Abstract
Infection with mycobacterium tuberculosis (MTB) can cause different outcomes in hosts with variant genetic backgrounds. Previously, we identified an intracellular pathogen resistance 1 (Ipr1) gene with the role of resistance of MTB infection in mice model. However, until now, its binding proteins have been little known even for its human homology, SP110. In this study, the homology for mouse Ipr1 in canines was found to have an extra domain structure, h.1.5.1. And 30 potential candidate proteins were predicted to bind canine Ipr1, which were characterized of the interacting structure with the h.1.5.1. Among them, MYBBP1A was verified to bind with both Ipr1 and eGFP-Ipr1 in mouse macrophage J774A.1 clone 21 cells using co-immunoprecipitation method. And with the constructed high-confidence Ipr1-involved network, we suggested that Ipr1 might be involved in apoptosis pathway via MYBBP1A.
Keywords: Ipr1, MYBBP1A, Lentivirus, PPI
Introduction
Tuberculosis remains a major cause of death in the world, although most countries have made great efforts to control this infectious disease. Hosts with variant genetic backgrounds have dramatically different outcomes following infection with mycobacterium tuberculosis (MTB). This holds true in both humans and experimental animal models [1]. By studying C3HeB/FeJ(C3H) mice and C57BL/6J(B6) mice, whose susceptibility to MTB infection varies, we previously identified a candidate gene, termed intracellular pathogen resistance 1 (Ipr1), within the sst1 locus of B6 mice [2, 3]. The observation that expression of the Ipr1 transgene in the Ipr1 negative macrophages limits the multiplication of Mycobacterium tuberculosis and switches the cell death pathway of the infected macrophages from necrosis to apoptosis confirms the role of Ipr1 in host resistance to MTB [2]. The homology of the Ipr1 protein in humans is SP110 (41% identity). Both proteins contain a putative SP100-like protein–protein interaction domain, chromatin binding SAND domain, nuclear localization signal (NLS), and a LXXLL nuclear receptor co-activator motif. Although there are many annotations about the Ipr1 protein, most of them are predicted from its sequence analysis. Furthermore, its binding proteins and mediated pathway are not sufficiently understood. A protein's binding partners and its involved networks are keys to describing its mediated pathway and to understanding the complex cellular biological processes involved [4-7].
There are many ways to predict PPIs. Homologous interaction, which means close protein homology could have similar binding protein partners, is a common in-silico method used to predict the PPI from other species [8]. Structure-based prediction is another method used to look for protein physical binding partner candidates [9]. The genomic threading database (GTD) contains structural assignments for the proteins encoded within the genomes of nine eukaryotes and 101 prokaryotes [10]. Also, the structural classification of protein–protein interfaces (SCOPPI) is a database containing all domain–domain interactions [11].
In the GTD, we attempted to find the special domain structures assigned to mouse Ipr1's homology in other species. With the structure-based prediction method, the candidate binding proteins for Ipr1's homology were collected. Based on the homologous interaction, in mice, Ipr1 might have similar binding candidates. Among those candidates, MYBBP1A was verified as a new Ipr1's binding protein with co-immunoprecipitation method in mouse macrophage J774A.1 clone 21 cells [12]. MYBBP1A, also known as P160, is first identified to bind with c-Myb, which is a transcription factor critical for hemopoietic cell proliferation and differentiation. Using protein interaction network analysis (PINA) platform [13], we built a high-confidence, Ipr1 and MYBBP1A involved PPI network in the mouse model, which is helpful for delineating the Ipr1 mediated pathway.
