Abstract
Many tumors continuously depend on the initiating oncogenes, but whether this extends to their downstream targets is unclear. In this issue of Genes & Development, Sodir and colleagues (pp. 907–916) demonstrate an essential role for endogenous Myc proteins in maintaining the tumor microenvironment, providing an unexpected molecular explanation for addiction to Myc.
Keywords: Myc inhibition, pancreas, tumor, microenvironment, therapeutics
A central tenet of molecular oncology states that tumors remain dependent on the activity of the oncogenic pathways that drive tumorigenesis (Weinstein 2002; Luo et al. 2009). This hypothesis forms the basis of many strategies for targeted tumor therapy. One central piece of evidence supporting the concept of oncogene “addiction” stems from mouse models using tunable expression of oncogenes. When expression of the oncogene is switched off in these models, many of the tumors regress.. Tumors induced by conditional expression of the MYC proto-oncogene exhibit this behavior. Turning MYC off in established tumors precipitates—albeit often only temporary—regression, sometimes leaving a small population of dormant tumor cells behind (Arvanitis and Felsher 2006). Multiple mechanisms, including apoptosis and senescence, have been documented to contribute to this regression. In contrast, the molecular mechanisms that establish oncogene addiction are incompletely understood.
The inducible transgenic models used in these experiments mimic the situation in Burkitt's lymphoma, in which translocations juxtapose MYC to strong enhancers that normally control the expression of immunoglobulin genes. This transcriptional deregulation initiates tumor development by inducing grossly elevated levels of Myc expression. In contrast, mutations in MYC genes are relatively rare in solid tumors, and, more frequently, a nonmutated MYC gene is expressed at elevated levels in tumor cells due to mutations in any one of a large number of upstream signaling pathways. Whether endogenous MYC genes are required for initiation and maintenance of most solid tumors—and, if so, whether tumor cells require elevated levels of MYC relative to normal cells—is much less clear. One example for a strict dependence appears to be the development of colon carcinoma, which is often driven by loss of the APC tumor suppressor gene. In a corresponding mouse model, deletion of MYC ablates tumor development (Sansom et al. 2007). Furthermore, downstream targets of Myc can be haploinsufficient for tumor formation but not normal development, suggesting that tumor cells indeed depend on elevated levels of Myc activity (Nilsson et al. 2005; Barna et al. 2008).
In addition to MYC, the genes encoding the closely related MYCN and MYCL proteins have also been implicated in the genesis of human tumors. Indeed, MYCN and MYC are partially redundant during embryonal development and in hematopoietic stem cells (Laurenti et al. 2008). Furthermore, individual neuroblastomas show enhanced expression of either MYC or MYCN but not both, suggesting that these genes can substitute for each other during tumorigenesis. In order to test the requirement for “Myc activity” rather than the involvement of a specific member of the MYC gene family in tumorigenesis, Soucek and Evan (Soucek et al. 2008) developed an elegant dominant-negative allele of Myc, termed OmoMyc. OmoMyc heterodimerizes with endogenous Myc proteins and blocks their association with a heterodimeric partner protein—termed Max—that is required for sequence-specific binding to DNA and the subsequent transactivation of target genes. However, OmoMyc does not block the interaction of Myc with another partner protein—termed Miz1—that is required for transrepression by Myc. The expression of OmoMyc in tissue culture consistently blocks transactivation, but not transrepression, by Myc (Soucek et al. 2002). In previously published work, Evan and colleagues (Soucek et al. 2008) used OmoMyc to suppress the development of KRAS-dependent lung carcinomas, confirming and extending the work on c-myc knockout mice. They showed that OmoMyc not only has a high therapeutic efficacy, but is also well tolerated in adult animals for an extended period of time, demonstrating the potential of inhibiting Myc as a therapeutic strategy.
In this issue of Genes & Development, Sodir et al. (2011) now use OmoMyc to show that insulinomas arising in β cells of mice that express a SV40 large T protein are similarly continuously dependent on Myc activity. This is surprising, since at least some cell-autonomous functions of Myc are likely to be nonessential in cells that express large T, because large T binds to and sequesters the pRb and p53 tumor suppressor proteins and thereby overrides many of the controls that control cell cycle progression and maintain chromosomal stability.
