartiles with that of the first quartile. The data are summarized in Appendix Table A5 (online only). Increasing GLI1 expression values were associated with worse outcomes for all end points, with a tendency toward a threshold effect at the highest quartile of values for metastases and progression. Patients in the highest quartile had an approximately 2.5-fold greater risk of metastasis and a 1.9-fold greater risk of death. CTNNB1 did not show any consistent association with any of the end points. As seen previously,34 increased EGFR was associated with greater failure risk for all three end points. Analysis of EGFR on a continuous scale and HRs by quartile indicated that patients with values above the median showed a large increment in risk of failure relative to those with values below the median.
Multivariable Models
Other covariates associated with one or more outcomes were age at diagnosis, stage components (tumor and lymph node involvement), and Karnofsky performance score. Treatment arm assignment was also included in multivariable models. HRs for each protein from models that included these covariates are summarized in Table 3. For GLI1 and EGFR, prognostic influence persisted after adjustment for other covariates, and results were similar to those in the univariate analyses. For EGFR, values greater than the median have a 1.76- to 2.05-fold greater failure risk for the three end points. For GLI1, patients with values in the upper quartile had significantly greater risk of failure for all end points (HR, 1.65 to 2.68), and the strongest correlation was seen with risk of metastasis. Again, CTNNB1 did not show a statistically significant prognostic effect on metastasis, disease progression, or survival.
Table 3.
Effects of GLI1, CTNNB1, and EGFR Individually, Adjusted for Other Patient and Tumor Characteristics, Including Age at Diagnosis, Stage Components (tumor and lymph node involvement), Karnofsky Performance Score, and Treatment Arm Assignment
| Characteristic | Regional-Distant Metastasis |
Progression |
Mortality |
|||
|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | HR | 95% CI | |
| GLI1 | ||||||
| Continuous* | 1.00 | 1.00 to 1.01 | 1.00 | 1.00 to 1.00 | 1.00 | 1.00 to 1.00 |
| Q1 | 1.00 | — | 1.00 | — | 1.00 | — |
| Q2 | 1.33 | 0.69 to 2.56 | 1.09 | 0.69 to 1.72 | 1.31 | 0.92 to 1.87 |
| Q3 | 1.07 | 0.55 to 2.10 | 0.95 | 0.60 to 1.51 | 1.44 | 1.02 to 2.03 |
| Q4 | 2.68 | 1.46 to 4.91 | 1.65 | 1.06 to 2.55 | 1.93 | 1.37 to 2.71 |
| CTNNB1 | ||||||
| Continuous* | 1.00 | 1.00 to 1.00 | 1.00 | 1.00 to 1.00 | 1.00 | 1.00 to 1.00 |
| Q1 | 1.00 | — | 1.00 | — | 1.00 | — |
| Q2 | 0.87 | 0.36 to 2.13 | 1.24 | 0.65 to 2.39 | 1.28 | 0.79 to 2.09 |
| Q3 | 0.88 | 0.36 to 2.14 | 1.18 | 0.60 to 2.29 | 1.06 | 0.62 to 1.80 |
| Q4 | 0.69 | 0.30 to 1.61 | 0.56 | 0.28 to 1.14 | 0.84 | 0.51 to 1.37 |
| EGFR | ||||||
| Continuous* | 1.01 | 1.00 to 1.02 | 1.01 | 1.00 to 1.02 | 1.01 | 1.00 to 1.02 |
| Q1 | 1.00 | — | 1.00 | — | 1.00 | — |
| Q2 | 1.54 | 0.82 to 2.92 | 1.35 | 0.80 to 2.27 | 1.28 | 0.88 to 1.86 |
| Q3 | 1.76 | 0.92 to 3.38 | 1.90 | 1.15 to 3.15 | 2.05 | 1.42 to 2.97 |
| Q4 | 1.93 | 1.01 to 3.71 | 2.21 | 1.34 to 3.64 | 1.77 | 1.22 to 2.59 |
Abbreviations: GLI1, glioma-associated oncogene family zinc finger 1; CTNNB1, beta-catenin; EGFR, epidermal growth factor receptor; HR, hazard ratio; Q, quartile.
Per 10-unit increment.
For models involving multiple proteins, initially GLI1 and CTNNB1 were considered. As was seen previously, CTNNB1 did not show meaningful and consistent associations with outcomes and was not considered further in models that evaluated markers simultaneously. In a model that combined GLI1 expression levels (n = 339 patients) and clinical data, high GLI1 values were associated with increased risk of failure for all end points (Table 4). Finally, GLI1 and EGFR were considered together. Because EGFR was evaluated among patients from only two treatment arms of the study, the sample of patients with both markers was limited to 149 patients. EGFR expression levels greater than the median value of 87 remained as a significant predictor of progression and survival (Table 4). High GLI1 expression levels were associated with nearly a three-fold greater risk of metastasis and remained modestly associated with progression and death. Because we had evidence that GLI1 expression was associated with metastasis, we further examined distant metastasis alone as an end point in the models with multiple markers. Again, GLI1 expression was strongly associated with distant metastasis (HR, 2.70; 95% CI, 1.47 to 4.96; P = .001); however, GLI1 expression was marginally significant when combined with EGFR expression (HR, 2.53; 95% CI, 0.99 to 6.48; P = .054). EGFR expression was not associated with distant metastasis (HR, 2.01; 95% CI, 0.81 to 5.20; P = .131), as was previously reported.42
Table 4.
