Abstract
Mitochondria are the major sites where energy is produced in the cell. Functions of organs such as the heart which has high energy demand are seriously affected by dysfunction of mitochondria. The functional changes in energy-dependent organs such as heart due to aging or any other cause are expected to be reflected in changes in expression of genes related to mitochondrial structure and function. Conversely, alteration of mitochondrial gene expression by any reason may also adversely affect function of organs such as heart that are energy-dependent. Molecular profiling of mitochondrial gene expression is therefore critical to understanding the mechanism of organ dysfunction. Mitochondrial structure and function are controlled by genes in the nuclear DNA and those in the mitochondrial DNA (mtDNA). The transcriptome from these two sources, together, contributing to the structure and function of mitochondria may be called mitoscriptome. This review elaborates on data gathered using a gene chip, RoMitochip, developed in our laboratory to study mitochondrial functional alteration in cardiomyocytes and left ventricular tissue following hypoxia or hemorrhagic injury. RoMitochip consists of probesets representing genes from nuclear DNA and mtDNA of both mice and rats. Our experiments using this chip in in vitro model of hypoxia and in vivo hemorrhagic injury model determined mitoscriptome signatures following hypoxia and hemorrhage, respectively. In addition, we also discuss past initiatives from other investigators that led to the development of microarray tools to profile mitoscriptome.
Keywords: Mitochondria; Microarray; Mitochip; Apoptosis, Custom chip; Injury
Mitochondria are organelles with individuality. A mitochondrion has its own DNA, though a smaller one compared to the nuclear genome, and originates from another mitochondrion [1]. The main function of mitochondria is generation of ATP, the energy molecule, by a very efficient energy producing bioprocess called oxidative phosphorylation. As mitochondria meet most of the energy demands of the cell, they are often called the “powerhouse” of a cell. Oxidative phosphorylation generates an estimated 26 of the 30 molecules of ATP that are formed when glucose is completely oxidized to carbon dioxide and water [2]. Therefore, any functional defect in mitochondria is likely to profoundly affect the cellular energetics and function. The number of mitochondria per cell may vary from one to several thousand, depending on the metabolic requirement of the tissue. Though a number of factors are necessary for mitochondrial DNA (mtDNA) maintenance, the mechanism by which mtDNA levels are controlled is unknown [3]. Apart from supplying metabolic energy to sustain cell function, mitochondria may also control cell death by initiating apoptosis. During hypoxic conditions, however, mitochondrial function is diminished as the oxidative phosphorylation in mitochondria decreases with reduced oxygen availability with uncoupling of glycolysis from mitochondrial oxidation.
Mitochondria in aging
Functional preservation of mitochondria is important for longevity and to minimize age-related diseases. Several studies have shown an age-dependent decline in mitochondrial function [4]. One of the theories still hotly debated in this context is the free radical theory of aging [5]. According to this theory, aging is due to sustained oxidative damage to cellular constituents over time by free radicals. Mitochondria are a major source of free radicals and due to the proximity to such reactive oxidation species (ROS) there could be increased ROS exposure and damage to mitochondrial proteins and DNA [6,7]. The mitochondrial oxidative damage by free radicals causes a vicious cycle that perpetuates more free radical production and further damage to mitochondria, leading to declining mitochondrial function with age. Mitochondria lack the extensive DNA repair systems compared to nuclear DNA [8]. Possibly owing to their purported origins as symbiotic bacteria, mitochondria are deficient in DNA-binding histones and interspersed introns [6,8]. These factors contribute to a higher rate of mtDNA mutation compared to nuclear DNA [9]. For example, mice expressing mtDNA polymerase with a mutated polymerase gamma subunit A demonstrated increased somatic mtDNA mutations associated with reduced lifespan and premature onset of aging-related phenotypes [10,11]. The reduced capacity for energy transduction upon aging leads to secondary dysregulation of cellular processes [12]. According to the Framingham Longevity Study of Coronary Heart Disease, longevity is more strongly associated with age of maternal death than that of paternal death, further implicating mitochondrial DNA (mtDNA) in aging [13]. It is suggested that declining bioengetic capacity may be the most important factor that compromises organ system functional reserve with aging [14].
