Abstract
In higher eukaryotes, the centromere is epigenetically specified by the histone H3 variant Centromere Protein-A (CENP-A). Deposition of CENP-A to the centromere requires histone chaperone HJURP (Holliday junction recognition protein). The crystal structure of an HJURP–CENP-A–histone H4 complex shows that HJURP binds a CENP-A–H4 heterodimer. The C-terminal β-sheet domain of HJURP caps the DNA-binding region of the histone heterodimer, preventing it from spontaneous association with DNA. Our analysis also revealed a novel site in CENP-A that distinguishes it from histone H3 in its ability to bind HJURP. These findings provide key information for specific recognition of CENP-A and mechanistic insights into the process of centromeric chromatin assembly.
Keywords: centromere, CENP-A, histone chaperone, HJURP, structure
Faithful inheritance of genetic information requires precise segregation of sister chromatids during mitosis, which entails the attachment of spindle microtubules to each chromatid at a specialized locus, known as the centromere (Cleveland et al. 2003; Henikoff and Dalal 2005; Allshire and Karpen 2008). In mammals, centromeres are specified by the location of nucleosomes containing Centromere Protein-A (CENP-A), which assembles into a nucleosome together with histones H4, H2A, and H2B without any known dependence of DNA sequence (Black and Cleveland 2011). Centromeric chromatin is interspersed with CENP-A- and histone H3-containing nucleosomes, and it forms a repressive heterochromatin structure through the concerted action of various epigenetic mechanisms, such as processes involving histone methylation and microRNA (Grewal and Rice 2004). The integrity of centromeric chromatin is crucial for the assembly of a large multiprotein complex that bridges the nucleosome to the microtubule attachment site on the outer kinetochore. Despite its importance, little is known about the molecular mechanism by which CENP-A is specifically recognized, targeted to the centromere, and assembled into the nucleosome.
Recently, a CENP-A-specific histone chaperone, HJURP (Holliday junction recognition protein), has been identified (Dunleavy et al. 2009; Foltz et al. 2009). It is distantly related to the yeast centromeric protein Scm3 (Camahort et al. 2007; Mizuguchi et al. 2007; Stoler et al. 2007; Sanchez-Pulido et al. 2009). HJURP interacts directly with CENP-A and histone H4, localizes CENP-A to the centromere in a cell cycle-dependent manner, and enables the deposition of newly synthesized CENP-A into the centromeric nucleosome. An approximately 80-amino-acid CENP-A-binding domain (CBD) at the N terminus of HJURP is necessary and sufficient for binding CENP-A (Shuaib et al. 2010), and a region encompassing loop-1 and helix α2 of the histone fold domain of CENP-A, known as the CENP-A targeting domain (CATD), is required for interaction with HJURP (Black et al. 2004; Foltz et al. 2009). Here we provide the structural basis for the recognition of CENP-A by HJURP, as well as mechanistic insights into the histone chaperone activity of HJURP.
Results and Discussion
Overall structure
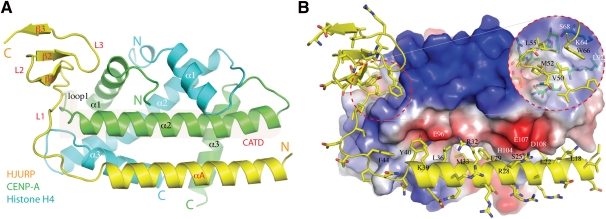
Human HJURP CBD was cocrystallized with CENP-A and histone H4. The 2.6 Å cocrystal structure shows that HJURP CBD forms a complex with a heterodimer of CENP-A and histone H4 (Fig. 1A). The ordered portion of HJURP CBD consists of amino acids 14–74, and the structure is composed of a long helix (αA) and a three-stranded anti-parallel β sheet, connected by a 15-residue loop (L1) (Fig. 1A). The structure of the CENP-A and histone H4 heterodimer, as expected, is similar to the ones in the CENP-A–H4 tetramer (Sekulic et al. 2010). Helix αA of HJURP packs against the central helix (α2) of the histone fold of CENP-A in an anti-parallel manner, with the hydrophobic side of the amphipathic αA facing CENP-A (Fig. 1B). Polar interactions are found between Glu 96 of CENP-A and three HJURP residues (Arg 32, Lys 39, and Tyr 40), between Glu 107 of CENP-A and Arg 28 of HJURP, and between His 104 and Asp 108 of CENP-A and Ser 25 of HJURP (Fig. 1B). Side chain interactions with CENP-A, involving amino acids from L1 and the β-sheet domain of HJURP, are mostly hydrophobic (Fig. 1B). HJURP also makes direct contacts with α3 of histone H4, primarily via hydrophobic and van der Waals interactions through HJURP residues located in the region encompassing the αA–L1 junction (Fig. 1A). Extensive interactions between HJURP and the CENP-A–H4 heterodimer bury a pairwise surface area of 2459 Å2, which is consistent with their stable association at even 1 M salt concentration during the purification process (see the Materials and Methods).