Materials and methods
Candidate proteins for IPR1 binding
Based on that two proteins with interacting domain structures might bind with each other [9, 14] and mouse Ipr1 might have similar binding partners as its homologies in other species, we blasted the mouse Ipr1 protein sequence in the GTD to collect its homologies' domain structures assigned by the GTD. SCOPPI was then used to identify the interacting domain pairs. The corresponding interacting domain name was then used as the keyword to search proteins characterized of query domain in the GTD. The search results might be the Ipr1's binding protein candidates. In order to identify more potential Ipr1 binding candidate partners, we processed the primary search results with the following steps: first, we discarded those with repeated gene names; second, we kept the candidate which has the structure assignment with certain confidence by GTD and has an alignment score of >85; third, according to the mouse Ipr1's cellular location and its role in biological processes, we kept the candidate whose associated GO (Gene Ontology) annotation terms were nucleus/its child term and were transcript/regulation of transcription/apoptosis/their child terms [15]. Finally, 30 more confident candidate Ipr1 binding proteins were obtained.
Construction of lentiviral vectors, preparation and transduction of lentivirus
Based on our previous construction of dual-promoter lentiviral vectors method [12], the pHAGE-CMV.Flag-MYBBP1A.UBC.dLNGFR.W was constructed. And the MYBBP1A cDNA was cloned by PCR from the J774A.1 macrophage cell line using the following primers: ATG GCG GAG ATG AAG AGC and TCA AGG TGT CTG CAC TCT C. Recombinant lentiviruses were produced through a five-plasmid transfection step, as previously reported [12]. The cell model J774A.1 clone 21, which could be induced to express the Ipr1 and/or the eGFP-Ipr, was described in Hui's paper [12]. Before transducing J774A.1 clone 21 cells, western blot analysis was used to test cells for expression of Ipr1 and eGFP-Ipr1 under the stimulation of 1 μg/ml Doxycycline (Dox) for 24 h and/or 100 U/ml IFN-g for 16 h. Immunoblotting was performed with anti-Ipr1 specific rabbit antiserum (Covance Research Products, Inc). The transduction method is also described in Hui's paper.
Nuclear extraction, co-immunoprecipitation, and immunoblotting
Before harvesting cells, we stimulated the transduced J774A.1 clone 21 with Dox (final con. 1 μg/ml) and/or IFN-g (final con. 100 U/ml) for 24 and 16 h, respectively. 2 × 107 cells were washed twice with PBS and then scraped in 2 ml of hypotonic buffer (containing 10 mM HEPES with pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.2% NP40). The cells were then kept on ice for 15 min with occasional vortexing, transferred to microfuge tubes and spun at 4°C, 14,000 rpm for 3 min. The pellet was washed with 500 μl of hypotonic buffer, and 400 μl of the prepared nuclear extraction buffer (containing 20 mM HEPES with pH 7.9, 420 mM NaCl, 1.5 mM MgCl2, 25% glycerol) was then added to suspend the pellet completely. The mixture was mixed for 2 h at 4°C and then spun at 4°C, 14,000 rpm for 10 min to remove debris. The supernatant was then nuclear extract. The concentration of total protein in the nuclear extract was determined with BAC protein assay kit (Pierce Biotechnology, Rockford, IL, USA). Co-immunoprecipitation was operated using an uMACS protein G kit (Miltenyi Biotec). Briefly, 50 μl of protein G Microbeads were incubated with an equal volume of 1% BSA IP buffer (150 mM NaCl, 1% Triton X-100, 50 mM Tris–HCl (pH 8.0)), containing 2 μg IgG or anti-flag monoclonal antibody (Sigma, St. Louis, MO), at 4°C overnight. A volume of 200 μg nuclear extract was diluted with nine volumes of the IP Buffer and immunoprecipitated with the above beads according to the manufacturer's manual. All buffers contained 1% protease inhibitors cocktail and 1% phosphatase inhibitor cocktail 1 and 2 (Sigma-Aldrich). The final elute was separated with 10% SDS-PAGE. Proteins were then transferred to PVDF membranes (Bio-Rad, Hercules, CA, USA) and immunoblotted with anti-Ipr1 specific rabbit antiserum (Covance Research Products, Inc, Denver, CO, USA). The membrane was developed with SuperSignal chemiluminescent substrate (Pierce Biotechnology).