Mitoscriptome and the mitochondrial structure
Alterations in the function of mitochondria are not solely due to changes in proteins or RNA encoded only in the mitochondrial genome. Most of the genetic information for the mitochondrial biogenesis and function resides in the nuclear genome [11,15,16]. Therefore, the expression profile of the transcripts on mitochondrial genomes and those on the nuclear genomes contributing to the mitochondrial structure and function is critical in understanding the role of mitochondria in health and disease. This subset of transcriptomes, of mtDNA and nuclear DNA origin, can be together called mitoscriptome.
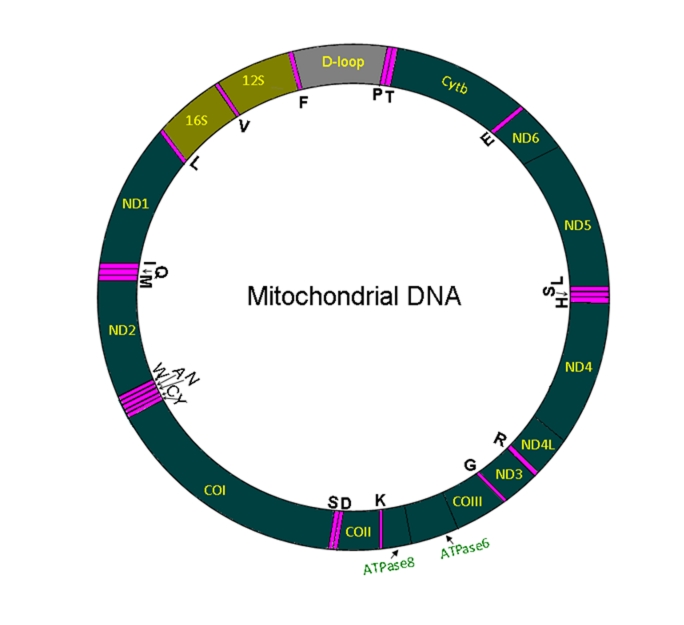
The mammalian mtDNA is a double-stranded circular DNA with about 16500 bp (Fig 1). Unlike genes on the nuclear DNA, the genes on the heavy (H) or light (L) strand of mtDNA are transcribed as a single polycistronic RNA, which is then cleaved and polyadenylated [17–19]. There are several factors that control the cleavage, polyadenylation and stability of each mRNA and therefore, despite a polycistronic transcription the transcript levels of each molecular entity vary [17, 20]. The mtDNA encodes for 37 transcripts: 13 polypeptides, 22 transfer RNA (tRNAs) and two ribosomal RNAs (rRNAs). The 13 polypeptides encoded on mtDNA are: seven subunits (ND1, ND2, ND3, ND4, ND4L, ND5 and ND6) of NADH dehydrogenase (Complex I), cytochrome b (complex III), three subunits (CO1, CO2 and CO3) of cytochrome c oxidase (complex IV) and two subunits of ATP synthase (ATP6 and ATP8). Profiling the mitoscriptome is important in understanding the molecular basis of diseases that may be related to mitochondrial dysfunction. Mitoscriptome profiling may also be important in the identification of valuable biomarkers in health and disease.
Figure 1:
Structure of mitochondrial DNA. Mammalian mitochondrial DNA (mtDNA) is a double-stranded circular DNA with approximately 16,000 base pairs. mtDNA is formed by a light strand and a heavy strand. Pink color bands denote tRNA coding sequences (there are 22 mitochondrial tRNAs, each are labeled by the respective amino acid code). Protein coding segments on mtDNA do not have introns and are transcribed by a single polycistronic mRNA from each strand. All protein coding sequences are marked in green color with respective gene name abbreviations. Abbreviations: Cyt b, Cytochrome b; ND, NADH dehydrogenase; CO, cytochrome c oxidase; and ATPase, ATP synthase. The two ribosomal (rRNAs) locations are marked, 12S and 16S RNAs. D-loop (grey color) region does not have coding sequences. The map is not on scale.
