Abstract
The Hippo pathway coordinates organ size and cell proliferation. The retinoblastoma family of proteins regulates progression through the G0/G1 phase of the cell cycle. Disruption of either pathway contributes to cancer formation. Three recent studies in Genes & Development reveal how cellular proliferation is coordinated between these pathways. Here we discuss the implications of these studies and the new questions that they raise.
Keywords: Hippo pathway, pRB pathway, DREAM complex, cell cycle, cellular senescence, phosphorylation
The regulation of cellular proliferation is critical to the development of most tissues and organs. At its most basic level, control of cell cycle exit allows for precise regulation of organ size, as cell numbers are one of its primary determinants. Perhaps more importantly, careful control of cell proliferation during differentiation allows specialized cells to make heterotypic interactions with appropriate neighbors, rather than inappropriate or homotypic interactions with adjacent cells that arise from aberrant cycling. In short, the intricate organization of specialized cells during tissue morphogenesis is highly dependent on the ability of its constituent cells to read cell cycle exit cues.
Cancer is often summarized as a disease of inappropriate cell division. The most fundamental characteristics of cancer cells, called the “hallmarks” of cancer, are intimately related to a cell's intrinsic ability to control proliferation (Hanahan and Weinberg 2011). Just as the response to regulatory signals that trigger cell cycle exit is essential in development, the loss of responsiveness to these signals allows quiescent or differentiated cells to revert to a more proliferative state and contribute to tumor development.
The balance between signals that induce proliferation or suppress it controls the cell cycle. Our knowledge of these signals and the molecular pathways that transmit them are only now being revealed. Two proliferative control pathways that are the focus of recent studies in Genes & Development from the Frolov (Nicolay et al. 2011), Dyson (Tschöp et al. 2011), and DeCaprio (Litovchick et al. 2011) laboratories are the Retinoblastoma (RB) pathway and the Hippo pathway. Both pathways are well known to regulate proliferation in development, and mutations that affect their integrity contribute to cancer (Classon and Harlow 2002; Pan 2010). In most cases where these pathways have been investigated in experimental paradigms of development or disease, their roles are readily detectable. For this reason, a broad question of interest in understanding cell cycle regulation is whether these distinctive proliferative control pathways coordinate their activities and, if so, how this is accomplished. Insight into how separate pathways cooperate in cell cycle decisions in G1 is essential to truly understand the control of proliferation in multicellular organisms.
The RB pathway in proliferative control and cancer
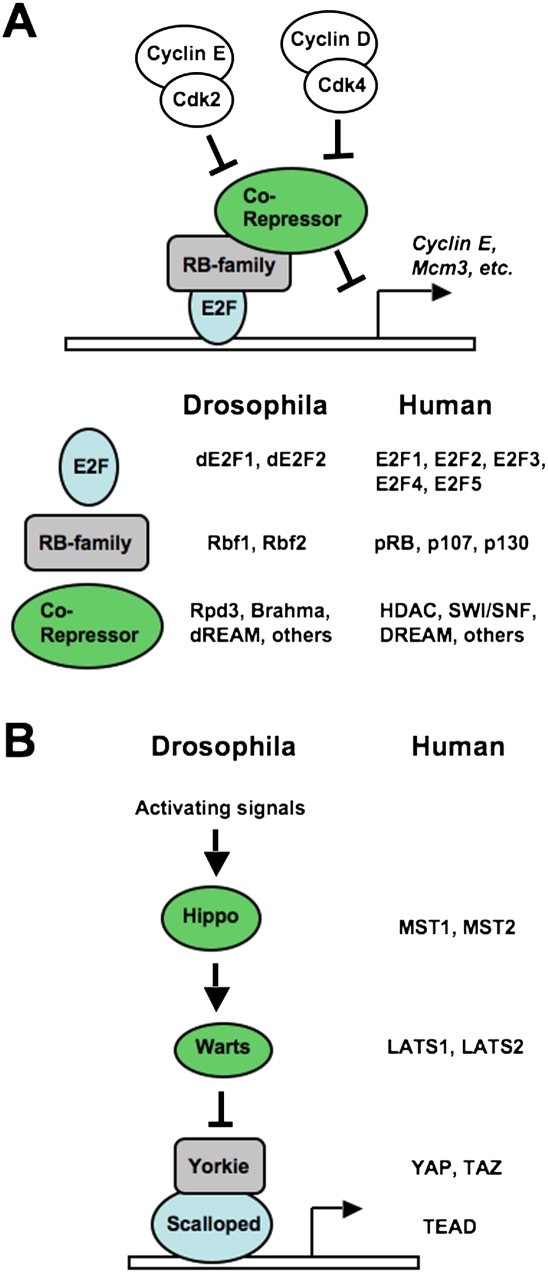