Construction of PPI network
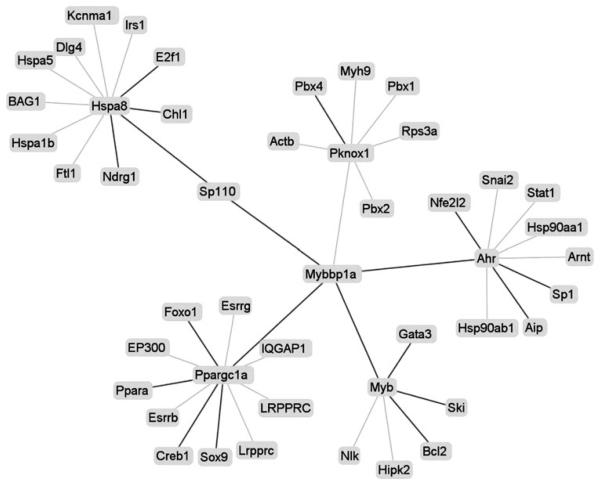
The PINA platform is a currently developed network construction tool that integrates PPI data from six databases: InAct, MINT, BioGRID, DIP, HPRD and MPact. Until now, Ipr1 has been verified to bind with two proteins, MYBBP1A and Hsc70 [12]. Using these three proteins in one PINA query, in mice model, Ipr1 had no interactors, MYBBP1A had only one and Hsc70 had 669 interactors. In order to construct high-confidence network, those PPIs without any evidence support or with wrong references were filtered out in the PINA platform. Thus, 651 Hsc70's interactors were discarded, which were examined to be from two wrong cited references [16, 17]. Additionally, protein interactions supported by experiment evidences from PUBMED were added to the network. In order to get more information from the network, both MYBBP1A's partners and their binding partners were listed in the network. According to the publications from PUBMED and our findings, AhR, Myb, PGC-1(Ppargc1a) and Ipr1 were added as the interactors of MYBBP1A [18-20], Aip, Nfr2 (Nfe212) and Sp1 were added as AhR's [21-23], Bcl2, Gata3 and Ski were added as Myb's [24-26], Pbx4 was added as Pknox1's [27] and Creb, Foxo1, Ppara and Sox9 were added as PGC-1's [28-31]. Since Hsc70 was a chaperone protein, which was involved in folding and trafficking of most proteins, only its binding partners were listed in the network. And E2f1, Chl1, Ndrg1 and Ipr1 were added as the interactors of Hsc70 based on the references from PUBMED [12, 32-34]. With the PINA, the function of network was analyzed with the cutoff P-value as 0.005.
Results and discussion
Candidates of the Ipr1's binding proteins
The GTD provides the predicted proteins' structure information in nine eukaryotes and 101 prokaryotes based on GenTHREADER method [6]. Using the mouse Ipr1 protein sequence for a BLAST search in the GTD, we found that the homology of Ipr1 in canines (gene: ENSCAFG0000 0010682) with the P-value of 2e-59 has an extra structure domain h.1.5.1, except for the common domains in other species. This domain was assigned to the middle of the canine Ipr1 sequence according to the alignment result, between the SP110 domain and the SAND domain. This sequence region of Ipr1 has never been annotated before, which attracted us to look for proteins that could bind with this domain structure. In the SCOPPI database, we found proteins with an h.1.5.1 domain could bind with other proteins with the same domain structure. Therefore, we used the domain of h.1.5.1 as the keyword to search for Ipr1's binding candidates in the GTD within the species of canis familiaris. We set the search results to list 2000 entries, and to arrange result sequences according to the alignment score. A higher alignment score means a more significant relationship between the query protein and template. The search results with an alignment score of >85 and with the certain confidence level totaled 1000 entries (listed in supplementary file 1). In order to identify more potential canine Ipr1 binding partners, we discarded those with repeated gene names. Since two domains are more likely to have similar GO terms if they are interacting [35], each candidate proteins' GO annotation terms were checked based on the Ipr1's cellular location and its role in biological processes. Those with a GO annotation term of nuclear and terms of transcription/regulation of transcription/apoptosis or child terms of these annotations were kept. Finally, based on the h.1.5.1 domain structure information, we identified 30 potential candidate proteins (listed in supplementary file 2), including the MYBBP1A (gene: ENSCAFG00000015266), which could bind with Ipr1 in canines. Since mouse Ipr1 was the homology of canine Ipr1, we predicted those 30 potential candidates might bind with Ipr1 in the mice.