Mitochondrial genomics by mitoscriptome profiling
The exact number of mammalian mitochondrial proteins is not known, though about 1200 proteins have been suggested to constitute this organelle [16]. Proteomic studies have reported approximately 600 mitochondrial proteins in the mouse and about 700 in the rat [16,21]. Though most, if not all, mitochondrial genes encoded in the nuclear genome are represented in the existing Affymetrix mouse and rat gene chips, the lack of representation of genes of mtDNA in these chips and the presence of over 30- to 50,000 thousand probe sets, necessitates the need to analyze several thousand genes. This increased workload demands time and computing power, making focused arrays an attractive alternative.
While the expression level of a small subset of genes can be probed using tools such as a PCR array, probing a greater abundance of several hundred or few thousand genes requires microarray technology. This may be achieved using glass slides, or more preferably, employing commercial printing technologies such as that used in Affymetrix chips. Mitochondrial arrays using both methods have been developed. While Affymetrix technology use short oligonucleotides, most other methods use longer cDNA based probe sets. Currently mitochondrial gene chip arrays are available for human, mouse and the rat, though none of them as yet is commercially available.
In the process of developing an integrated systems biological approach to understand the role of mitochondrial function in health and disease, we developed a custom gene chip to profile the mitoscriptome of the rat and mouse. This rodent mitochondrial gene chip, RoMitoChip, incorporates genes from the nuclear genome contributing to the mitochondrial structure and function as well as the genes on the mtDNA [20,22]. By incorporating the well-annotated mitochondria-related genes of both these species into the same chip, one can interrogate either rat or the mouse mitoscriptome using the same gene chip [20, 22]. Included in this chip are 1088 probesets from the mouse and 419 from the rat. The genes corresponding to these probe sets were selected from existing databases such as the Rat Genome Database, Mouse Genome Database, NetAffx, RatMap database and Mitop2. The probesets were designed using Affymetrix custom algorithm and the 11 micron chip was printed on the Affymetrix platform.
In our laboratory, the RoMitochip was first used to address changes in mitochondrial gene expression in cardiomyocytes subjected to hypoxia in vitro [20]. Mouse cardiomyocytes were subjected to normoxia or hypoxia (1% oxygen) for 8 or 24 hours and the mitoscriptome was profiled using the RoMitochip. Among the most upregulated genes following hypoxia were Aldoc, Bnip3, Hk1, and Pdk1, which are known to have hypoxia responsive element (HRE) binding domains on their promoters and regulated directly by hypoxia-inducible factor (Hif)-1α. The oxygen sensing transcription factor Hif-1 binds to HRE on multiple chromosomal loci, and initiates transcription of several genes during hypoxia. The specific role of Hif-1 in aging process remains controversial [23–25]. It has been reported that transgenic expression of non-degradable Hif-1 increases lifespan. In contrary to this, Zhang et al observed that deletion of Hif-1 increases lifespan [25]. Another study found that the Hif-1 loss-of-function mutant extends lifespan under rich nutrient conditions but fails to show lifespan extension under dietary restriction [23]. Leiser et al suggest that Hif-1 modulates longevity and healthspan in a temperature-dependent manner and further proposes that the reported discrepancy in previous study results on the role of Hif-1 in altering longevity could be due to the disparate models used [26]. Though these studies suggest an important role for Hif-1 in worm aging, whether as a positive or negative regulator of longevity, further studies are required to understand the specific role of Hif-1 in the context of aging [27]. Nevertheless, the hypoxic response is emerging as a promising new avenue in aging research.
Hypoxia is a secondary factor in most injuries, and it is known that injury outcome worsens with aging. When young adult (6 months) and old (22 months) Fisher 344 rats were subjected to either hemorrhagic injury or sham operation, we found a significant decrease in +dP/dt (a measure of cardiac contractility) in the aged group [22]. In addition, aging in the absence of injury also demonstrated a significant decline in cardiac contractility [22]. Using our custom mitochondrial gene chip, RoMitochip, we found a decreased alteration in mitochondrial gene expression in these rats with age, following hemorrhagic injury. Among the genes that were altered with hemorrhagic injury, we found c-myc, a pleotropic transcription factor, to be the most upregulated gene in both 6- and 22-month-old rats [22]. When 142 probesets were significantly altered (39 up and 103 down) in 6-month old rats, only 66 were altered (30 up and 36 down) in 22-month old rats following hemorrhage. The marked decrease in the extent of gene expression changes in the post-hemorrhage mitoscriptome profile of aged animals may be due to cellular senescence. It has been noted that though mitochondria can adapt in response to a wide range of intra- and extra-cellular stimuli, the plasticity decreases with age [4]. The expression of c-myc and cardiac cell death promoting gene Bnip3 were increased, and the expression of mitochondrial biogenesis factors Pgc1-α and Ppara were decreased following hemorrhage. This is consistent with a previous report suggesting that c-myc activation leads to downregulation of Pgc-1α as well as the downstream target Cpt-1 implicated in fatty acid oxidation [28].