Figure 1.
Overall structure. (A) A ribbon diagram of the complex structure. CENP-A is shown in green, histone H4 is shown in cyan, and HJURP is shown in yellow. Secondary structure elements in CENP-A, H4, and HJURP are labeled in black, white, and red letters, respectively. The shaded area indicates the CATD of CENP-A. (B) Overview of interactions between HJURP and the CENP-A–H4 heterodimer. The structure is viewed from the same direction as in A, and the heterodimer is shown in a surface representation with electrostatic potential distribution; white, blue, and red regions indicate neutral areas, positively charged areas, and negatively charged areas, respectively. The side chains of HJURP are shown in a stick model (carbon, yellow; nitrogen, blue; oxygen, red; sulfur, gold) superimposed on the ribbon representation of the main chain. An inset enclosed in the red dashed circle on the right indicates an orthogonal view of an interaction region (enclosed in the left circle) involving amino acids in the L1 and β-sheet domain. CENP-A residues involved in the interactions in this region are also shown in a stick model. (Green) Carbon. White and black letters label selected CENP-A and HJURP residues, respectively.
HJURP binds a heterodimer of CENP-A and histone H4
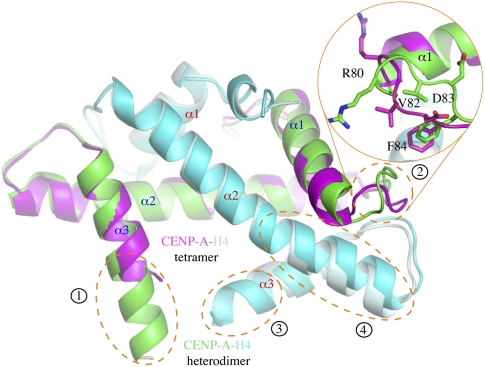
CENP-A and histone H4 normally exist as an equimolar tetramer. The crystal structure of the CENP-A–H4 tetramer shows that two CENP-A–H4 heterodimers dimerize through amino acids located on helix α3 and the C-terminal portion of α2, mainly via hydrophobic interactions (Sekulic et al. 2010). Structural superposition shows that the CENP-A–H4 heterodimer superimposes with the ones from the tetrameric structure with a 0.96 Å root-mean-squared deviation (RMSD) of Cα positions. The main differences between the two are that (1) the complete C terminus of CENP-A is visible in the current structure, while the last six residues were disordered in the tetrameric structure; (2) an N-terminal segment of L1 (amino acids 77–85) has significantly different main chain and side chain conformations (the largest deviation of the Cα position occurs on Asp 83 and is 4.7 Å apart); (3) four additional residues at the C-terminal end of histone H4 became ordered in the current structure; and (4) the C-terminal half of α2 of histone H4 is bent ∼10° toward α1 of H3 (Fig. 2).
Figure 2.
Structural comparison of heterodimeric and tetrameric CENP-A–H4 complexes. The CENP-A–H4 complex from the cocrystal structure with HJURP is colored green (CENP-A) and cyan (histone H4), and the same complex from the CENP-A–H4 tetramer is shown in magenta (CENP-A) and pale cyan (histone H4). The structures are viewed from a direction opposite to that in Figure 1A. For viewing clarity, the HJURP molecule is not shown. Four regions of main differences are enclosed in orange dashed-line circles, and the regions are numbered according to the order in which they were referenced in the text. An inset encircled with a solid orange line represents an enlarged view of region 2, and relevant amino acids are shown in a stick model.