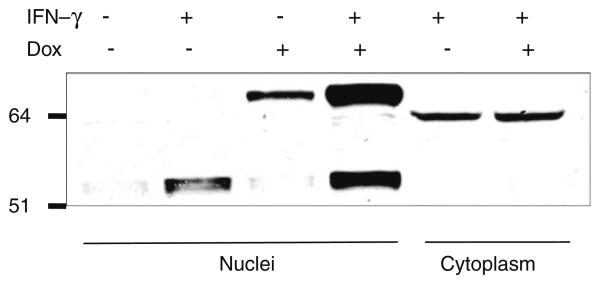
Verification of the Ipr1 binding protein
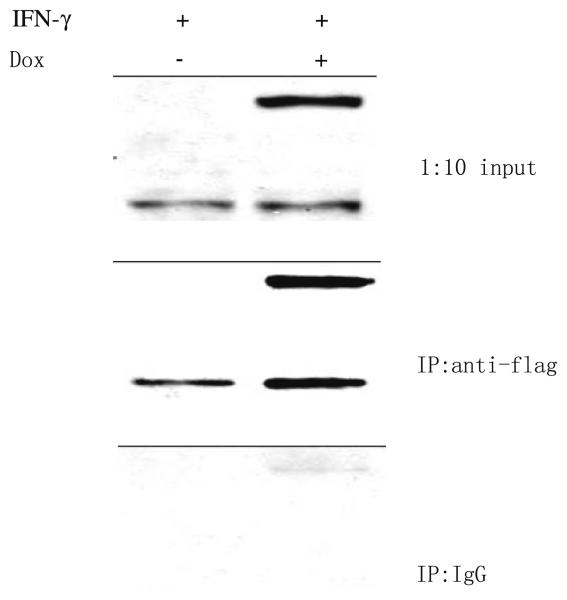
Among 30 Ipr1's potential binding candidates, only MYBBP1A has been verified with co-immunoprecipitation so far. To verify the mouse Ipr1's binding protein, the J774A.1 clone 21 cells were first tested with Western blot (Fig. 1). In the J774A.1 clone 21 cells, endogeneous Ipr1 was mainly expressed under IFN-g stimulation; eGFP-Ipr1 could be induced to express with DOX; when stimulated with IFN-g and DOX simultaneously, cells could express both Ipr1 and eGFP-Ipr1. We then transduced the J774A.1 clone 21 cells with lentivirus containing pHAGE-CMV.Flag-MYBBP1A. UBC.dLNGFR.W vector. After cells were stimulated with IFN-g and/or Dox, nuclear extracts were isolated from those cells and immunoprecipitated with anti-flag monoclonal antibody. The final eluted proteins were analyzed with Western blot, using anti-Ipr1-specific antiserum (Fig. 2). Both endogeneous Ipr1 and extrogenous eGFP-Ipr1 were found to bind with Flag-MYBBP1A. It was noted that eGFP was a protein with 238 Aa which might affect Ipr1's binding on its N-terminus in eGFP-Ipr1 fusion proteins. Thus, it seemed that MYBBP1A might bind with Ipr1 on its middle part, since Ipr1's C-terminus was the SAND domain responsible for chromatin binding. This inference was consistent with that the h.1.5.1 structure domain was assigned to the canine Ipr1's middle part by GTD. And it needed further experiments of mapping the interaction sites of Ipr1 and MYBBP1A, like [36, 37].