Pgc-1α co-activates the coordinated expression of genes involved in mitochondrial biogenesis, energy production and defense systems against reactive oxygen species (ROS). These include genes encoding Tfam, ATP synthase, and uncoupling proteins such as Ucp2. Pgc-1α acts with transcription factors such as Ppar-γ, retinoid receptor (RXR)-α and nuclear respiratory factor-1 (Nrf-1). Zingarelli and coworkers have previously shown that Ppar expression decreases following hemorrhagic injury and when its expression was augmented using specific agonist, liver apoptosis was significantly decreased [29]. The Pgc-1 family of transcriptional coactivators control fatty acid oxidation and mitochondrial biogenesis in the adult heart. Hemorrhage causes a complex injury, but is known to reduce cardiac function and organ blood flow, and induce whole body hypoxia [30, 31]. Low oxygen levels will force intracellular stabilization of Hif-1α, thereby promoting transcription of Hif-1-inducible genes. Previous experiments by us and others have clearly demonstrated the effect of hypoxia in promoting glycolysis while reducing ATP production through decreased mitochondrial oxidation [20,32]. The upregulation of c-myc following hemorrhage is suggestive of its role in promoting glycolytic processes in the ensuing hypoxic condition. Additionally, c-myc protein level has been shown to increase markedly after hypoxia and ischemia [33]. c-myc is also known to activate p53 and promote apoptosis [34,35]. In a cancer model, it was hypothesized that the c-myc transgene causes cell death and probably also inhibition of proliferation, which may be related to its inhibition of cyclin D1 and other oncogenes (e.g., Bcl-2) and induction of p53 and other tumor suppressor genes [34]. Hif and c-myc act on multiple targets to regulate carbon metabolism and act in concert to fine tune adaptive responses to hypoxic environment [36]. Ahuja et al recently reported that in response to pathological stress such as hemodynamic load or ischemia, Myc regulates the increased metabolic energy demand by enhancing glycolysis and downregulating fatty acid oxidation genes [28]. But they also observed that Myc activation attenuates ischemia-induced left ventricular function and increases mitochondrial biogenesis [28]. On the other hand both left ventricular function and mitochondrial biogenesis were decreased following hemorrhage, indicating disparate regulation of Myc-induced pathways in these conditions [22, 30, 37]. However, the upregulation of c-myc in our experiment was consistent with the observed decreased expression of genes negatively regulated by c-myc, such as Cpt1 (−2.3 fold) and Gpam (−2.1 fold). Nevertheless, additional studies are required to further elucidate the role of c-myc in mitochondrial biogenesis regulation in aging and injury (Fig 2). Our studies demonstrate the significance of mitoscriptome profiling in aging and injury, as we were able to obtain valuable information pertaining to both aging and injury in relation to mitochondrial functional alteration.
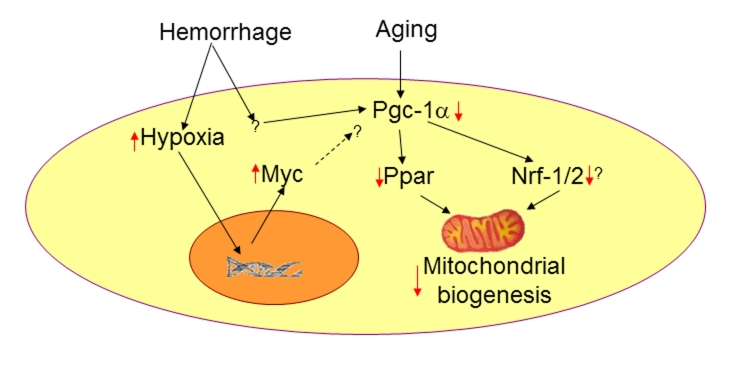
Figure 2:
A model for hemorrhage and age-induced mitochondrial dysfunction. Hemorrhage causes a complex injury including whole body hypoxia. Upregulation of the transcription factor Myc and downregulation of Pgc-1α have been observed following hemorrhagic injury. The specific role of c-myc in mitochondrial biogenesis following hemorrhage is not known. However, it is possible that an enhanced Myc expression is one of the factors responsible for declined Pgc-1α expression and resulting mitochondrial dysfunction following hemorrhage. Aging is also known to downregulate Pgc-1α [49] and decrease mitochondrial biogenesis and function [4]. Red arrows denote the effect due to hemorrhage or aging.