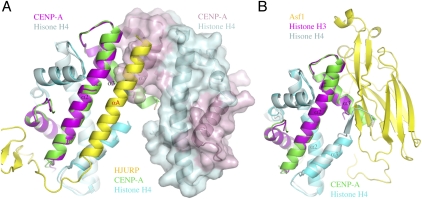
Structural superposition shows that HJURP prevents tetramer formation through two means: (1) HJURP blocks the self-association region of CENP-A located at the C-terminal portion of α2, which is involved in interaction with helix α3 of another CENP-A molecule in the tetramer; and (2) the C-terminal end of CENP-A α3, which became ordered upon HJURP binding, adopts a conformation that will bump into helices α2 and α3 of a neighboring CENP-A (Figs. 2, 3A). We also confirmed that HJURP forms a heterotrimer with CENP-A and H4 in solution by multiangle light scattering (MALS) (see the Supplemental Material). The binding of HJURP to a CENP-A–H4 heterodimer is reminiscent of that between chaperone Asf1 and a histone H3–H4 heterodimer (English et al. 2006; Natsume et al. 2007). Superposition of the two structures shows that the two histone heterodimers overlay well, with a 0.83 Å RMSD of Cα positions, but HJURP and Asf1 have a different structure and interact with their cognate histone heterodimers differently (Fig. 3B).
Figure 3.
HJURP binding prevents the formation of a CENP-A–histone H4 tetramer. (A) The structure of the HJURP–CENP-A–H4 complex is superimposed with the structure of the CENP-A–H4 tetramer (PDB ID: 3NQJ). (Left) The heterodimer from the tetramer structure superimposed with the HJURP complex is shown in a ribbon representation (CENP-A, magenta; H4, pale cyan), and the ribbon representation of the other heterodimer (CENP-A, light pink; H4, pale cyan) is overlaid with a surface representation. The HJURP complex is colored the same as in Figure 1A. (B) Histone chaperone Asf1 employs a different mechanism to block histone H3–H4 tetramer formation. The structure of the Asf1 complex (PDB ID: 2HUE) is shown in a ribbon diagram. (Yellow) Asf1; (magenta) H3; (pale cyan) H4. The H3–H4 dimer is aligned with the CENP-A–H4 heterodimer from the HJURP complex, the latter is positioned the same as in A but with HJURP removed.
HJURP prevents spontaneous association of DNA
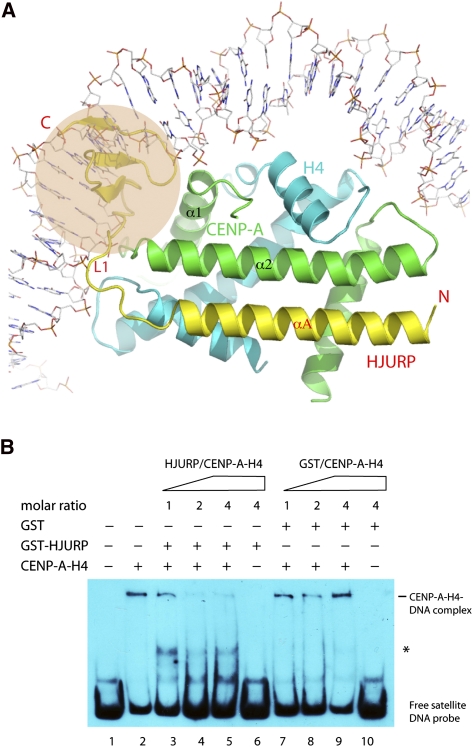
In addition to its role in keeping the CENP-A–H4 complex in a dimeric form, HJURP also appears to compete with DNA for binding to the histone complex. As seen in Figure 1B, the convex side of the CENP-A–H4 heterodimer is enriched with positively charged residues. The positively charged area coincides with the path of nucleosomal DNA when the CENP-A–H4 heterodimer is superimposed with a nucleosomal H3–H4 complex (Fig. 4A; Luger et al. 1997). It is immediately clear that the β-sheet domain and the L1 loop of HJURP block the wrapping of DNA (Fig. 4A). Interestingly, a segment of the L1 loop approximately follows the trajectory of one strand of the DNA backbone for 4 bases, and the main chain carbonyl groups take the place of the DNA backbone phosphates in hydrogen bonding to H3/CENP-A. Furthermore, the outer surface of this region of HJURP is negatively charged, which repels the negatively charged DNA.