Fig. 1.
Ipr1's expression in mouse J774A.1 clone 21 cells. Nuclear extracts and cytoplasm were separated from J774A.1 clone 21 cells stimulated with Dox and/or IFN-g. Anti-Ipr1 specific rabbit antiserum was used for Western blot
Fig. 2.
Binding of MYBBP1A with Ipr1 and eGFP-Ipr1. Mouse J774A.1 clone 21 cells were transduced with lentivirus containing the vector of pHAGE-CMV.Flag-MYBBP1A.UBC.dLNGFR.W. After stimulated with IFN-g and/or DOX, transduced cells were processed into cytoplasm and nuclei. Nuclear extracts were incubated with either anti-Flag antibody or IgG isotype-matched control, and then precipitated with Protein G Microbeads. Eluate from immunoprecipitation and 1:10 amount of nuclear extract (input) were separated on 10% SDS-PAGE and then immunoblotted using anti-Ipr1 specific rabbit antiserum
Construction of protein–protein interaction network
Despite there are 10 GO annotation terms associated with mouse Ipr1, however, its mediated pathway is still not clear. With the Ipr1 involved network, we could not only know Ipr1's detailed function also its mediated pathway. Till now, it has been verified that Ipr1 has two binding proteins: MYBBP1A and Hsc70. Interestedly, both proteins could translocate between cytoplasm and nuclear, but had different functions. With these proteins, we constructed Ipr1 involved network in mouse model. And only the interactions supported by experimental evidences in PINA or from PUBMED were used to construct high-confidence network. Finally, a network with 46 protein nodes and 45 interacting edges was built in mice model (Fig. 3). Using the network function analysis tool, we found that Ipr1 might have the detailed functions (listed in the supplementary file 3), such as: sequence-specific DNA binding. And interestedly, both positive transcription regulation and negative transcription regulation were found in network function analysis result. It suggested that Ipr1 might regulate some genes' transcription with positive effects while regulate other genes with negative effects.
Fig. 3.
PINA-generated visualization of the high-confidence network for Ipr1 in mice. There were 46 interaction nodes and 45 interaction edges. Bold lines indicated the added interaction
Since Hsc70 is a cytoplasmic chaperone protein and oxidative stress but not apoptosis induces its translocation into the nucleus [38, 39], it is more likely that Ipr1 tends to bind with MYBBP1A in the nuclear. And furthermore, MYBBP1A could promote the apoptosis through regulation of its interacting proteins: Ahr and Myb, which were reported to mediate cells apoptosis [40, 41]. Therefore, it suggested that Ipr1 might be involved in apoptosis pathway via MYBBP1A. This needs further biochemical analysis results to support.
In the end, MYBBP1A was found to be a new Ipr1's binding protein in mice for the first time. And it was suggested that Ipr1 might be involved in apoptosis pathway via MYBBP1A, which needs the support of evidences.
Supplementary Material
Acknowledgement
We gratefully acknowledge Dr. Gustavo Mostoslavsky for his advices on the construction of lentivirus vectors and Dr. Jianmin Wu for his advices on the construction of PPI networks. This work was supported by grants AI49421 and P01 AI056296 from the National Institutes of Health and by the National Basic Research Program of China (Grant No. 2009CB918700).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11033-010-0042-1) contains supplementary material, which is available to authorized users.