Multiple Marker Models Including Clinical and Laboratory Data
| Model | Regional-Distant Metastasis |
Progression |
Mortality |
||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | |
| Without EGFR (n = 339) | 86 Events | 160 Events | 280 Events | ||||||
| N2 vN0-N1 | 2.56 | 1.54 to 4.25 | < .001 | 1.30 | 0.91 to 1.84 | .145 | 1.45 | 1.11 to 1.90 | .006 |
| N3 v N0-N1 | 2.84 | 1.31 to 6.16 | .008 | 1.54 | 0.89 to 2.66 | .120 | 1.90 | 1.24 to 2.91 | .003 |
| T4 v T1-T3 | 0.89 | 0.51 to 1.56 | .688 | 1.83 | 1.29 to 2.60 | .001 | 1.50 | 1.13 to 1.97 | .005 |
| KPS < 90 v 90+ | 0.76 | 0.44 to 1.30 | .307 | 0.80 | 0.55 to 1.16 | .231 | 1.31 | 1.00 to 1.71 | .051 |
| Age 65+ v < 65 years | 1.39 | 0.86 to 2.25 | .182 | 1.44 | 1.02 to 2.03 | .040 | 1.83 | 1.41 to 2.37 | < .001 |
| GLI1 Q4 v Q1-Q3 | 2.45 | 1.55 to 3.86 | < .001 | 1.60 | 1.13 to 2.27 | .008 | 1.58 | 1.21 to 2.07 | < .001 |
| With EGFR (n = 149) | 32 Events | 65 Events | 125 Events | ||||||
| N2 v N0-N1 | 2.33 | 1.02 to 5.33 | .046 | 1.15 | 0.66 to 2.00 | .615 | 1.21 | 0.81 to 1.81 | .345 |
| N3 v N0-N1 | 2.02 | 0.53 to 7.69 | .301 | 1.55 | 0.69 to 3.47 | .289 | 1.81 | 1.00 to 3.26 | .049 |
| T4 v T1-T3 | 2.07 | 0.87 to 4.97 | .102 | 2.62 | 1.45 to 4.75 | .002 | 1.41 | 0.93 to 2.13 | .102 |
| KPS < 90 v 90+ | 0.72 | 0.31 to 1.67 | .446 | 0.81 | 0.46 to 1.42 | .458 | 1.42 | 0.97 to 2.09 | .075 |
| GLI1 Q4 v Q1-Q3 | 2.93 | 1.40 to 6.10 | .004 | 1.68 | 0.97 to 2.90 | .065 | 1.46 | 0.98 to 2.16 | .061 |
| EGFR Q3-Q4 v Q1-Q2 | 1.83 | 0.89 to 3.79 | .103 | 2.17 | 1.28 to 3.67 | .004 | 1.70 | 1.18 to 2.46 | .005 |
Abbreviations: HR, hazard ratio; EGFR, epidermal growth factor receptor; N, TNM stage; T, TNM stage; KPS, Karnofsky performance score; GLI1, glioma-associated oncogene family zinc finger 1; Q, quartile.
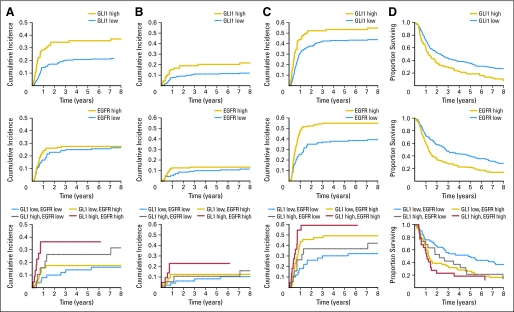
To illustrate risk of failure over time according to the protein values, we computed cumulative metastasis and risk of disease progression for patients with GLI1 values in the highest quartile and EGFR values above the median compared with patients with lower values (Fig 2). For OS, distinct patterns of prognosis over time by GLI1 and EGFR were also seen. By considering EGFR and GLI1 together, we defined a low-risk group (EGFR below the median and GLI1 below the fourth quartile) and a high-risk group (EGFR at the median or higher and GLI1 in the fourth quartile). For all four end points, the low-risk and high-risk patients have substantially different prognosis through 8 years of follow-up.
Fig 2.
Cumulative incidence functions (A-C) and Kaplan-Meier survival curves (D) according to risk groups defined by quartiles (Q) of glioma-associated oncogene family zinc finger 1 (GL1) expression (low, Q1 to Q3 v high, Q4) and the median of epidermal growth factor receptor (EGFR) expression (low, ≤ median v high, > median), and a combination of the groups by both GL1 and EGFR expression levels; (A) time to regional-distant metastasis, (B) time to distant metastasis, (C) time to disease progression, and (D) overall survival.
DISCUSSION
Identification of novel therapeutic targets and agents is critical for improving clinical management of HNSCC. Recent studies have shown that the Hh pathway provides an important oncogenic signaling in numerous epithelial neoplasms; however, it has not been evaluated in HNSCC. In this study, we examined a potential involvement of the Hh pathway in HNSCC by assessing the expression levels of a key downstream effector protein, GLI1.
We found that GLI1 activation, determined by nuclear GLI1 expression, is frequent and is associated with metastasis and poor survival in HNSCC. This is consistent with the data for esophageal SCC in which GLI1 activation has been associated with lymph node metastasis and tumor progression after chemoradiotherapy.43,44 Furthermore, high GLI1 activation was associated with distant metastasis. Because of the differences between treatments for patients with distant metastases and for those with regional metastases, identification of patients with a high risk of distant metastasis would have significant implications in treatment decisions. Currently, patients with locally advanced HNSCC are treated with induction chemotherapy to decrease the risk of distant metastasis and provide better disease control.45 However, distant metastasis is relatively rare in HNSCC, and there is no predictive biomarker for selecting patients with a significant risk. Whether and to what extent current chemotherapy regimens can prevent distant metastasis has not been clearly resolved because there are no clinical trial data available for comparing the distant metastasis rates between patients treated with induction chemotherapy followed by concurrent chemoradiotherapy and those treated with concurrent chemoradiotherapy alone. Predictive biomarkers to select patients with high risk of distant metastasis and identification of targeted agents that can prevent distant metastasis would provide clinical benefits to HNSCC patients.
One limitation of our study is that the mechanism of GLI1 activation and its biologic role have not been examined, and the mechanistic studies that use cell line models are currently ongoing. GLI1 can be activated through both canonical pathways and noncanonical pathways. Known activating mutations in canonical Hh pathway proteins that associated with Hh-targeting agents has not been examined in HNSCC. EGFR expression, which may activate GLI1 via a noncanonical pathway through RAS/MEK or AKT, was not associated with GLI1 expression in our data, suggesting that metastasis and poor survival seen with GLI1 may be independent of EGFR signaling. This lack of interaction was substantiated by the finding that GLI1 and EGFR were independently prognostic determinants in our multivariable statistical model. Furthermore, although most of the Hh signaling studies performed in the context of cancer1,46,47 have focused on direct cell autonomous promotion of proliferation, differentiation, EMT, and tumorigenesis, the Hh signaling in development is typically mediated through paracrine effects. A recent study by Yauch et al48 reported that Hh-GLI activation was required in the tissue mesenchyme surrounding pancreatic cancer cells to support tumor growth by paracrine effects. The role of paracrine signaling in HNSCC is under active investigation.