E2F transcription factors were some of the earliest pRB (Retinoblastoma protein)-interacting proteins to be discovered. Ectopic expression of E2Fs induces cell cycle advancement from G1 to S phase, and expression of pRB can block this effect, thus offering a simple binary switch mechanism that can inhibit cell proliferation (Dyson 1998). In G1, pRB binds and inhibits E2Fs. Upon mitogenic signaling, cyclin-dependent kinases phosphorylate and inactivate pRB, releasing E2Fs to transcribe cell cycle target genes such as Cyclin E and Mcm3 that induce progression into S phase (Fig. 1A). While mutation of the human RB1 gene is the underlying cause of retinoblastoma, and disruption of other components in its regulatory pathway is a requirement in cancer (Sherr and McCormick 2002), the loss of function of the mouse Rb1 gene reveals a less than essential role in proliferative control in development. When Rb1 is deleted in the embryo proper, many tissues and organs develop normally and exit the cell cycle appropriately (Wu et al. 2003). Furthermore, chimeric mice composed of a mixture of wild-type and Rb1 knockout cells revealed that pRB-deficient cells can contribute to most tissues and organs. Surprisingly, this results in only mild hyperplasia, and this is observed in only a few developmental contexts (Williams et al. 1994). Conversely, deletion of Rb1 along with its two related family members, p107 and p130, results in early embryonic lethality and massively deregulated proliferation. In only a few instances do these triple-knockout cells display any ability to control the cell cycle (Wirt et al. 2010). The concept of a shared role for RB family proteins in proliferative control is of great importance, but has also been a key impediment to our understanding of how G1-to-S-phase progression is regulated.
Figure 1.
Conservation of the RB and Hippo signaling pathways in humans and fruit flies. (A) Illustration of the RB pathway from cyclin-dependent kinases to RB family proteins and E2Fs. The names of family members from Drosophila melanogaster and Homo sapiens are indicated. Note that RB family proteins recruit corepressor complexes to E2F promoters using their pocket domain. In the case of p130 (humans) and Rbf2 (fruit fly), the corepressor includes the MuvB core components that assemble into the DREAM and dREAM complexes, respectively. (B) Components of the Hippo pathway are similarly diagrammed, with the names of fruit fly and human counterparts included.
All RB family proteins possess the growth-suppressing pocket domain and can bind to E2F transcription factors in order to inhibit transcription of cell cycle genes (Dyson 1998). However, the expression pattern of these proteins is different in that p130 is the most abundant in quiescent or differentiated cells and p107 predominates in proliferating cells. The expression of pRB is less dynamic, as it displays a mild elevation of expression during proliferation. Remarkably, deletion of any one member of the family results in up-regulation of the others. In particular, loss of p130 leads to increased expression of p107 in quiescence, even though its expression is normally quite low at this stage of the cell cycle (Hurford et al. 1997). For these reasons, it has been challenging to delineate boundaries between the respective roles for pRB, p107, and p130 in cell cycle control.