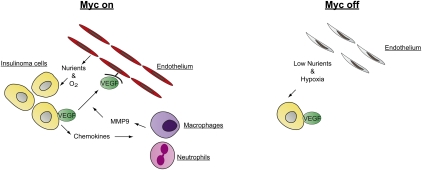
The mouse model used here is the exemplar to demonstrate an “angiogenic switch” during tumor development: The term refers to the ability of a subset of early-stage malignant lesions to acquire the ability to promote the formation of new blood vessels and develop fully vascularized tumors (Hanahan and Folkman 1996). This ability is essential to sustain the oxygen and nutrient supply of tumors that grow beyond a certain minimal size. Molecularly, tumors that undergo the angiogenic switch begin to secrete proangiogenic factors (such as VEGF) and repress anti-angiogenic factors, most notably thrombospondin. Sodir et al. (2011) show that expression of OmoMyc in insulinoma cells leads to apoptosis of endothelial cells and disrupts the oxygen supply of the tumor cells, demonstrating an essential role for endogenous MYC in tumor maintenance. Furthermore, continuous Myc activity is required for recruitment of neutrophils and tumor macrophages, essential components of the tumor microenvironment. Both cells produce matrix metalloproteinase MMP-9, which in turn increases the availability of VEGF that is secreted by the tumor cells but initially is retained in an inactive form in the extracellular matrix. Molecular analyses show that secretion of a broad spectrum of cytokines depends on ongoing Myc activity (Fig. 1). Taken together, the data show that endogenous Myc function is required to maintain the tumor microenvironment—in particular, the blood supply of growing tumors—and strongly support a non-cell-autonomous role of Myc in tumor formation. These findings significantly extend previous work that links expression of Myc to the tumor microenvironment and angiogenesis. For example, previous studies had shown that endogenous c-myc is required to express VEGF and suppress thrombospondin in murine embryonic stem cells and derived teratomas (Baudino et al. 2002). Similarly, in Myc-driven lymphomas, thrombospondin is required for tumor regression and collapse of tumor angiogenesis when MYC expression is turned off (Giuriato et al. 2006; Rakhra et al. 2010).
Figure 1.
Model for the interplay between Myc and the microenvironment. When endogenous Myc is active (Myc on), insulinoma cells secrete VEGF and chemokines. These chemokines serve as chemoattractants for inflammatory cells, such as neutrophils and macrophages. These cells then secrete MMP9, leading to an increased availability of VEGF, up-regulated cell proliferation, and the survival of endothelial cells. In turn, these synergistic actions provide the tumor with sufficient oxygen and nutrients for expansion. Turning off Myc in the tumor cells (Myc off) leads to a first wave of apoptosis in the endothelial cell population and, subsequently, tumor regression due to hypoxia-induced apoptosis.
Like all major steps forward, the current work raises a number of important questions. For example, how is Myc recruited to direct the genetic program that promotes the angiogenic switch? One possibility is that the elevated levels of Myc present in many tumor cells regulate genes that are not regulated by physiological levels of Myc (Yustein et al. 2010; Kress et al. 2011). However, in the insulinoma cells studied here, Myc is not overtly up-regulated, suggesting that genetic factors that are present in tumor cells alter the transcriptional properties of Myc. For example, the Arf tumor suppressor protein associates with both Myc and Miz1 and alters the target gene spectrum of the complex. This leads to, among other changes, the induction of Egr1, a potently proangiogenic transcription factor (Herkert et al. 2010; Boone et al. 2011). Similarly, low oxygen supply leads to stabilization of the Hif1α transcription factor, which can both repress Myc expression and cooperate with Myc in the transcriptional activation of genes (Gordan et al. 2007). The relationship between Hif1α and Myc is reminiscent of that between Myc and TGFβ: TGFβ represses Myc expression in primary cells, but synergizes with deregulated Myc in promoting metastasis once its cytostatic function is disabled during tumor progression (Smith et al. 2009). Similarly, the angiogenic switch may reflect a change in Myc activity that allows cells to overcome the repressive effects of Hif1α on Myc expression and allow the synergistic activation of joint target genes.