In recent years, much effort has been directed at the development of small molecule inhibitors of the Hh pathway as a potential cancer treatment.49–53 However, most of the currently available small molecule inhibitors, including cyclopamine and GDC-0449, appear to be limited by poor in vivo bioavailability. In addition, current inhibitors mostly target upstream receptors such as SMO48,54 while the downstream GLI transcription factors can be activated by a noncanonical pathway independent of Hh-Smo signaling.23–26 Noncanonical pathway activation of GLI has been proposed as one of the resistance mechanisms. Further understanding of the Hh-GLI pathway and its role in development of distant metastasis may present ways to apply these novel agents in the prevention and control of distant metastasis in HNSCC.
In summary, we show that GLI1 is frequently activated in and associated with metastasis in HNSCC. Further understanding of regulatory mechanisms, oncogenic significance, and important pathway crosstalk, including Hh/GLI, WNT/CTNNB1, and EGFR will improve therapeutic strategies. Additionally, risk classification based on this analysis needs a validation in independent cohorts through future studies in HNSCC.
Acknowledgment
Supported by Grants No. U10 CA21661 from the Radiation Therapy Oncology Group, No. U10 CA37422 to the Community Clinical Oncology Program from the National Cancer Institute, the Damon Runyon Clinical Investigator Award (CI-28-05; C.H.C.), No. 1 R01 DE017982-01 from National Institutes of Health (C.H.C.), and by 2006 Formula Grant No. 4100037703 from the Pennsylvania Department of Health.
Glossary Terms
- AKT:
A transforming serine-threonine kinase involved in cell survival.
- AQUA (automated quantitative protein expression analysis):
This technology overcomes limitations associated with traditional “brown stain” immunohistochemistry (IHC). In traditional IHC, protein expression is reported on a quantized scale such as 0, 1, 2, 3. Biologic material rarely is expressed in such neatly quantized packets, but rather is expressed on a continuous scale. AQUA is a method of computerized interpretation of fluorescence IHC images that allows protein expression to be automatically assigned to subcellular compartment and expressed on a continuous scale.
- beta-catenin:
Originally identified as a component of cell-cell adhesion complexes composed of cadherins and actin, β-catenin has now been shown to be a downstream signaling molecule in the Wnt signaling pathway.
- Canonical pathway:
A core pathway established for a given molecule in the cell in which molecular interactions occur in a linear and stepwise manner. Although clustering expression data groups functionally related genes, they do not order pathway components according to physical or regulatory relationships. Software is now available for linking significant genes in one's experiments with a world collection of biologic networks created from millions of individually modeled relationships between genes, proteins, complexes, cells, and tissues. The software allows a view of one's data, integrated in biologic networks according to different biologic context and identifies canonical and noncanonical pathways that connect molecules within a biologic network.
- EGFR (epidermal growth factor receptor):
Also known as HER-1, EGFR belongs to a family of receptors (HER-2, HER-3, HER-4 are other members of the family) and binds to the EGF, TGF-α, and other related proteins, leading to the generation of proliferative and survival signals within the cell. It also belongs to the larger family of tyrosine kinase receptors and is generally overexpressed in several solid tumors of epithelial origin.
- EMT (epithelial-to-mesenchymal transition):
Cellular changes that occur in epithelial cells to lose epithelial cell junction proteins and to gain mesenchymal phenotypes by expressing proteins such as vimentin and fibronectin.
- GLI1 (glioma-associated oncogene family zinc finger 1):
GLI1 is a transcription factor which is activated by the hedgehog signaling cascade.
- PTCH1 (patched 1):
PTCH1 is a receptor for three hedgehog ligands (sonic, desert and Indian hedgehogs). It represses the activity of smoothened (SMO). Upon the binding of hedgehog ligands, it releases the inhibition on SMO activity.
- SHH (sonic hedgehog):
SHH is one of the ligands for the PATCHED1 protein (PTCH1). Binding of SHH to PTCH1 is one of the early steps in activation of the hedgehog signaling cascade.
- SMO (smoothened):
SMO is a receptor that is inhibited by PTCH1. When one of the hedgehog ligands binds PTCH1, PTCH1 releases the inhibition of SMO and SMO activates the hedgehog signal transduction cascade. Downstream of SMO is a family of transcription factors, GLI, which are key proteins to mediate the hedgehog signal. The hedgehog signaling pathway is important in cell proliferation and differentiation during embryonic development and tumorigenesis.
Appendix
Table A1.
Characteristics of Patients With and Without GLI1, CTNNB1, and EGFR Expression Data
| Characteristic | GLI1 |
CTNNB1 |
EGFR |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No |
Yes |
No |
Yes |
No |
Yes |
|||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | |
| Treatment | ||||||||||||