The identification and characterization of DP, RB, E2F, and MuvB (DREAM) complexes is of great importance for understanding the biology of RB family proteins. The Caenorhabditis elegans Lin35 gene resembles human RB1 and functions in cooperation with E2F-like genes as well as others in the synMuvB complementation group, such as Lin9, Lin37, Lin52, and Lin55 (van den Heuvel and Dyson 2008). The Drosophila Rbf, E2f, and Myb (dREAM) complex was identified through biochemical purification and also contains fruit fly orthologs of the synMuvB components LIN9, LIN37, LIN52, LIN54, and RBBP4 (Korenjak et al. 2004; Lewis et al. 2004). Last, the orthologous human DREAM complex was copurified with p130 (Litovchick et al. 2007). Intriguingly, analysis of DREAM association with mammalian pocket proteins demonstrated that it is found exclusively with p130, and only under conditions of p130 loss did it associate with p107 (Litovchick et al. 2007). This suggests that DREAM-mediated repression of E2F transcription is specific for p130 and mechanistically distinct from other RB family members. To emphasize that DREAM contains p130 in our discussion of its function, we refer to it as p130–DREAM throughout this perspective. The unique inclusion of p130 in DREAM raises the question of whether it can be regulated separately from other pocket protein–E2F complexes in cell cycle control.
The role of Hippo in proliferation and development
The Hippo pathway has emerged recently as a critical regulator of proliferation in development. It was discovered largely through genetic screens for overproliferation of cells in developing Drosophila embryos (Zhao et al. 2010). The Hippo pathway is an evolutionarily conserved signaling cascade that senses cell density and in turn can suppress proliferation (Fig. 1B). In this way, loss of function of this pathway leads to the appearance of hyperplastic tissue and dramatically enlarged organs (Pan 2010). In transgenic and knockout mice, these overgrowth phenotypes cause a strong predisposition to cancer, although naturally occurring mutations of Hippo components are relatively rare in human cancer (Pan 2010).
The extracellular origin of signals that control the activity of the Hippo pathway is largely unknown. The first transmembrane protein discovered to exert influence on Hippo is encoded by the Drosophila fat gene (Grusche et al. 2010). How Fat signals, and the immediate downstream targets that it requires for activating Hippo, is an area of intensive investigation. Three components that are downstream from Fat include Expanded (ex), Merlin (mer), and Kibra (kb). However, it is not clear how they communicate with Fat or how they signal to the next component downstream, Hippo. The hippo gene encodes a kinase (abbreviated as hpo; MST1/2 in mammals) that phosphorylates and activates the Warts kinase (wts; LATS1/2 in mammals), which in turn phosphorylates the transcriptional cofactor Yorkie (yki; YAP/TAZ in mammals). In the absence of phosphorylation, Yorkie translocates to the nucleus, where it cooperates with DNA-binding factors such as Scalloped (sd; TEAD in mammals) to induce transcription of genes that advance the cell cycle (Pan 2010). In this way, signals that activate the Hippo pathway suppress Yorkie-dependent transcription and inhibit proliferation. In contrast, mutations that inactivate the negative regulation of Yorkie cause dramatic overgrowth phenotypes in many tissues and organs in fruit flies and mice alike. The extent of the overgrowth phenotype has been an important guiding principle in Hippo signaling. Components such as expanded and merlin are thought to act in parallel because individual mutation of these genes causes relatively mild overgrowth phenotypes. However, fruit flies bearing mutations in both genes appear more comparable with the single-mutation phenotypes of core components such as warts or hippo (Zhao et al. 2010). Transgenic expression of Yorkie also serves to model deregulation of the pathway, as this recapitulates the overgrowth phenotype observed from loss of hippo or warts. Taken together, these data suggest that Yorkie is the primary target of growth regulation by the Hippo pathway, and this has led to a focused search for targets of Yorkie transcription (Pan 2010).