References
- 1.Kramnik I. Genetic dissection of host resistance to mycobacterium tuberculosis: The sst1 locus and the Ipr1 gene. Curr Top Microbiol Immunol. 2008;321:123–148. doi: 10.1007/978-3-540-75203-5_6. [DOI] [PubMed] [Google Scholar]
- 2.Pan H, Yan BS, Rojas M, Shebzukhov YV, Zhou HW, Kobzik L, Higgins DE, Daly MJ, Bloom BR, Kramnik I. Ipr1 gene mediates innate immunity to tuberculosis. Nature. 2005;434:767–772. doi: 10.1038/nature03419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apt A, Kramnik I. Man and mouse TB: contradictions and solutions. Tuberculosis. 2009;89:195–198. doi: 10.1016/j.tube.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, Li Y, Lin J, Liang Q, Sheng X, Wu J, Huang R, Liu S. Connexin43 interacts with Caveolin-3 in the heart. Mol Biol Rep. 2009;6 doi: 10.1007/s11033-009-9584-5. doi: 10.1007/s11033-009-9584-5. [DOI] [PubMed] [Google Scholar]
- 5.Zheng D, Sun Y, Gu S, Ji C, Zhao W, Xie Y, Mao Y. LNX (Ligand of Numb-protein X) interacts with RhoC, both of which regulate AP-1-mediated transcriptional activation. Mol Biol Rep. 2009;8 doi: 10.1007/s11033-009-9754-5. doi: 10.1007/s11033-009-9754-5. [DOI] [PubMed] [Google Scholar]
- 6.Yang L, Liu N, Hu X, Zhang W, Wang T, Li H, Zhang B, Xiang S, Zhou J, Zhang J. CK2 phosphorylates TNFAIP1 to affect its subcellular localization and interaction with PCNA. Mol Biol Rep. 2009;10 doi: 10.1007/s11033-009-9863-1. doi: 10.1007/s11033-009-9863-1. [DOI] [PubMed] [Google Scholar]
- 7.Borutinskaite VV, Magnusson KE, Navakauskiene R. alpha-Dystrobrevin distribution and association with other proteins in human promyelocytic NB4 cells treated for granulocytic differentiation. Mol Biol Rep. 2010;1 doi: 10.1007/s11033-010-9965-9. doi: 10.1007/s11033-010-9965-9. [DOI] [PubMed] [Google Scholar]
- 8.Park J, Bolser D. Conservation of protein interaction network in evolution. Genome Inform. 2001;12:135–140. [PubMed] [Google Scholar]
- 9.Aloy P, Russell RB. InterPreTS: protein interaction prediction through tertiary structure. Bioinformatics. 2003;19:161–162. doi: 10.1093/bioinformatics/19.1.161. [DOI] [PubMed] [Google Scholar]
- 10.McGuffin LJ, Street SA, Bryson K, Sorensen SA, Jones DT. The genomic threading database: a comprehensive resource for structural annotations of the genomes from key organisms. Nucleic Acids Res. 2004;32:D196–D199. doi: 10.1093/nar/gkh043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter C, Henschel A, Kim WK, Schroeder M. SCOPPI: a structural classification of protein–protein interfaces. Nucleic Acids Res. 2006;34:D310–D314. doi: 10.1093/nar/gkj099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan H, Mostoslavsky G, Eruslanov E, Kotton DN, Kranmik I. Dual-promoter lentiviral system allows inducible expression of noxious proteins in macrophages. J Immunol Methods. 2008;329:31–44. doi: 10.1016/j.jim.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jianmin Wu J, Kristian Ovaska, Jukka Westermarck, Tomi PM, Hautaniemi S. Integrated network analysis platform for protein–protein interactions. Nat Methods. 2009;6:75–77. doi: 10.1038/nmeth.1282. [DOI] [PubMed] [Google Scholar]
- 14.Lu L, Lu H, Skolnick J. MULTIPROSPECTOR: an algorithm for the prediction of protein–protein interactions by multimeric threading. Proteins. 2002;49:350–364. doi: 10.1002/prot.10222. [DOI] [PubMed] [Google Scholar]