There were several past initiatives to study mitoscriptome using focused gene expression profiling. Alesci et al developed a human mitochondrial gene chip (hMitChip) to study the mitochondrial involvement in glucocorticoid-induced myopathy [38, 39]. The initial version of the chip, hMitChip2, contained 501 nuclear DNA-encoded genes but mtDNA-encoded genes were not represented. In order to correct this deficiency, they constructed an enhanced hMiChip3 with 37 mtDNA-encoded genes and 1098 nuclear DNA-encoded and mitochondria-related genes [40]. Unlike the RoMitochip, which used small oligonucleotide probesets, hMitChip3 was created as a cDNA microarray. Their custom database associated 645 molecular functions with 946 hMitChip3 genes, 612 biological processes with 930 genes, 172 cellular components with 869 genes, 107 biological chemistry pathways with 476 genes, 23 reactome events with 227 genes, 320 genetic disorders with 237 genes, and 87 drugs targets with 55 genes [40, 41]. The hMitChip3 can be used with either the one- or two-fluorescent approach according to users’ preferences [40]. In experiments where only one dye is used, the user may label control and test targets with the same dye and hybridize them on to two different microarrays, where as in two-dye approaches, one dye is used to label target sequences from the control sample and the other dye is used to label the test sample. In the latter, one microarray unit is hybridized with a mixture of the two dyes. The fluorescent detectors can separately quantify hybridization signals due to each dye, and therefore distinctively recognize targets from each sample [42].
A mouse mitochondria-specific microarray, MitoChip, was developed by Desai et al to measure transcripts of mitochondria-associated genes in various diseases and drug-induced toxicities in the mouse [43]. The original chip was made by printing oligonucleotides on poly-L-lysine-coated glass slides. The mouse array had 542 oligonucleotides that represented genes from the mitochondrial and nuclear genomes associated with mitochondrial structure and functions [43]. Using this gene chip, the expression of mitochondrial genes was measured in the liver of both p53 heterozygous and wild-type C3B6F1 female mice exposed to antiretroviral agents, Zidovudine (AZT) and Lamivudine (3TC). They found a significant effect of AZT and 3TC combination treatment on expression levels of approximately 317 genes on the MitoChip. They also observed a high correlation between microarray and real-time PCR results [43].
In another initiative where one of the authors of this manuscript (R.R.) was involved, a human mitochondrial gene chip (huMITOchip) was created using a technology similar to that was used for the RoMitochip [44]. The huMITOchip contains 4,774 probe sets identical to the Affymetrix U 133 plus 2.0 chip covering genes affecting mitochondrial, lipid, cytokine, apoptosis, and muscle function transcripts. Unlike some other gene chips described above, the huMITOchip has 51 probe sets that interrogate 37 genes of the mitochondrial genome [44]. The chip was validated by comparison with the Affymetrix U 133 gene chip and profiling the expression change in mitoscriptome following treatment of the muscle cell line CCL136 with IFN-γ. The validation of the huMITOchip with the Affymetrix U 133 Plus 2.0, such as with the stimulation of muscle cells with a proinflammatory marker, adds to early reliability and validity of this gene array [44].
Age, environment and mitochondria-mediated disease-associated changes on mitochondrial function as well as drug-induced toxicities may have their imprint in the mitoscriptome. Therefore profiling mitochondrial gene expression may be of paramount importance in understanding the fundamental basis of mitochondrial dysfunction and disease. The gene array technology is not without drawbacks. One of the major impediments in this line of study is the associated cost, which would prohibit the use of such tools in smaller laboratories. However, in high throughput screening to evaluate mitochondrial pathway specific gene expression changes, using chips such as the RoMitochip may be more practical, considering the fact that the use of whole genome chips entails analysis of expression levels of thousands of genes. A drawback is the continuously evolving gene annotations that makes such chips obsolete after few years, unless newer versions are developed, dropping some genes and adding others with newly established significance.