Transcriptional targets of Yorkie include a number of genes in Drosophila that are not conserved in higher eukaryotes, implying that a substantial divergence in transcriptional regulation of cell proliferation by this pathway has occurred between species. However, given that a common feature of cell cycle control by the RB and Hippo pathways is transcriptional control, an important question remains as to whether these pathways regulate common or distinct targets in cell cycle regulation.
The convergence of Hippo and RB pathway signaling in Drosophila melanogaster
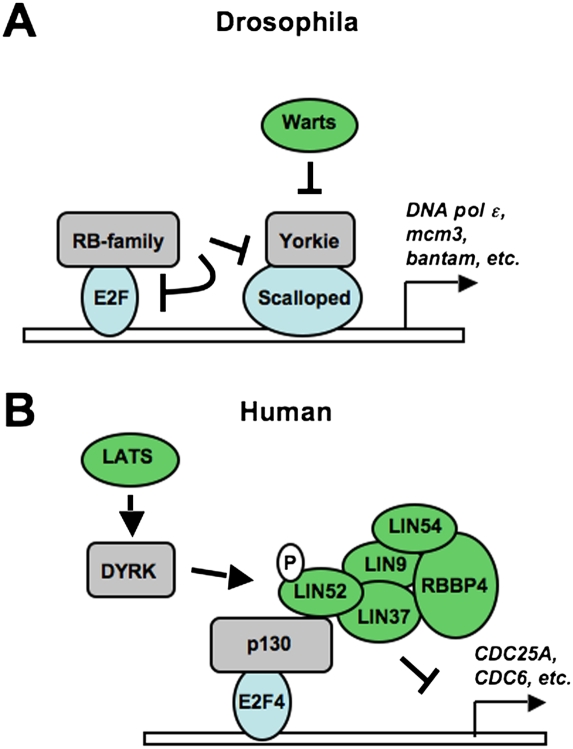
Both the Hippo and RB pathways control proliferation, and both have undeniable links to cancer. The ability of these pathways to work together was first investigated in the developing eye of the fruit fly, where it was discovered that rbf and warts mutations synergize to create a deficiency in cell cycle exit during differentiation (Nicolay et al. 2010). Nicolay et al. (2011) presented key evidence that these pathways collaborate at a transcriptional level in the February 15, 2011, issue of Genes & Development. In this study, they used a DNA microarray approach to identify common classes of target genes that are deregulated in response to rbf and warts mutations, alone or in combination. This revealed some similarity in the patterns of gene expression that are changed by these mutants. Their data also revealed a significant increase in cell cycle genes that are deregulated by both mutations in combination, compared with either single mutation. Inspection of promoter sequences of these common target genes revealed binding sites for both dE2F transcription factors and Scalloped. Transcriptional reporter assays further revealed a synergistic activation of these target genes by dE2F1 and Yorkie/Scalloped. In addition, chromatin immunoprecipitation experiments showed that dE2F1 and Scalloped occupy many common cell cycle promoters in relatively close proximity. Nicolay et al. (2011) also demonstrated that Yorkie-induced proliferation in the eye requires dE2F1, and that Rbf proteins can contribute to the suppression of transcription and proliferation by Yorkie. Surprisingly, Yorkie-induced proliferation is suppressed by Rbf even in the absence of dE2F1. This demonstrates that the RB and Hippo pathways not only converge to regulate common genes, but also cooperate with and interregulate one another at these target promoters (Fig. 2A).
Figure 2.
Mechanisms of coordination between RB and Hippo pathways in cell cycle control. The convergence of Hippo signaling on E2F transcription that was described in recent studies is summarized. (A) A diagram of how Rbf regulates dE2F and Yki/Sd cooperation at E2F target genes in growth-arrested cells. (B) The signaling cascade from LATS to DYRK and p130–DREAM is illustrated to explain how DREAM is assembled to silence E2F target gene expression in growth-arrested cells.