| Standard | 188 | 25.8 | 78 | 23.0 | 234 | 25.9 | 32 | 19.5 | 112 | 14.6 | 154 | 51.3 |
| Hypofractionation | 161 | 22.1 | 100 | 29.5 | 212 | 23.5 | 49 | 29.9 | 261 | 34.0 | — | — |
| Split-course accelerated fractionation | 186 | 25.5 | 88 | 26.0 | 220 | 24.3 | 54 | 32.9 | 274 | 35.7 | — | — |
| Accelerated fractionation with concomitant boost | 194 | 26.6 | 73 | 21.5 | 238 | 26.3 | 29 | 17.7 | 121 | 15.8 | 146 | 48.7 |
| Sex | ||||||||||||
| Male | 588 | 80.7 | 263 | 77.6 | 719 | 79.5 | 132 | 80.5 | 625 | 81.4 | 226 | 75.3 |
| Female | 141 | 19.3 | 76 | 22.4 | 185 | 20.5 | 32 | 19.5 | 143 | 18.6 | 74 | 24.7 |
| KPS | ||||||||||||
| 60 | 32 | 4.4 | 16 | 4.7 | 39 | 4.3 | 9 | 5.5 | 34 | 4.4 | 14 | 4.7 |
| 70 | 73 | 10.0 | 48 | 14.2 | 94 | 10.4 | 27 | 16.5 | 82 | 10.7 | 39 | 13.0 |
| 80 | 168 | 23.0 | 75 | 22.1 | 200 | 22.1 | 43 | 26.2 | 174 | 22.7 | 69 | 23.0 |
| 90 | 341 | 46.8 | 153 | 45.1 | 426 | 47.1 | 68 | 41.5 | 362 | 47.1 | 132 | 44.0 |
| 100 | 115 | 15.8 | 47 | 13.9 | 145 | 16.0 | 17 | 10.4 | 116 | 15.1 | 46 | 15.3 |
| Age, years | ||||||||||||
| ≤ 55 | 227 | 31.1 | 109 | 32.2 | 280 | 31.0 | 56 | 34.1 | 247 | 32.2 | 89 | 29.7 |
| 56-65 | 276 | 37.9 | 119 | 35.1 | 335 | 37.1 | 60 | 36.6 | 275 | 35.8 | 120 | 40.0 |
| ≥ 66 | 226 | 31.0 | 111 | 32.7 | 289 | 32.0 | 48 | 29.3 | 246 | 32.0 | 91 | 30.3 |
| Primary site | ||||||||||||
| Oral cavity | 77 | 10.6 | 33 | 9.7 | 95 | 10.5 | 15 | 9.1 | 75 | 9.8 | 35 | 11.7 |
| Oropharynx | 455 | 62.4 | 191 | 56.3 | 546 | 60.4 | 100 | 61.0 | 480 | 62.5 | 166 | 55.3 |
| Hypopharynx | 97 | 13.3 | 43 | 12.7 | 122 | 13.5 | 18 | 11.0 | 99 | 12.9 | 41 | 13.7 |
| Larynx | 100 | 13.7 | 72 | 21.2 | 141 | 15.6 | 31 | 18.9 | 114 | 14.8 | 58 | 19.3 |
| T stage | ||||||||||||
| T1 | 52 | 7.1 | 10 | 2.9 | 56 | 6.2 | 6 | 3.7 | 39 | 5.1 | 23 | 7.7 |
| T2 | 204 | 28.0 | 83 | 24.5 | 250 | 27.7 | 37 | 22.6 | 208 | 27.1 | 79 | 26.3 |
| T3 | 257 | 35.3 | 148 | 43.7 | 329 | 36.4 | 76 | 46.3 | 296 | 38.5 | 109 | 36.3 |
| T4 | 215 | 29.5 | 98 | 28.9 | 268 | 29.6 | 45 | 27.4 | 225 | 29.3 | 88 | 29.3 |
| Tx | 1 | 0.1 | — | — | 1 | 0.1 | — | — | — | — | 1 | 0.3 |
| N stage | ||||||||||||
| N0 | 159 | 21.8 | 79 | 23.3 | 198 | 21.9 | 40 | 24.4 | 181 | 23.6 | 57 | 19.0 |
| N1 | 134 | 18.4 | 78 | 23.0 | 179 | 19.8 | 33 | 20.1 | 153 | 19.9 | 59 | 19.7 |
| N2a | 77 | 10.6 | 26 | 7.7 | 92 | 10.2 | 11 | 6.7 | 72 | 9.4 | 31 | 10.3 |
| N2b | 137 | 18.8 | 63 | 18.6 | 167 | 18.5 | 33 | 20.1 | 142 | 18.5 | 58 | 19.3 |
| N2c | 129 | 17.7 | 60 | 17.7 | 158 | 17.5 | 31 | 18.9 | 130 | 16.9 | 59 | 19.7 |
| N3 | 93 | 12.8 | 33 | 9.7 | 110 | 12.2 | 16 | 9.8 | 90 | 11.7 | 36 | 12.0 |
Abbreviations: GLI1, glioma-associated oncogene family zinc finger 1; CTNNB1, beta-catenin; EGFR, epidermal growth factor receptor; KPS, Karnofsky performance score.
Table A2.
GLI1 Quartiles by Patient and Disease Characteristics
| Characteristic | GLI1 Quartiles |
All |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Q1 |
Q2 |
Q3 |
Q4 |
|||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | |
| Treatment | ||||||||||
| Standard | 20 | 23.8 | 17 | 20.7 | 17 | 19.3 | 24 | 28.2 | 78 | 23.0 |
| Hypofractionation | 25 | 29.8 | 22 | 26.8 | 32 | 36.4 | 21 | 24.7 | 100 | 29.5 |
| Split-course accelerated fractionation | 23 | 27.4 | 22 | 26.8 | 20 | 22.7 | 23 | 27.1 | 88 | 26.0 |
| Accelerated fractionation with concomitant boost | 16 | 19.0 | 21 | 25.6 | 19 | 21.6 | 17 | 20.0 | 73 | 21.5 |
| Sex | ||||||||||
| Male | 64 | 76.2 | 62 | 75.6 | 72 | 81.8 | 65 | 76.5 | 263 | 77.6 |
| Female | 20 | 23.8 | 20 | 24.4 | 16 | 18.2 | 20 | 23.5 | 76 | 22.4 |
| KPS | ||||||||||
| 60 | 7 | 8.3 | 2 | 2.4 | 3 | 3.4 | 4 | 4.7 | 16 | 4.7 |
| 70 | 8 | 9.5 | 7 | 8.5 | 17 | 19.3 | 16 | 18.8 | 48 | 14.2 |
| 80 | 16 | 19.0 | 15 | 18.3 | 24 | 27.3 | 20 | 23.5 | 75 | 22.1 |
| 90 | 44 | 52.4 | 41 | 50.0 | 32 | 36.4 | 36 | 42.4 | 153 | 45.1 |
| 100 | 9 | 10.7 | 17 | 20.7 | 12 | 13.6 | 9 | 10.6 | 47 | 13.9 |
| Age, years | ||||||||||
| ≤ 55 | 20 | 23.8 | 32 | 39.0 | 25 | 28.4 | 32 | 37.6 | 109 | 32.2 |
| 56-65 | 28 | 33.3 | 27 | 32.9 | 35 | 39.8 | 29 | 34.1 | 119 | 35.1 |
| ≥ 66 | 36 | 42.9 | 23 | 28.0 | 28 | 31.8 | 24 | 28.2 | 111 | 32.7 |
| Primary site | ||||||||||
| Oral cavity | 8 | 9.5 | 6 | 7.3 | 12 | 13.6 | 7 | 8.2 | 33 | 9.7 |
| Oropharynx | 47 | 56.0 | 47 | 57.3 | 47 | 53.4 | 50 | 58.8 | 191 | 56.3 |
| Hypopharynx | 10 | 11.9 | 12 | 14.6 | 10 | 11.4 | 11 | 12.9 | 43 | 12.7 |
| Larynx | 19 | 22.6 | 17 | 20.7 | 19 | 21.6 | 17 | 20.0 | 72 | 21.2 |
| T stage | ||||||||||
| T1 | — | — | 5 | 6.1 | 3 | 3.4 | 2 | 2.4 | 10 | 2.9 |
| T2 | 31 | 36.9 | 16 | 19.5 | 15 | 17.0 | 21 | 24.7 | 83 | 24.5 |
| T3 | 33 | 39.3 | 40 | 48.8 | 40 | 45.5 | 35 | 41.2 | 148 | 43.7 |
| T4 | 20 | 23.8 | 21 | 25.6 | 30 | 34.1 | 27 | 31.8 | 98 | 28.9 |
| N stage | ||||||||||
| N0 | 19 | 22.6 | 23 | 28.0 | 23 | 26.1 | 14 | 16.5 | 79 | 23.3 |
| N1 | 18 | 21.4 | 20 | 24.4 | 16 | 18.2 | 24 | 28.2 | 78 | 23.0 |
| N2a | 6 | 7.1 | 6 | 7.3 | 6 | 6.8 | 8 | 9.4 | 26 | 7.7 |
| N2b | 17 | 20.2 | 13 | 15.9 | 17 | 19.3 | 16 | 18.8 | 63 | 18.6 |
| N2c | 15 | 17.9 | 15 | 18.3 | 17 | 19.3 | 13 | 15.3 | 60 | 17.7 |
| N3 | 9 | 10.7 | 5 | 6.1 | 9 | 10.2 | 10 | 11.8 | 33 | 9.7 |
Abbreviations: GLI1, glioma-associated oncogene family zinc finger 1; Q, quartile; KPS, Karnofsky performance score.