Figure 4.
DNA-binding effect of HJURP. (A) Position of the nucleosomal DNA with one of the H3–H4 heterodimers (PDB code: 1KX5) aligned with the HJURP complex. The shaded area indicates the β-sheet and L1 regions that obstruct the potential path of nucleosomal DNA in a CENP-A-containing nucleosome. (B) HJURP prevents spontaneous DNA binding. (Lane 1) Free 208-bp α-satellite DNA probe. (Lane 2) Spontaneous binding of CENP-A–H4 to DNA. (Lanes 3–5) Increasing amount of GST-HJURP reduces binding to DNA. (Lanes 6, 10) GST-HJURP or GST alone do not bind DNA. (Lanes 7–9) GST does not affect DNA binding. An asterisk indicates the position of a protein–DNA complex formed with the addition of HJURP and the CENP-A–H4 complex. Please note that the intensity of this band is stronger at a 1:1 than at higher HJURP-to-CENP-A–H4 molar ratios.
Our in vitro experiments are consistent with the structural observation (Fig. 4B). In the experiment, addition of purified CENP-A–H4 tetramer to 208-base-pair (bp) α-satellite DNA forms slow-migrating bands—which correspond to large, nonspecific protein–DNA complexes (Fig. 4B, lane 2)—while GST-HJURP CBD alone does not show any DNA-binding activities (Fig. 4B, lane 6). Preincubation of increasing amounts of GST-HJURP CBD with the CENP-A–H4 tetramer (at molar ratios of 1:1, 2:1, and 4:1, with a fixed amount of the tetramer) before adding to the DNA resulted in a decreasing amount of the slow-migrating bands (Fig. 4B, lanes 3–5), while no such effect was observed by adding GST (Fig. 4B, lanes 7–9). Interestingly, concomitant with the decrease of the slow-migrating bands, a fast-migrating band (Fig. 4B, indicated with an asterisk on the right) emerges, and the intensity of this band is stronger at a 1:1 GST-HJURP CBD-to-CENP-A–H4 tetramer molar ratio than at higher concentrations of GST-HJURP CBD. The identity of this protein–DNA complex remains unknown, but its property is reminiscent of the previously suggested CENP-A–H4 tetrasomes (Shuaib et al. 2010). Thus, disruption of the nonspecific association of the CENP-A–H4 tetramer with DNA may promote nucleosome assembly. This property of histone chaperones has been reported for NAP1, which promotes nucleosome assembly by the disassembly of nonproductive histone–DNA interactions (Andrews et al. 2010). From a functional perspective, the ability of HJURP to prevent spontaneous binding of DNA to a CENP-A–H4 complex provides a means to ensure assembly of CENP-A-containing nucleosomes only in an appropriate environment.
Specificity for HJURP recognition
A critical question concerning CENP-A is what features distinguish it from histone H3. It was shown previously that the CATD of CENP-A serves as a cis-acting element that confers the CENP-A specificity of centromere targeting and nucleosome assembly (Black et al. 2004). In particular, a chimera protein of histone H3 carrying the CATD of CENP-A (H3CATD) was able to bind HJURP in vitro and in vivo (Foltz et al. 2009; Shuaib et al. 2010). Interestingly, among the 22 amino acids of CATD that are different between CENP-A and histone H3, only two residues in loop-1 (Asn 85 and Ala 88) and two residues in αA (Gln 89 and His 104) of CENP-A make direct interactions with HJURP. The two loop-1 residues are unlikely to be key factors for CENP-A's ability to interact with HJURP, as their histone H3 counterparts (Arg 83 and Ser 86, respectively) can interact with HJURP equally well or better, judging from the number of hydrogen bonds between histone H3 and HJURP residues in the model with a superimposed histone H3–H4 heterodimer. Gln 89 of CENP-A interacts with Phe 44 of HJURP via hydrophobic and van der Waals interactions (Fig. 5A). The corresponding residue in histone H3, Ser 87, is unable to make such interactions. His 104, which corresponds to Gly 102 in histone H3, stacks with the phenyl ring of Phe 29 and makes one hydrogen bond with Ser 25 of HJURP (Fig. 5A). The extra interactions provided by Gln 89 and His 104 make the binding of CENP-A to HJURP better than that of histone H3, but our analysis did not find any histone H3 residues in loop-1 and helix α2 that will prevent histone H3 from binding to HJURP.