The introductions for the RB and Hippo pathways above highlight the extensive conservation of these molecular mechanisms for transcriptional and growth control between organisms such as fruit flies and humans (Fig. 1). Puzzlingly, the conservation of this mechanism ends with transcriptional regulation by Yorkie and its mammalian orthologs, YAP/TAZ. As explained by Nicolay et al. (2011), the regulation of key cell cycle targets such as Cyclin E, Cyclin A, and Cyclin B by Yorkie is not mirrored by mammalian YAP/TAZ. This leaves in question how the mammalian RB and Hippo pathways communicate. Conservation of the function of these pathways suggests they will, but the available data lead to the conclusion that the mechanism must be different in mammals compared with Drosophila.
The interaction of the RB and Hippo pathways in mammals
Two studies that appeared in the April 15, 2011, issue of Genes & Development investigated the functions of RB family proteins and discovered clear mechanisms of cooperation with the Hippo pathway in mammalian cell cycle control (Litovchick et al. 2011; Tschöp et al. 2011). These reports offer a substantial advancement in our broader understanding of cell proliferation by providing mechanistic insight into cell cycle withdrawal.
In the study by Tschöp et al. (2011), the authors investigate the mechanism of pRB-dependent growth arrest. They take advantage of the sensitivity of RB1-null human osteoblast-like osteosarcoma SaOS-2 cells to arrest in response to pRB re-expression and perform an shRNA screen for kinases that are required for pRB-dependent proliferative control. In addition, they also screen for kinases that are necessary for markers of senescence at later stages of this pRB-dependent arrest. These screens offer the generality that stress-responsive kinases are the most commonly required companions of pRB in suppressing cell proliferation. Intriguingly, the senescence screen showed a strong requirement for the mammalian Warts kinase homolog LATS2, providing the first evidence in mammalian cells that pRB-induced senescence is impacted by components of the Hippo pathway. Phenotypically, the LATS2 requirement in senescence is not completely related to supporting pRB function per se. Surprisingly, SaOS-2 cells arrest in G1 in response to expression of exogenous pRB, regardless of whether LATS2 is depleted by shRNA. Despite this, it was noted that LATS2 was required for complete repression of E2F target genes by pRB. More importantly, the role for LATS2 appears to be critical in establishing downstream characteristics of senescent cells, including inducing senescence-associated β-galactosidase (SA-βGal) activity and stimulating the formation of senescence-associated heterochromatic foci (SAHF). Tschöp et al. (2011) further investigated the effect of LATS2 depletion on SAHF formation and SA-βGal expression in a number of different cell systems and paradigms for senescence. Using these assays, they came to the same conclusion: LATS2 is required for many of the defining characteristics of senescence.
By mining expression microarray data sets, Tschöp et al. (2011) discovered that LATS2 mRNA levels are inversely correlated with those of known E2F target genes. Furthermore, depletion of LATS2 expression by shRNA in senescence leads to elevated levels of E2F targets. Given that YAP/TAZ are known to be the focus of Hippo signaling, Tschöp et al. (2011) sought to investigate whether they can regulate E2F target gene expression. However, unlike the results published by Nicolay et al. (2011) studying the fruit fly Rbf and Hippo pathways, Tschöp et al. (2011) were unable to detect elevated E2F target gene expression when YAP was ectopically expressed in cells. Similarly, coexpression of YAP failed to enhance E2F1-dependent activation of transcription. These data suggest that LATS2 signaling has targets other than YAP/TAZ that are required for silencing E2F target genes.
A logical candidate that represses E2F transcription independently of pRB during senescence is the p130–DREAM complex. To this end, Tschöp et al. (2011) used chromatin immunoprecipitation experiments to investigate the occupancy of E2F target gene promoters by p130 and DREAM components. As expected p130, LIN9, and LIN54 associate with E2F-responsive promoters in a pRB-dependent manner. Furthermore, depletion of LATS2 expression reduces the assembly of this complex at E2F-responsive promoters in senescence. These experiments elegantly demonstrate that the mammalian Hippo pathway contributes to E2F-regulated gene silencing in senescence. Given the past challenges in delineating independent roles for pRB and family members like p130, this study reveals an intriguing separation of function whereby pRB is the initial brake that allows for cell cycle exit. In turn, the p130–DREAM complex is then enabled by LATS2 to repress E2F-dependent transcription, which leads to the characteristics of a fully senescent cell.