Table A3.
CTNNB1 Quartiles by Patient and Disease Characteristics
| Characteristic | CTNNB1 Quartiles | All |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Q1 |
Q2 |
Q3 |
Q4 |
|||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | |
| Treatment | ||||||||||
| Standard | 6 | 14.6 | 8 | 19.5 | 9 | 22.5 | 9 | 21.4 | 32 | 19.5 |
| Hypofractionation | 7 | 17.1 | 14 | 34.1 | 14 | 35.0 | 14 | 33.3 | 49 | 29.9 |
| Split-course accelerated fractionation | 21 | 51.2 | 14 | 34.1 | 10 | 25.0 | 9 | 21.4 | 54 | 32.9 |
| Accelerated fractionation with concomitant boost | 7 | 17.1 | 5 | 12.2 | 7 | 17.5 | 10 | 23.8 | 29 | 17.7 |
| Sex | ||||||||||
| Male | 32 | 78.0 | 34 | 82.9 | 32 | 80.0 | 34 | 81.0 | 132 | 80.5 |
| Female | 9 | 22.0 | 7 | 17.1 | 8 | 20.0 | 8 | 19.0 | 32 | 19.5 |
| KPS | ||||||||||
| 60 | 3 | 7.3 | 3 | 7.3 | 1 | 2.5 | 2 | 4.8 | 9 | 5.5 |
| 70 | 7 | 17.1 | 6 | 14.6 | 7 | 17.5 | 7 | 16.7 | 27 | 16.5 |
| 80 | 11 | 26.8 | 12 | 29.3 | 15 | 37.5 | 5 | 11.9 | 43 | 26.2 |
| 90 | 15 | 36.6 | 17 | 41.5 | 12 | 30.0 | 24 | 57.1 | 68 | 41.5 |
| 100 | 5 | 12.2 | 3 | 7.3 | 5 | 12.5 | 4 | 9.5 | 17 | 10.4 |
| Age, years | ||||||||||
| ≤ 55 | 9 | 22.0 | 20 | 48.8 | 16 | 40.0 | 11 | 26.2 | 56 | 34.1 |
| 56-65 | 18 | 43.9 | 11 | 26.8 | 15 | 37.5 | 16 | 38.1 | 60 | 36.6 |
| ≥ 66 | 14 | 34.1 | 10 | 24.4 | 9 | 22.5 | 15 | 35.7 | 48 | 29.3 |
| Primary site | ||||||||||
| Oral cavity | 4 | 9.8 | 4 | 9.8 | 4 | 10.0 | 3 | 7.1 | 15 | 9.1 |
| Oropharynx | 26 | 63.4 | 27 | 65.9 | 20 | 50.0 | 27 | 64.3 | 100 | 61.0 |
| Hypopharynx | 6 | 14.6 | 5 | 12.2 | 2 | 5.0 | 5 | 11.9 | 18 | 11.0 |
| Larynx | 5 | 12.2 | 5 | 12.2 | 14 | 35.0 | 7 | 16.7 | 31 | 18.9 |
| T stage | ||||||||||
| T1 | 2 | 4.9 | 1 | 2.4 | 2 | 5.0 | 1 | 2.4 | 6 | 3.7 |
| T2 | 11 | 26.8 | 5 | 12.2 | 5 | 12.5 | 16 | 38.1 | 37 | 22.6 |
| T3 | 21 | 51.2 | 25 | 61.0 | 17 | 42.5 | 13 | 31.0 | 76 | 46.3 |
| T4 | 7 | 17.1 | 10 | 24.4 | 16 | 40.0 | 12 | 28.6 | 45 | 27.4 |
| N stage | ||||||||||
| N0 | 11 | 26.8 | 10 | 24.4 | 10 | 25.0 | 9 | 21.4 | 40 | 24.4 |
| N1 | 10 | 24.4 | 10 | 24.4 | 3 | 7.5 | 10 | 23.8 | 33 | 20.1 |
| N2a | 1 | 2.4 | 3 | 7.3 | 3 | 7.5 | 4 | 9.5 | 11 | 6.7 |
| N2b | 11 | 26.8 | 9 | 22.0 | 7 | 17.5 | 6 | 14.3 | 33 | 20.1 |
| N2c | 7 | 17.1 | 5 | 12.2 | 11 | 27.5 | 8 | 19.0 | 31 | 18.9 |
| N3 | 1 | 2.4 | 4 | 9.8 | 6 | 15.0 | 5 | 11.9 | 16 | 9.8 |
Abbreviations: CTNNB1, beta-catenin; Q, quartile; KPS, Karnofsky performance score.
Table A4.