With the emergence of ever-increasing significance for mitochondrial biology in health and disease [7, 45–48], mitoscriptome profile information will likely be useful in developing diagnostic methods and new therapeutic strategies. The high relevance of energetics to the maintenance of cellular structure and function places mitoscriptome analysis as an important piece of the puzzle in the understanding of aging and age-related studies.
Acknowledgments
This work was supported by NIH grants AG 031440(RR), GM 39519 (IC), and UAB HSF GEF Scholar Award (RR).
Reference
- [1].Lane N. Mitochondrial disease: powerhouse of disease. Nature. 2006;440:600–602. doi: 10.1038/440600a. [DOI] [PubMed] [Google Scholar]
- [2].Berg JM, Tymoczko JL, Stryer L. Biochemistry. W.H. Freeman and Company; New York: 2006. Oxidative Phosphorylation; pp. 502–540. [Google Scholar]
- [3].Moraes CT. What regulates mitochondrial DNA copy number in animal cells? Trends Genet. 2001;17:199–205. doi: 10.1016/s0168-9525(01)02238-7. [DOI] [PubMed] [Google Scholar]
- [4].Seo AY, Joseph AM, Dutta D, Hwang JC, Aris JP, Leeuwenburgh C. New insights into the role of mitochondria in aging: mitochondrial dynamics and more. J Cell Sci. 2010;123:2533–2542. doi: 10.1242/jcs.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- [6].Vaidya AB, Mather MW. Mitochondrial evolution and functions in malaria parasites. Annu Rev Microbiol. 2009;63:249–267. doi: 10.1146/annurev.micro.091208.073424. [DOI] [PubMed] [Google Scholar]
- [7].Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- [8].Wiesner RJ, Zsurka G, Kunz WS. Mitochondrial DNA damage and the aging process: facts and imaginations. Free Radic Res. 2006;40:1284–1294. doi: 10.1080/10715760600913168. [DOI] [PubMed] [Google Scholar]
- [9].Hiona A, Sanz A, Kujoth GC, Pamplona R, Seo AY, Hofer T, Someya S, Miyakawa T, Nakayama C, Samhan-Arias AK. Mitochondrial DNA mutations induce mitochondrial dysfunction, apoptosis and sarcopenia in skeletal muscle of mitochondrial DNA mutator mice. PLoS One. 2010;5:e11468. doi: 10.1371/journal.pone.0011468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly Y, Gidlof S, Oldfors A, Wibom R. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- [11].Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- [12].Huss JM, Kelly DP. Mitochondrial energy metabolism in heart failure: a question of balance. J Clin Invest. 2005;115:547–555. doi: 10.1172/JCI200524405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Brand FN, Kiely DK, Kannel WB, Myers RH. Family patterns of coronary heart disease mortality: the Framingham Longevity Study. J Clin Epidemiol. 1992;45:169–174. doi: 10.1016/0895-4356(92)90009-c. [DOI] [PubMed] [Google Scholar]
- [14].Muravchick S, Levy RJ. Clinical implications of mitochondrial dysfunction. Anesthesiology. 2006;105:819–837. doi: 10.1097/00000542-200610000-00029. [DOI] [PubMed] [Google Scholar]
- [15].Graham RM, Frazier DP, Thompson JW, Haliko S, Li H, Wasserlauf BJ, Spiga MG, Bishopric NH, Webster KA. A unique pathway of cardiac myocyte death caused by hypoxia-acidosis. J Exp Biol. 2004;207:3189–3200. doi: 10.1242/jeb.01109. [DOI] [PubMed] [Google Scholar]
- [16].Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- [17].Nagao A, Hino-Shigi N, Suzuki T. Measuring mRNA decay in human mitochondria. Methods Enzymol. 2008;447:489–499. doi: 10.1016/S0076-6879(08)02223-4. [DOI] [PubMed] [Google Scholar]