Regulation of p130–DREAM assembly in senescence
The discovery that the mammalian Hippo pathway can regulate the activity of the p130–DREAM complex and, subsequently, E2F-dependent transcription raises the obvious question of how this is accomplished on a mechanistic level. A study by Litovchick et al. (2011) that was also published in the April 15, 2011, issue of Genes & Development offers a very complete answer as to how assembly of the p130–DREAM complex is regulated and how this impacts on cell cycle control to induce senescence.
This study is based on a proteomic analysis of p130-, E2F4-, and Myb-containing complexes. In a previous report, this approach revealed the composition of the DREAM complex in humans (Litovchick et al. 2007). In this new study, Litovchick et al. (2011) mined their data set of peptides for phosphorylation sites on the constituent subunits of DREAM. By comparing the frequency of phospho-peptide identification between each immunoprecipitation (performed with different antibodies against individual components of the p130–DREAM complex and Myb), they were able to identify phosphorylation of LIN52 on Ser 28 as being an exclusive post-translational modification to the DREAM complex when bound to p130. Taking advantage of the slower migration in SDS-PAGE induced by Ser 28 phosphorylation, Litovchick et al. (2011) independently verified that p130 exclusively bound DREAM containing phosphorylated LIN52. Furthermore, by depleting endogenous LIN52 and re-expressing a serine-to-alanine LIN52 mutant, they established that phosphorylation of LIN52 is necessary and sufficient to regulate DREAM binding to p130.
Given the elegant regulation of p130 and DREAM interactions by a single phosphorylation event, Litovchick et al. (2011) then sought to identify the kinase responsible. To do this, they again referred to their mass spectrometry data and identified DYRK1A as a candidate that copurifies with p130–DREAM. The DYRKs (dual-specificity tyrosine phosphorylation-regulated kinases) are a family of kinases that autophosphorylate a tyrosine residue in their activation loop and catalyze the phosphorylation of serine/threonine residues on numerous substrates (Becker and Joost 1999), including many transcription factors. Members of this kinase family from lower eukaryotes are Yak1 in Saccharomyces cerevisiae, Pom1 in Schizosaccharomyces pombe, YakA in Dictyostelium, and minibrain in Drosophila. Although strains with mutations in these proteins display different phenotypic abnormalities, all are involved in cell cycle regulation or in controlling the transition from cell growth to differentiation (Tejedor and Hammerle 2011). Mammalian DYRKs include DYRK1A, DYRK1B, DYRK2, DYRK3, and DYRK4 (Becker and Joost 1999). DYRK1A is the most extensively characterized member of this family, and it shares all of the characteristic motifs of its catalytic domain with the other family members. In humans, DYRK1A is located in the “Down syndrome-critical region” on chromosome 21q22.2, and is thought to play a role in the aberrant brain development, early onset neurodegeneration, neuronal loss, and dementia associated with this disease (Tejedor and Hammerle 2011; Wegiel et al. 2011). The human and rodent DYRK1A genes are ubiquitously expressed in tissues of adult and fetal origin.
Litovchick et al. (2011) provide experimental evidence that DYRK1A-dependent assembly of p130–DREAM is required for serum starvation-induced exit from the cell cycle and senescence. In addition, expression of a LIN52 S28A mutant interferes with entry into senescence and SA-βGal expression. Taken together, this study provides compelling evidence that DYRK1A is a novel regulator of E2F target gene expression through its essential role in controlling the assembly of p130–DREAM complexes. Through phosphorylation of just one site on the DREAM complex, it has a clear impact on growth arrest and senescence.