EGFR Quartiles by Patient and Disease Characteristics
| Characteristic | EGFR Quartiles |
All |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Q1 |
Q2 |
Q3 |
Q4 |
|||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | |
| Treatment | ||||||||||
| Standard | 38 | 50.7 | 44 | 58.7 | 30 | 40.0 | 42 | 56.0 | 154 | 51.3 |
| Accelerated fractionation with concomitant boost | 37 | 49.3 | 31 | 41.3 | 45 | 60.0 | 33 | 44.0 | 146 | 48.7 |
| Sex | ||||||||||
| Male | 52 | 69.3 | 58 | 77.3 | 61 | 81.3 | 55 | 73.3 | 226 | 75.3 |
| Female | 23 | 30.7 | 17 | 22.7 | 14 | 18.7 | 20 | 26.7 | 74 | 24.7 |
| KPS | ||||||||||
| 60 | 4 | 5.3 | 3 | 4.0 | 5 | 6.7 | 2 | 2.7 | 14 | 4.7 |
| 70 | 4 | 5.3 | 10 | 13.3 | 11 | 14.7 | 14 | 18.7 | 39 | 13.0 |
| 80 | 17 | 22.7 | 16 | 21.3 | 18 | 24.0 | 18 | 24.0 | 69 | 23.0 |
| 90 | 41 | 54.7 | 35 | 46.7 | 26 | 34.7 | 30 | 40.0 | 132 | 44.0 |
| 100 | 9 | 12.0 | 11 | 14.7 | 15 | 20.0 | 11 | 14.7 | 46 | 15.3 |
| Age, years | ||||||||||
| ≤ 55 | 19 | 25.3 | 21 | 28.0 | 28 | 37.3 | 21 | 28.0 | 89 | 29.7 |
| 56-65 | 32 | 42.7 | 24 | 32.0 | 27 | 36.0 | 37 | 49.3 | 120 | 40.0 |
| ≥ 66 | 24 | 32.0 | 30 | 40.0 | 20 | 26.7 | 17 | 22.7 | 91 | 30.3 |
| Primary site | ||||||||||
| Oral cavity | 4 | 5.3 | 7 | 9.3 | 14 | 18.7 | 10 | 13.3 | 35 | 11.7 |
| Oropharynx | 47 | 62.7 | 40 | 53.3 | 39 | 52.0 | 40 | 53.3 | 166 | 55.3 |
| Hypopharynx | 11 | 14.7 | 9 | 12.0 | 8 | 10.7 | 13 | 17.3 | 41 | 13.7 |
| Larynx | 13 | 17.3 | 19 | 25.3 | 14 | 18.7 | 12 | 16.0 | 58 | 19.3 |
| T stage | ||||||||||
| T1 | 9 | 12.0 | 9 | 12.0 | 2 | 2.7 | 3 | 4.0 | 23 | 7.7 |
| T2 | 27 | 36.0 | 22 | 29.3 | 17 | 22.7 | 13 | 17.3 | 79 | 26.3 |
| T3 | 21 | 28.0 | 26 | 34.7 | 29 | 38.7 | 33 | 44.0 | 109 | 36.3 |
| T4 | 18 | 24.0 | 18 | 24.0 | 26 | 34.7 | 26 | 34.7 | 88 | 29.3 |
| Tx | — | — | — | — | 1 | 1.3 | — | — | 1 | 0.3 |
| N stage | ||||||||||
| N0 | 12 | 16.0 | 15 | 20.0 | 15 | 20.0 | 15 | 20.0 | 57 | 19.0 |
| N1 | 9 | 12.0 | 15 | 20.0 | 15 | 20.0 | 20 | 26.7 | 59 | 19.7 |
| N2a | 10 | 13.3 | 7 | 9.3 | 9 | 12.0 | 5 | 6.7 | 31 | 10.3 |
| N2b | 19 | 25.3 | 12 | 16.0 | 15 | 20.0 | 12 | 16.0 | 58 | 19.3 |
| N2c | 16 | 21.3 | 16 | 21.3 | 13 | 17.3 | 14 | 18.7 | 59 | 19.7 |
| N3 | 9 | 12.0 | 10 | 13.3 | 8 | 10.7 | 9 | 12.0 | 36 | 12.0 |
Abbreviations: EGFR, epidermal growth factor receptor; Q, quartile; KPS, Karnofsky performance score.
Table A5.
Univariate Evaluation of GLI1, CTNNB1, and EGFR in Relation to the End Points
| Model | Regional-Distant Metastasis |
Progression |
Mortality |
|||
|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | HR | 95% CI | |
| GLI1 | ||||||
| Continuous* | 1.003 | 1.001 to 1.005 | 1.001 | 1.000 to 1.003 | 1.002 | 1.001 to 1.003 |
| Q1 | 1.000 | — | 1.000 | — | 1.000 | — |
| Q2 | 1.198 | 0.628 to 2.288 | 1.016 | 0.648 to 1.593 | 1.104 | 0.781 to 1.562 |
| Q3 | 1.117 | 0.575 to 2.169 | 1.040 | 0.663 to 1.632 | 1.389 | 0.992 to 1.945 |
| Q4 | 2.548 | 1.407 to 4.613 | 1.670 | 1.085 to 2.570 | 1.871 | 1.336 to 2.619 |
| CTNNB1 | ||||||
| Continuous* | 1.000 | 1.000 to 1.000 | 1.000 | 1.000 to 1.000 | 1.000 | 1.000 to 1.000 |
| Q1 | 1.000 | — | 1.000 | — | 1.000 | — |
| Q2 | 0.932 | 0386 to 2.254 | 1.391 | 0.733 to 2.639 | 1.251 | 0.776 to 2.015 |
| Q3 | 1.172 | 0.506 to 2.712 | 1.689 | 0.910 to 3.134 | 1.099 | 0.671 to 1.800 |
| Q4 | 0.792 | 0.349 to 1.799 | 0.701 | 0.354 to 1.389 | 0.864 | 0.536 to 1.393 |
| EGFR | ||||||
| Continuous* | 1.008 | 0.999 to 1.017 | 1.012 | 1.005 to 1.019 | 1.010 | 1.005 to 1.015 |
| Q1 | 1.000 | — | 1.000 | — | 1.000 | — |
| Q2 | 1.312 | 0.699 to 2.463 | 1.295 | 0.773 to 2.170 | 1.279 | 0.886 to 1.846 |
| Q3 | 1.538 | 0.812 to 2.913 | 1.984 | 1.213 to 3.245 | 2.002 | 1.398 to 2.867 |
| Q4 | 1.685 | 0.895 to 3.170 | 2.288 | 1.412 to 3.710 | 1.743 | 1.207 to 2.519 |
Abbreviations: GLI1, glioma-associated oncogene family zinc finger 1; CTNNB1, beta-catenin; EGFR, epidermal growth factor receptor; HR, hazard ratio; Q, quartile.