- [18].Fernandez-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp Physiol. 2003;88:41–56. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- [19].Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410:103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- [20].Jian B, Wang D, Chen D, Voss J, Chaudry I, Raju R. Hypoxia induced alteration of mitochondrial genes in cardiomyocytes- role of Bnip3 and Pdk1. Shock. 2010;34:169–175. doi: 10.1097/SHK.0b013e3181cffe7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Forner F, Foster LJ, Campanaro S, Valle G, Mann M. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol Cell Proteomics. 2006;5:608–619. doi: 10.1074/mcp.M500298-MCP200. [DOI] [PubMed] [Google Scholar]
- [22].Jian B, Yang S, Chen D, Zou L, Chatham JC, Chaudry I, Raju R. Aging influences cardiac mitochondrial gene expression and cardiovascular function following hemorrhage injury. Molecular Medicine. 2011;17:5–6. doi: 10.2119/molmed.2010.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen D, Thomas EL, Kapahi P. HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000486. doi: 10.1371/journal.pgen.1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mehta R, Steinkraus KA, Sutphin GL, Ramos FJ, Shamieh LS, Huh A, Davis C, Chandler-Brown D, Kaeberlein M. Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science. 2009;324:1196–1198. doi: 10.1126/science.1173507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang Y, Shao Z, Zhai Z, Shen C, Powell-Coffman JA. The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PLoS One. 2009;4:e6348. doi: 10.1371/journal.pone.0006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Leiser SF, Begun A, Kaeberlein M. HIF-1 modulates longevity and healthspan in a temperature-dependent manner. Aging Cell. 2011;10:318–326. doi: 10.1111/j.1474-9726.2011.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Leiser SF, Kaeberlein M. The hypoxia-inducible factor HIF-1 functions as both a positive and negative modulator of aging. Biol Chem. 2010;391:1131–1137. doi: 10.1515/BC.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ahuja P, Zhao P, Angelis E, Ruan H, Korge P, Olson A, Wang Y, Jin ES, Jeffrey FM, Portman M. Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice. J Clin Invest. 2010;120:1494–1505. doi: 10.1172/JCI38331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zingarelli B, Chima R, O’Connor M, Piraino G, Denenberg A, Hake PW. Liver apoptosis is age-dependent and is reduced by activation of peroxisome proliferator activated receptor-{gamma} in hemorrhagic shock. Am J Physiol Gastrointest Liver Physiol. 2010;298:G133–41. doi: 10.1152/ajpgi.00262.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hsieh YC, Choudhry MA, Yu HP, Shimizu T, Yang S, Suzuki T, Chen J, Bland KI, Chaudry IH. Inhibition of cardiac PGC-1alpha expression abolishes ERbeta agonist-mediated cardioprotection following trauma-hemorrhage. FASEB J. 2006;20:1109–1117. doi: 10.1096/fj.05-5549com. [DOI] [PubMed] [Google Scholar]
- [31].Shimizu T, Szalay L, Hsieh YC, Suzuki T, Choudhry MA, Bland KI, Chaudry IH. A role of PPAR-gamma in androstenediol-mediated salutary effects on cardiac function following trauma-hemorrhage. Ann Surg. 2006;244:131–138. doi: 10.1097/01.sla.0000217709.00863.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sakamoto T, Niiya D, Seiki M. Targeting the warburg effect, which arises in tumor cells expressing membrane-type-1 matrix metalloproteinase. J. Biol. Chem. 2011. (epub). [DOI] [PMC free article] [PubMed]
- [33].Rosano GM, Fini M, Caminiti G, Barbaro G. Cardiac metabolism in myocardial ischemia. Curr Pharm Des. 2008;14:2551–2562. doi: 10.2174/138161208786071317. [DOI] [PubMed] [Google Scholar]