Figure 5.
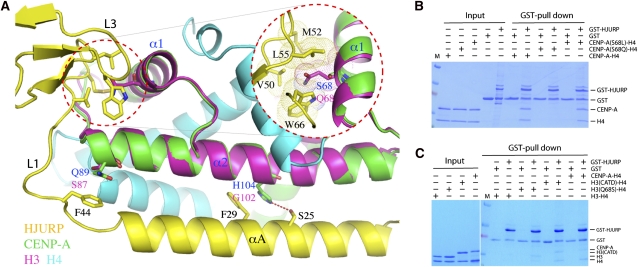
Determinants of CENP-A specificity. (A) His 104 and Gln 89, shown in the CATD of CENP-A (α2 and loop-1), make CENP-A-specific contacts with HJURP. Histone H3 (magenta) of a H3–H4 heterodimer (PBD ID: 1KX5) is superimposed with CENP-A. CENP-A residues making specific contacts, H3 residues in corresponding positions, and HJURP residues involved in the interactions are shown in a stick model. Selected residues of CENP-A, H3, and HJURP are labeled with blue, magenta, and black letters, respectively. An inset encircled with a red dashed line shows the interaction site of Ser 68 of CENP-A located outside of the CATD and surrounding HJURP residues. The atomic radii of side chain atoms of the Ser 68 counterpart in H3, Gln 68, and HJURP residues are shown in a dot model to indicate unfavorable packing of Gln 68. (B) GST pull-down experiments showing the effect of HJURP binding to CENP-A with Ser 68 mutations. (C) GST pull-downs with the H3–H4, H3(Q68S)–H4, H3CATD–H4, and CENP-A–H4 complexes. To lower the background level of H3–H4 binding, 0.05% NP-40 in the washing buffer was used, compared with 0.025% NP-40 in B.
It is clear that the CATD of CENP-A is important for HJURP binding, but it seems insufficient for distinguishing CENP-A from histone H3. We searched for CENP-A residues outside of the CATD region that may be important for differentiating the abilities of CENP-A and histone H3 to bind HJURP. We found that Ser 68 on helix α1 of CENP-A is situated in a shallow hydrophobic pocket formed by Val 50, Met 52, Leu 55, and Trp 66 (Fig. 5A). The corresponding residue in histone H3 is Gln 68, which has a bigger size and is difficult to fit into the hydrophobic pocket without making steric clashes with the HJURP residues. We predicted that a S68Q mutation would result in a weak binding of CENP-A to HJURP, while changing Ser 68 to a leucine, which is hydrophobic and has a smaller side chain than glutamine, should be benign to HJURP binding. Indeed, GST pull-down experiments confirmed our prediction (Fig. 5B). To further test the significance of the newly discovered CENP-A site for HJURP binding, we mutated the corresponding residue in histone H3, Gln 68, to a serine and tested its binding to HJURP. Indeed, GST pull-down experiments show that Q68S gained the ability to bind HJURP, and robust binding of H3CATD to HJURP was also detected in a parallel experiment (Fig. 5C). Thus, we conclude that Ser 68 of CENP-A is an important determinant for CENP-A specificity due to its small size, and the glutamine in histone H3 has an adverse effect on HJURP binding. However, the CATD in the H3CATD chimera protein appears to be able to overcome the unfavorable effect of Gln 68.