Per 10-unit increment.
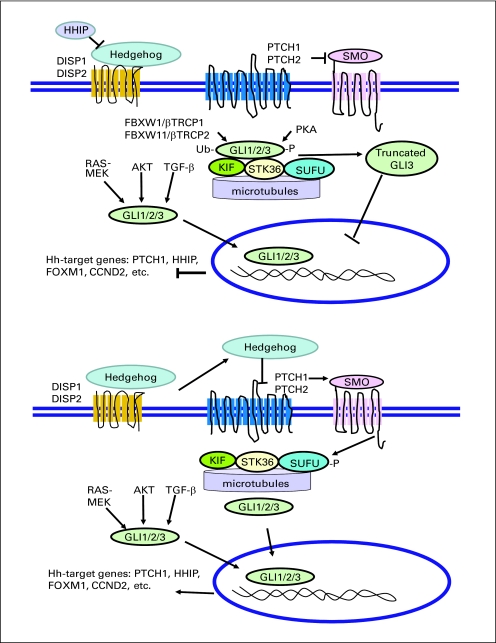
Fig A1.
Schematic diagram of hedgehog (Hh) -GLI signaling pathway based on published reviews.19,55,56 HHIP, hedgehog interacting protein; PTCH1, patched 1; SMO, smoothened; DISP1, dispatched homolog 1; FBXW1/β-TRCP, beta-transducin repeat containing; GLI1, glioma-associated oncogene family zinc finger 1; KIF, Costal-2 homologs; STK36, serine/threonine kinase 36; SUFU, suppressor of fused homolog; FOXM1, forkhead box M1; CCND2, cyclin D2.
Footnotes
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
This article's content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or of the Pennsylvania Department of Health.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: None Stock Ownership: None Honoraria: None Research Funding: Anthony Magliocco, HistoRx Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Christine H. Chung, Quynh-Thu Le,K. Kian Ang
Provision of study materials or patients: M. Elizabeth Hammond, K. Kian Ang
Collection and assembly of data: Christine H. Chung, Alexander C. Klimowicz, Stephanie K. Petrillo, Anthony Magliocco, Andy Trotti, Jay S. Cooper, K. Kian Ang
Data analysis and interpretation: Christine H. Chung, James J. Dignam, M. Elizabeth Hammond, Richard Jordan, Sharon Spencer, Quynh-Thu Le, K. Kian Ang
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Ingham PW. Hedgehog signaling: A tale of two lipids. Science. 2001;294:1879–1881. doi: 10.1126/science.1064115. [DOI] [PubMed] [Google Scholar]
- 2.Chiang C, Litingtung Y, Lee E, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 3.Fietz MJ, Concordet JP, Barbosa R. The hedgehog gene family in Drosophila and vertebrate development. Dev Suppl. 1994:43–51. [PubMed] [Google Scholar]
- 4.Nüsslein-Volhard C, Lohs-Schardin M, Sander K, et al. A dorso-ventral shift of embryonic primordia in a new maternal-effect mutant of Drosophila. Nature. 1980;283:474–476. doi: 10.1038/283474a0. [DOI] [PubMed] [Google Scholar]
- 5.Berman DM, Karhadkar SS, Maitra A, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 6.Thayer SP, di Magliano MP, Heiser PW, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman DM, Karhadkar SS, Hallahan AR, et al. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- 8.Stecca B, Ruiz i Altaba A. Brain as a paradigm of organ growth: Hedgehog-Gli signaling in neural stem cells and brain tumors. J Neurobiol. 2005;64:476–490. doi: 10.1002/neu.20160. [DOI] [PubMed] [Google Scholar]
- 9.Gailani MR, Ståhle-Bäckdahl M, Leffell DJ, et al. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat Genet. 1996;14:78–81. doi: 10.1038/ng0996-78. [DOI] [PubMed] [Google Scholar]
- 10.Gailani MR, Bale AE. Developmental genes and cancer: Role of patched in basal cell carcinoma of the skin. J Natl Cancer Inst. 1997;89:1103–1109. doi: 10.1093/jnci/89.15.1103. [DOI] [PubMed] [Google Scholar]
- 11.Stecca B, Mas C, Ruiz i Altaba A. Interference with HH-GLI signaling inhibits prostate cancer. Trends Mol Med. 2005;11:199–203. doi: 10.1016/j.molmed.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez P, Hernández AM, Stecca B, et al. Inhibition of prostate cancer proliferation by interference with SONIC HEDGEHOG-GLI1 signaling. Proc Natl Acad Sci U S A. 2004;101:12561–12566. doi: 10.1073/pnas.0404956101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watkins DN, Berman DM, Baylin SB. Hedgehog signaling: Progenitor phenotype in small-cell lung cancer. Cell Cycle. 2003;2:196–198. [PubMed] [Google Scholar]
- 14.Watkins DN, Berman DM, Burkholder SG, et al. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 15.Schneider S, Thurnher D, Kloimstein P, et al. Expression of the Sonic hedgehog pathway in squamous cell carcinoma of the skin and the mucosa of the head and neck. Head Neck. 2011;33:244–250. doi: 10.1002/hed.21437. [DOI] [PubMed] [Google Scholar]
- 16.Chuang PT, McMahon AP. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;397:617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- 17.Jiang J, Struhl G. Protein kinase A and hedgehog signaling in Drosophila limb development. Cell. 1995;80:563–572. doi: 10.1016/0092-8674(95)90510-3. [DOI] [PubMed] [Google Scholar]
- 18.Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature. 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- 19.Katoh Y, Katoh M. Hedgehog signaling pathway and gastrointestinal stem cell signaling network (review) Int J Mol Med. 2006;18:1019–1023. [PubMed] [Google Scholar]
- 20.Pan Y, Bai CB, Joyner AL, et al. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 22.Lemjabbar-Alaoui H, Dasari V, Sidhu SS, et al. Wnt and Hedgehog are critical mediators of cigarette smoke-induced lung cancer. PLoS One. 2006;1:e93. doi: 10.1371/journal.pone.0000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stecca B, Mas C, Clement V, et al. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci U S A. 2007;104:5895–5900. doi: 10.1073/pnas.0700776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji Z, Mei FC, Xie J, et al. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J Biol Chem. 2007;282:14048–14055. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- 25.Riobó NA, Lu K, Ai X, et al. Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc Natl Acad Sci U S A. 2006;103:4505–4510. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dennler S, André J, Alexaki I, et al. Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007;67:6981–6986. doi: 10.1158/0008-5472.CAN-07-0491. [DOI] [PubMed] [Google Scholar]