DYRKs, potential components of the Hippo pathway
The Tschöp et al. (2011) study demonstrates a role for p130–DREAM in pRB-induced senescence, and the Litovchick et al. (2011) study reveals that assembly of p130–DREAM requires phosphorylation by DYRK1A. Together, they both demonstrate an essential role for p130–DREAM in suppressing E2F target gene expression during senescence. The linkage between these observations begins with the finding that members of the DYRK family are essential for pRB-induced senescence in the kinome screen performed by Tschöp et al. (2011). In addition, DYRK1A contains putative consensus phosphorylation sites for the LATS2 kinase. This suggests a linear pathway of assembly in which LATS is directly upstream of DYRK1A. Using an in vitro kinase assay system, the DeCaprio and Dyson groups (Tschöp et al. 2011) collaborated to demonstrate that LATS2 can indeed phosphorylate and activate DYRK1A, and this in turn stimulates its ability to phosphorylate LIN52. These data suggest that DYRK kinases may be new members of the Hippo signaling pathway that link it to p130-dependent regulation of proliferation (Fig. 2B). Information regarding in vivo regulation of DYRK activity in mammalian cells is scarce, and the novel observation by Tschöp et al. (2011) that LATS2 can directly phosphorylate DYRK1A and stimulate its activity represents the first description of a cellular mechanism for regulating DYRK1A beyond alterations in expression.
These experiments demonstrate how DYRK1A can block proliferation through p130–DREAM. Is this activity fully duplicated by the paralogous DYRK1B kinase? The Litovchick et al. (2011) study clearly demonstrates that DYRK1B can also interact with and phosphorylate LIN52 in vitro, but they were unable to demonstrate a specific role for endogenous DYRK1B in vivo. Although exogenous overexpression of DYRK1B was shown to rescue the effect of depleting DYRK1A, it remains to be determined whether this is physiologically relevant. Should this turn out to be the case, control of DYRK1B may be independent of LATS2, as the putative consensus phosphorylation site for this kinase is not present in DYRK1B.
Activation of DYRK1A by LATS2 presumably leads to phosphorylation of other targets beyond LIN52 Ser 28. Are these substrates solely confined to regulating cell proliferation? Given that altered DYRK1A dosage has effects on neuronal differentiation, placing it downstream from HIPPO—a regulator of neural progenitor cell number (Cao et al. 2008)—may suggest a broader function for this kinase signaling cascade.
DYRK1A and DYRK1B are currently considered to be prosurvival kinases (Aranda et al. 2011). Each has been reported to be expressed at elevated levels in multiple cancer types and to inhibit apoptosis through a variety of direct or indirect mechanisms. Given that DYRK1A, and potentially DYRK1B, promotes cell cycle arrest via p130–DREAM, this concept may require further investigation. In some instances, high levels of these DYRKs could confer a selective survival advantage to tumor cells by enhancing their ability to attain quiescence during unfavorable growth conditions, which might be interpreted as inhibition of apoptosis. In other instances, high level expression of these DYRKs may not actually have any functional consequence, but may merely reflect a futile attempt to regain control of the cell cycle in cells with a nonfunctional RB pathway, akin to that observed for p16 expression in cells deficient for pRB (von Knebel Doeberitz 2001).
Final thoughts
In conclusion, these three studies collectively offer exciting insight into the coordination of proliferation through two key cell cycle regulatory pathways. They demonstrate the convergence of signaling between the Hippo and RB pathways at the level of transcriptional control. They also reveal important details of the mechanisms in fruit flies and mammals that allow these pathways to communicate. This work also raises important new questions about signaling by the Hippo pathway in growth arrest and, potentially, other paradigms such as differentiation, stress response, cell survival, and cancer.
Acknowledgments
Research in our laboratories into the RB pathway and DYRK kinases is supported by the Canadian Institutes of Health Research.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2050411.