The second issue raised by these findings is how, exactly, OmoMyc acts in insulinoma cells to block angiogenesis. Clearly, OmoMyc itself is not a therapeutic agent, but serves as a tool that models the efficacy of future strategies that interfere with Myc function in tumorigenesis. But, as described before, OmoMyc is not simply a dominant-negative allele, since it does not block all functions of Myc. OmoMyc's astounding efficacy in this and the previously described tumor models may therefore depend on a particular way of inactivating Myc function. In order to design and evaluate strategies that use small molecules to interfere with Myc function for tumor therapy, the data presented here establish an urgent need to further delineate how OmoMyc works on a cell-biological and mechanistic level.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2053311.
References
- Arvanitis C, Felsher DW 2006. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin Cancer Biol 16: 313–317 [DOI] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D 2008. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456: 971–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudino TA, McKay C, Pendeville-Samain H, Nilsson JA, Maclean KH, White EL, Davis AC, Ihle JN, Cleveland JL 2002. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes & Dev 16: 2530–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone DN, Qi Y, Li Z, Hann SR 2011. Egr1 mediates p53-independent c-Myc-induced apoptosis via a noncanonical ARF-dependent transcriptional mechanism. Proc Natl Acad Sci 108: 632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuriato S, Ryeom S, Fan AC, Bachireddy P, Lynch RC, Rioth MJ, van Riggelen J, Kopelman AM, Passegue E, Tang F, et al. 2006. Sustained regression of tumors upon MYC inactivation requires p53 or thrombospondin-1 to reverse the angiogenic switch. Proc Natl Acad Sci 103: 16266–16271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, Simon MC 2007. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell 12: 108–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Folkman J 1996. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell 86: 353–364 [DOI] [PubMed] [Google Scholar]
- Herkert B, Dwertmann A, Herold S, Abed M, Naud JF, Finkernagel F, Harms GS, Orian A, Wanzel M, Eilers M 2010. The Arf tumor suppressor protein inhibits Miz1 to suppress cell adhesion and induce apoptosis. J Cell Biol 188: 905–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TR, Cannell IG, Brenkman AB, Samans B, Gaestel M, Roepman P, Burgering BM, Bushell M, Rosenwald A, Eilers M 2011. The MK5/PRAK kinase and Myc form a negative feedback loop that is disrupted during colorectal tumorigenesis. Mol Cell 41: 445–457 [DOI] [PubMed] [Google Scholar]
- Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, Knoepfler PS, Cheng PF, MacDonald HR, Eisenman RN, et al. 2008. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell 3: 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ 2009. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson JA, Keller UB, Baudino TA, Yang C, Norton S, Old JA, Nilsson LM, Neale G, Kramer DL, Porter CW, et al. 2005. Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell 7: 433–444 [DOI] [PubMed] [Google Scholar]
- Rakhra K, Bachireddy P, Zabuawala T, Zeiser R, Xu L, Kopelman A, Fan AC, Yang Q, Braunstein L, Crosby E, et al. 2010. CD4+ T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell 18: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR 2007. Myc deletion rescues Apc deficiency in the small intestine. Nature 446: 676–679 [DOI] [PubMed] [Google Scholar]
- Smith AP, Verrecchia A, Faga G, Doni M, Perna D, Martinato F, Guccione E, Amati B 2009. A positive role for Myc in TGFβ-induced Snail transcription and epithelial-to-mesenchymal transition. Oncogene 28: 422–430 [DOI] [PubMed] [Google Scholar]
- Sodir NM, Swigart LB, Karnezis AN, Hanahan D, Evan GI, Soucek L 2011. Endogenous Myc maintains the tumor microenvironment. Genes Dev (this issue). doi: 10.1101/gad.2038411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Jucker R, Panacchia L, Ricordy R, Tato F, Nasi S 2002. Omomyc, a potential Myc dominant negative, enhances Myc-induced apoptosis. Cancer Res 62: 3507–3510 [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI 2008. Modelling Myc inhibition as a cancer therapy. Nature 455: 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB 2002. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science 297: 63–64 [DOI] [PubMed] [Google Scholar]
- Yustein JT, Liu YC, Gao P, Jie C, Le A, Vuica-Ross M, Chng WJ, Eberhart CG, Bergsagel PL, Dang CV 2010. Induction of ectopic Myc target gene JAG2 augments hypoxic growth and tumorigenesis in a human B-cell model. Proc Natl Acad Sci 107: 3534–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]