- 15.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, Gene Ontology C Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins MO, Husi H, Yu L, Brandon JM, Anderson CNG, Blackstock WP, Choudhary JS, Grant SGN. Molecular characterization and comparison of the components and multi-protein complexes in the postsynaptic proteome. J Neurochem. 2006;97:16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- 17.Abul-Husn NS, Bushlin I, Moron JA, Jenkins SL, Dolios G, Wang R, Iyengar R, Ma'ayan A, Devi LA. Systems approach to explore components and interactions in the presynapse. Proteomics. 2009;9:3303–3315. doi: 10.1002/pmic.200800767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones LC, Okino ST, Gonda TJ, Whitlock JP. Myb-binding protein 1a augments AhR-dependent gene expression. J Biol Chem. 2002;277:22515–22519. doi: 10.1074/jbc.M200740200. [DOI] [PubMed] [Google Scholar]
- 19.Fan M, Rhee J, St-Pierre J, Handschin C, Puigserver P, Lin JD, Jaeger S, Erdjument-Bromage H, Tempst P, Spiegelman BM. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1 alpha: modulation by p38 MAPK. Genes Dev. 2004;18:278–289. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tavner FJ, Simpson R, Tashiro S, Favier D, Jenkins NA, Gilbert DJ, Copeland NG, Macmillan EM, Lutwyche J, Keough RA, Ishii S, Gonda TJ. Molecular cloning reveals that the p160 myb-binding protein is a novel, predominantly nucleolar protein which may play a role in transactivation by Myb. Mol Cell Biol. 1998;18:989–1002. doi: 10.1128/mcb.18.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell DR, Poland A. Binding of aryl hydrocarbon receptor (AhR) to AhR-interacting protein—the role of hsp90. J Biol Chem. 2000;275:36407–36414. doi: 10.1074/jbc.M004236200. [DOI] [PubMed] [Google Scholar]
- 22.Shin S, Wakabayashi N, Misra V, Biswal S, Lee GH, Agoston ES, Yamamoto M, Kensler TW. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol Cell Biol. 2007;27:7188–7197. doi: 10.1128/MCB.00915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F, Wang WL, Safe S. Regulation of constitutive gene expression through interactions of Sp1 protein with the nuclear aryl hydrocarbon receptor complex. Biochemistry. 1999;38:11490–11500. doi: 10.1021/bi982578f. [DOI] [PubMed] [Google Scholar]
- 24.Peng S, Lalani S, Leavenworth JW, Ho IC, Pauza ME. c-Maf interacts with c-Myb to down-regulate Bcl-2 expression and increase apoptosis in peripheral CD4 cells. Eur J Immunol. 2007;37:2868–2880. doi: 10.1002/eji.200636979. [DOI] [PubMed] [Google Scholar]
- 25.Maurice D, Hooper J, Lang G, Weston K. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. EMBO J. 2007;26:3629–3640. doi: 10.1038/sj.emboj.7601801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nomura T, Tanikawa J, Akimaru H, Kanei-Ishii C, IchikawaIwata E, Khan MM, Ito H, Ishii S. Oncogenic activation of c-Myb correlates with a loss of negative regulation by TIF1 beta and Ski. J Biol Chem. 2004;279:16715–16726. doi: 10.1074/jbc.M313069200. [DOI] [PubMed] [Google Scholar]
- 27.Yoshioka H, Geyer CB, Hornecker JL, Patel KT, McCarrey JR. In vivo analysis of developmentally and evolutionarily dynamic protein-DNA interactions regulating transcription of the Pgk2 gene during mammalian spermatogenesis. Mol Cell Biol. 2007;27:7871–7885. doi: 10.1128/MCB.00990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herzig S, Long FX, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 29.Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong HJ, Accili D, Spiegelman BM. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1 alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 30.Finck BN, Gropler MC, Chen ZJ, Leone TC, Croce MA, Harris TE, Lawrence JC, Kelly DP. Lipin 1 is an inducible amplifier of the hepatic PGC-1 alpha/PPAR alpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 31.Kawakami Y, Tsuda M, Takahashi S, Taniguchi N, Esteban CR, Zemmyo M, Furumatsu T, Lotz M, Belmonte JCI, Asahara H. Transcriptional coactivator PGC-1 alpha regulates chondrogenesis via association with Sox9. Proc Natl Acad Sci USA. 2005;102:2414–2419. doi: 10.1073/pnas.0407510102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garimella R, Liu X, Qiao W, Liang XY, Zuiderweg ERP, Riley MI, Van Doren SR. Hsc70 contacts helix III of the J domain from polyomavirus T antigens: Addressing a dilemma in the chaperone hypothesis of how they release E2F from pRb. Biochemistry. 2006;45:6917–6929. doi: 10.1021/bi060411d. [DOI] [PubMed] [Google Scholar]
- 33.Leshchyns'ka I, Sytnyk V, Richter M, Andreyeva A, Puchkov D, Schachner M. The adhesion molecule CHL1 regulates uncoating of clathrin-coated synaptic vesicles. Neuron. 2006;52:1011–1025. doi: 10.1016/j.neuron.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 34.Sugiki T, Taketomi Y, Kikuchi-Yanoshita R, Murakami M, Kudo I. Association of N-myc downregulated gene 1 with heat-shock cognate protein 70 in mast cells. Biol Pharm Bull. 2004;27:628–633. doi: 10.1248/bpb.27.628. [DOI] [PubMed] [Google Scholar]
- 35.Liu M, Chen XW, Jothi R. Knowledge-guided inference of domain–domain interactions from incomplete protein–protein interaction networks. Bioinformatics. 2009;25:2492–2499. doi: 10.1093/bioinformatics/btp480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van den Berk LC, van Ham MA, te Lindert MM, Walma T, Aelen J, Vuister GW, Hendriks WJ. The interaction of PTP-BL PDZ domains with RIL: an enigmatic role for the RIL LIM domain. Mol Biol Rep. 2004;31:203–215. doi: 10.1007/s11033-005-1407-8. [DOI] [PubMed] [Google Scholar]
- 37.Jiayu W, Zhu H, Ming X, Xiong W, Songbo W, Bocui S, Wensen L, Jiping L, Keying M, Zhongyi L, Hongwei G. Mapping the interaction site of prion protein and Sho. Mol Biol Rep. 2009;8 doi: 10.1007/s11033-009-9722-0. doi: 10.1007/s11033-009-9722-0. [DOI] [PubMed] [Google Scholar]
- 38.Lagunas L, Bradbury CM, Laszlo A, Hunt CR, Gius D. Indomethacin and ibuprofen induce Hsc70 nuclear localization and activation of the heat shock response in HeLa cells. Biochem Biophys Res Commun. 2004;313:863–870. doi: 10.1016/j.bbrc.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 39.Dastoor Z, Dreyer JL. Nuclear translocation and aggregate formation of heat shock cognate protein 70 (Hsc70) in oxidative stress and apoptosis. J Cell Sci. 2000;113:2845–2854. doi: 10.1242/jcs.113.16.2845. [DOI] [PubMed] [Google Scholar]
- 40.Chung JY, Kim JY, Kim WR, Lee SG, Kim YJ, Park JE, Hong YP, Chun YJ, Park YC, Oh S, Yoo KS, Yoo YH, Kim JM. Abundance of aryl hydrocarbon receptor potentiates benzo[a]-pyrene-induced apoptosis in Hepa1c1c7 cells via CYP1A1 activation. Toxicology. 2007;235:62–72. doi: 10.1016/j.tox.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 41.Selvakumaran M, Lin HK, Sjin RTT, Reed JC, Liebermann DA, Hoffman B. The novel primary response gene MYD118 and the protooncogenes Myb, Myc, and Bcl-2 modulate transforming growth factor-beta-1-induced apoptosis of myeloidleukemia cells. Mol Cell Biol. 1994;14:2352–2360. doi: 10.1128/mcb.14.4.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.