- [34].Wang C, Tai Y, Lisanti MP, Liao DJ. c-Myc induction of programmed cell death may contribute to carcinogenesis: A perspective inspired by several concepts of chemical carcinogenesis. Cancer Biol. Ther. 2011;11 doi: 10.4161/cbt.11.7.14688. (epub). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lindstrom MS, Wiman KG. Myc and E2F1 induce p53 through p14ARF-independent mechanisms in human fibroblasts. Oncogene. 2003;22:4993–5005. doi: 10.1038/sj.onc.1206659. [DOI] [PubMed] [Google Scholar]
- [36].Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hsieh YC, Yu HP, Suzuki T, Choudhry MA, Schwacha MG, Bland KI, Chaudry IH. Upregulation of mitochondrial respiratory complex IV by estrogen receptor-beta is critical for inhibiting mitochondrial apoptotic signaling and restoring cardiac functions following trauma-hemorrhage. J Mol Cell Cardiol. 2006;41:511–521. doi: 10.1016/j.yjmcc.2006.06.001. [DOI] [PubMed] [Google Scholar]
- [38].Alesci A, Su YA, Chrousos GP. hMidas and hMitChip: new opportunities in mitochondrial bioinformatics and genomic medicine. In: Long RAS, Lee DJ, Nutter B, Zhang M, editors. Seventeenth IEEE Symposium on Computer-Based Medical Systems. IEEE ComputerSociety Press; Los Alamitos, CA: 2004. pp. 329–334. [Google Scholar]
- [39].Manoli I, Le H, Alesci S, McFann KK, Su YA, Kino T, Chrousos GP, Blackman MR. Monoamine oxidase-A is a major target gene for glucocorticoids in human skeletal muscle cells. FASEB J. 2005;19:1359–1361. doi: 10.1096/fj.04-3660fje. [DOI] [PubMed] [Google Scholar]
- [40].Bai X, Wu J, Zhang Q, Alesci S, Manoli I, Blackman MR, Chrousos GP, Goldstein AL, Rennert OM, Su YA. Third-generation human mitochondria-focused cDNA microarray and its bioinformatic tools for analysis of gene expression. Biotechniques. 2007;42:365–375. doi: 10.2144/000112388. [DOI] [PubMed] [Google Scholar]
- [41].Johnston DS, Su YA, Alesci S. Mitochondrial gene profiling: translational perspectives. Pharmacogenomics. 2009;10:1645–1655. doi: 10.2217/pgs.09.112. [DOI] [PubMed] [Google Scholar]
- [42].Dobbin K, Shih JH, Simon R. Questions and answers on design of dual-label microarrays for identifying differentially expressed genes. J Natl Cancer Inst. 2003;95:1362–1369. doi: 10.1093/jnci/djg049. [DOI] [PubMed] [Google Scholar]
- [43].Desai VG, Lee T, Delongchamp RR, Moland CL, Branham WS, Fuscoe JC, Leakey JE. Development of mitochondria-specific mouse oligonucleotide microarray and validation of data by real-time PCR. Mitochondrion. 2007;7:322–329. doi: 10.1016/j.mito.2007.02.004. [DOI] [PubMed] [Google Scholar]
- [44].Voss JG, Raju R, Logun C, Danner RL, Munson PJ, Rangel Z, Dalakas MC. A focused microarray to study human mitochondrial and nuclear gene expression. Biol Res Nurs. 2008;9:272–279. doi: 10.1177/1099800408315160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rains JL, Jain SK. Oxidative stress, insulin signaling, and diabetes. Free Radic Biol Med. 2011;50:567–575. doi: 10.1016/j.freeradbiomed.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mochel F, Haller RG. Energy deficit in Huntington disease: why it matters. J Clin Invest. 2011;121:493–499. doi: 10.1172/JCI45691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Rocha M, Apostolova N, Hernandez-Mijares A, Herance R, Victor VM. Oxidative stress and endothelial dysfunction in cardiovascular disease: mitochondria-targeted therapeutics. Curr Med Chem. 2010;17:3827–3841. doi: 10.2174/092986710793205444. [DOI] [PubMed] [Google Scholar]
- [48].Polster BM, Fiskum G. Mitochondrial mechanisms of neural cell apoptosis. J Neurochem. 2004;90:1281–1289. doi: 10.1111/j.1471-4159.2004.02572.x. [DOI] [PubMed] [Google Scholar]
- [49].Turdi S, Fan X, Li J, Zhao J, Huff AF, Du M, Ren J. AMP-activated protein kinase deficiency exacerbates aging-induced myocardial contractile dysfunction. Aging Cell. 2010;9:592–606. doi: 10.1111/j.1474-9726.2010.00586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]