- 27.Kasper M, Schnidar H, Neill GW, et al. Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol Cell Biol. 2006;26:6283–6298. doi: 10.1128/MCB.02317-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh Y, Katoh M. WNT antagonist, SFRP1, is Hedgehog signaling target. Int J Mol Med. 2006;17:171–175. [PubMed] [Google Scholar]
- 29.Chung CH, Parker JS, Ely K, et al. Gene expression profiles identify epithelial-to-mesenchymal transition and activation of nuclear factor-kappaB signaling as characteristics of a high-risk head and neck squamous cell carcinoma. Cancer Res. 2006;66:8210–8218. doi: 10.1158/0008-5472.CAN-06-1213. [DOI] [PubMed] [Google Scholar]
- 30.Von Hoff DD, LoRusso PM, Rudin CM, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med. 2009;361:1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 31.Rudin CM, Hann CL, Laterra J, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Low JA, de Sauvage FJ. Clinical experience with Hedgehog pathway inhibitors. J Clin Oncol. 2010;28:5321–5326. doi: 10.1200/JCO.2010.27.9943. [DOI] [PubMed] [Google Scholar]
- 33.Fu KK, Pajak TF, Trotti A, et al. A Radiation Therapy Oncology Group (RTOG) phase III randomized study to compare hyperfractionation and two variants of accelerated fractionation to standard fractionation radiotherapy for head and neck squamous cell carcinomas: First report of RTOG 9003. Int J Radiat Oncol Biol Phys. 2000;48:7–16. doi: 10.1016/s0360-3016(00)00663-5. [DOI] [PubMed] [Google Scholar]
- 34.Chung CH, Zhang Q, Hammond E. Integrating epidermal growth factor receptor assay with clinical parameters improves risk classification for relapse and survival in head-and-neck squamous cell carcinoma. Int J Radiat Oncol Biol Phys. doi: 10.1016/j.ijrobp.2010.05.024. [epub ahead of print on August 21, 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Psyrri A, Yu Z, Weinberger PM, et al. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin Cancer Res. 2005;11:5856–5862. doi: 10.1158/1078-0432.CCR-05-0420. [DOI] [PubMed] [Google Scholar]
- 36.Cox DR. Regression model and life tables. J R Stat Soc B. 1972;34:187–220. [Google Scholar]
- 37.Prentice RL, Kalbfleisch JD, Peterson AV, Jr, et al. The analysis of failure times in the presence of competing risks. Biometrics. 1978;34:541–554. [PubMed] [Google Scholar]
- 38.Therneau TM, Grambsch P, Fleming TR. Martingale-based residuals for survival models. Biometrika. 1990;77:147–160. [Google Scholar]
- 39.Korn EL, Dorey FJ. Applications of crude incidence curves. Stat Med. 1992;11:813–829. doi: 10.1002/sim.4780110611. [DOI] [PubMed] [Google Scholar]
- 40.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:467–481. [Google Scholar]
- 41.Chang IM, Gelman R, Pagano M. Corrected group prognostic curves and summary statistics. J Chronic Dis. 1982;35:669–674. doi: 10.1016/0021-9681(82)90019-4. [DOI] [PubMed] [Google Scholar]
- 42.Ang KK, Berkey BA, Tu X, et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 2002;62:7350–7356. [PubMed] [Google Scholar]
- 43.Mori Y, Okumura T, Tsunoda S, et al. Gli-1 expression is associated with lymph node metastasis and tumor progression in esophageal squamous cell carcinoma. Oncology. 2006;70:378–389. doi: 10.1159/000098111. [DOI] [PubMed] [Google Scholar]
- 44.Yoshikawa R, Nakano Y, Tao L, et al. Hedgehog signal activation in oesophageal cancer patients undergoing neoadjuvant chemoradiotherapy. Br J Cancer. 2008;98:1670–1674. doi: 10.1038/sj.bjc.6604361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Posner MR, Hershock DM, Blajman CR, et al. Cisplatin and fluorouracil alone or with docetaxel in head and neck cancer. N Engl J Med. 2007;357:1705–1715. doi: 10.1056/NEJMoa070956. [DOI] [PubMed] [Google Scholar]
- 46.Ingham PW, McMahon AP. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 47.van den Brink GR. Hedgehog signaling in development and homeostasis of the gastrointestinal tract. Physiol Rev. 2007;87:1343–1375. doi: 10.1152/physrev.00054.2006. [DOI] [PubMed] [Google Scholar]
- 48.Yauch RL, Gould SE, Scales SJ, et al. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455:406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 49.Frank-Kamenetsky M, Zhang XM, Bottega S, et al. Small-molecule modulators of Hedgehog signaling: Identification and characterization of Smoothened agonists and antagonists. J Biol. 2002;1:10. doi: 10.1186/1475-4924-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hosoya T, Arai MA, Koyano T, et al. Naturally occurring small-molecule inhibitors of hedgehog/GLI-mediated transcription. Chembiochem. 2008;9:1082–1092. doi: 10.1002/cbic.200700511. [DOI] [PubMed] [Google Scholar]
- 51.Chen JK, Taipale J, Cooper MK, et al. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams JA. Hedgehog signaling pathway as a target for therapeutic intervention in basal cell carcinoma. Drug News Perspect. 2003;16:657–662. doi: 10.1358/dnp.2003.16.10.829296. [DOI] [PubMed] [Google Scholar]
- 53.Rahnama F, Shimokawa T, Lauth M, et al. Inhibition of GLI1 gene activation by Patched1. Biochem J. 2006;394:19–26. doi: 10.1042/BJ20050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taipale J, Chen JK, Cooper MK, et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 55.Chari NS, McDonnell TJ. The sonic hedgehog signaling network in development and neoplasia. Adv Anat Pathol. 2007;14:344–352. doi: 10.1097/PAP.0b013e3180ca8a1d. [DOI] [PubMed] [Google Scholar]
- 56.Lauth M, Toftgård R. Non-canonical activation of GLI transcription factors: Implications for targeted anti-cancer therapy. Cell Cycle. 2007;6:2458–2463. doi: 10.4161/cc.6.20.4808. [DOI] [PubMed] [Google Scholar]