References
- Aranda S, Laguna A, de la Luna S 2011. DYRK family of protein kinases: evolutionary relationships, biochemical properties, and functional roles. FASEB J 25: 449–462 [DOI] [PubMed] [Google Scholar]
- Becker W, Joost HG 1999. Structural and functional characteristics of Dyrk, a novel subfamily of protein kinases with dual specificity. Prog Nucleic Acid Res Mol Biol 62: 1–17 [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH 2008. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev 22: 3320–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classon M, Harlow E 2002. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer 2: 910–917 [DOI] [PubMed] [Google Scholar]
- Dyson N 1998. The regulation of E2F by pRB-family proteins. Genes Dev 12: 2245–2262 [DOI] [PubMed] [Google Scholar]
- Grusche FA, Richardson HE, Harvey KF 2010. Upstream regulation of the hippo size control pathway. Curr Biol 20: R574–R582 doi: 10.1016/j.cub.2010.05.023 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2011. Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Hurford RK Jr, Cobrinik D, Lee MH, Dyson N 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev 11: 1447–1463 [DOI] [PubMed] [Google Scholar]
- Korenjak M, Taylor-Harding B, Binné UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A 2004. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 119: 181–193 [DOI] [PubMed] [Google Scholar]
- Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR 2004. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev 18: 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA 2007. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell 26: 539–551 [DOI] [PubMed] [Google Scholar]
- Litovchick L, Florens LA, Swanson SK, Washburn MP, DeCaprio JA 2011. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev 25: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay BN, Bayarmagnai B, Moon NS, Benevolenskaya EV, Frolov MV 2010. Combined inactivation of pRB and hippo pathways induces dedifferentiation in the Drosophila retina. PLoS Genet 6: e1000918 doi: 10.1371/journal.pgen.1000918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay BN, Bayarmagnai B, Islam AB, Lopez-Bigas N, Frolov MV 2011. Cooperation between dE2F1 and Yki/Sd defines a distinct transcriptional program necessary to bypass cell cycle exit. Genes Dev 25: 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D 2010. The hippo signaling pathway in development and cancer. Dev Cell 19: 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, McCormick F 2002. The RB and p53 pathways in cancer. Cancer Cell 2: 103–112 [DOI] [PubMed] [Google Scholar]
- Tejedor FJ, Hammerle B 2011. MNB/DYRK1A as a multiple regulator of neuronal development. FEBS J 278: 223–235 [DOI] [PubMed] [Google Scholar]
- Tschöp K, Conery AR, Litovchick L, DeCaprio JA, Settleman J, Harlow E, Dyson N 2011. A kinase shRNA screen links LATS2 and pRB tumor suppressor. Genes Dev 25: 814–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ 2008. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 9: 713–724 [DOI] [PubMed] [Google Scholar]
- von Knebel Doeberitz M 2001. New molecular tools for efficient screening of cervical cancer. Dis Markers 17: 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegiel J, Gong CX, Hwang YW 2011. The role of DYRK1A in neurodegenerative diseases. FEBS J 278: 236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BO, Schmitt EM, Remington L, Bronson RT, Albert DM, Weinbert RA, Jacks T 1994. Extensive contribution of Rb-deficient cells to adult chimeric mice with limited histopathological consequences. EMBO J 13: 4251–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirt SE, Adler AS, Gebala V, Weimann JM, Schaffer BE, Saddic LA, Viatour P, Vogel H, Chang HY, Meissner A, et al. 2010. G1 arrest and differentiation can occur independently of Rb family function. J Cell Biol 191: 809–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, de Bruin A, Saavedra HI, Starovic M, Trimboli A, Yang Y, Opavska J, Wilson P, Thompson JC, Ostrowski MC, et al. 2003. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421: 942–947 [DOI] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL 2010. The Hippo–YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev 24: 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]