Structural implications
It is striking that the Ser 68 site outside of the CENP-A CATD plays a critical role for HJURP recognition, as previous studies have shown that the H3CATD chimera protein recapitulated essential functions of CENP-A, and our in vitro results also confirmed direct binding of H3CATD to HJURP. Surprisingly, analyses of the HJURP-CENP-A–H4 and H3–H4 structures revealed no hindrance of HJURP binding in the histone H3 region corresponding to CATD. One scenario, which we called a “yin-yang” model, is that CATD provides major binding affinities for HJURP, and the Ser 68 site serves as a principal determinant of CENP-A specificity. In this model, the histone H3 region corresponding to the CATD of CENP-A can interact with HJURP, perhaps suboptimally, but Gln 68 pushes away HJURP; the latter force wins and there is no binding. In H3CATD, the artificially introduced CATD can overcome the energy barrier caused by the unfavorable contact of Gln 68, resulting in the binding of HJURP. Less intuitive in this mode is how the CATD overpowers Gln 68 in H3CATD, as the S68Q mutant of CENP-A with a native CATD loses the ability to bind HJURP. It should be pointed out that structural change may play an important role in the resolution of this puzzle, as it is evident from the deuterium exchange experiments that a non-CATD region in helix α1 of H3CATD has a different conformation from the corresponding region in histone H3 or CENP-A (Black et al. 2004). It is also evident from structural comparisons that the α1–L1 region is the most variable region among the structures of histone H3–H4 and CENP-A–H4 complexes. Thus, it is possible that introduction of a CATD into H3 resulted in an environment that remedied the adverse effect of Gln 68. A cocrystal structure of HJURP CBD in complex with the H3CATD–H4 complex should clarify the role of Gln 68 in H3CATD, and, together with in vivo experiments, will provide further tests of the “yin-yang” model of centromere targeting proposed here.
The discovery that HJURP binds a heterodimeric form of the CENP-A–H4 complex also has profound implications in understanding the molecular mechanism of assembly of CENP-A-containing nucleosomes. The precise model of CENP-A-containing nucleosomes is still a matter of debate (Henikoff and Furuyama 2010; Black and Cleveland 2011). A closely relevant point here is whether a centromeric nucleosome contains a CENP-A–H4 heterodimer or heterotetramer (Dimitriadis et al. 2010; Sekulic et al. 2010). The structural result cannot distinguish whether a CENP-A nucleosome is octameric or a hemisome. However, it does point out that, if CENP-A nucleosomes were octameric, additional processes or regulations would be required to ensure that the CENP-A-containing nucleosomes are predominantly homotypic, as heterotypic nucleosomes with both CENP-A and histone H3 are a minority species (Foltz et al. 2006).
In summary, our structural and biochemical analyses of the HJURP CBD–CENP-A–histone H4 complex have provided novel insights into the specificity of CENP-A recognition by HJURP, and advanced our understanding of the histone chaperone activities of HJURP in preventing the formation of a CEN-A–histone H4 tetramer and modulating the DNA-binding activity of the CENP-A–histone H4 complex. The results of this study should facilitate in-depth analyses of the molecular mechanism of centromeric chromatin assembly.
Materials and methods
Protein expression, purification, and crystallization
The CBD encompassing the first 80 amino acids of human HJURP was expressed as a polyhistidine- and sumo-tagged fusion protein from a pET28a-smt3-HJURP (1- to 80-amino-acid) plasmid. Human CENP-A and histone H4 were coexpressed using a pCDFDUET (CENPA + H4) plasmid. To purify the complex HJURP–CBD with CENP-A and histone H4, Escherichia coli cells overexpressing HJURP–CBD and CENP-A–H4 were mixed and lysed together in a buffer containing 20 mM Tris-HCl (pH 8.0), 1 M NaCl, and 1 mM PMSF. The trimeric complex was first purified using a HisTrap FF column (GE Healthcare), followed by cleavage of the polyhistidine-sumo tag with the sumo protease for 2 h at room temperature. The his-tagged sumo was removed by passing the protein sample through the HisTrap FF column one more time. The protein complex was further purified on a Superdex200 sizing column (GE Healthcare). The eluted fractions were analyzed by SDS-PAGE, and highly purified fractions were pooled and concentrated to ∼8 mg/mL for crystallization. The HJURP–CENP-A–H4 complex was crystallized by hanging-drop vapor diffusion at 16°C, and the crystals used for data collection grew in a condition with 2 M ammonium sulfate.
X-ray data collection and structure determination
Diffraction data were collected at beamline BL17U of Shanghai Synchrotron Radiation Facility (SSRF) using a Quantum 315r detector (ADSC). A cryoprotectant with a mixture of crystallization well solution and 15% glycerol was used for data collection at liquid nitrogen temperature. A 2.6 Å native data set was collected at a wavelength of 0.9788 Å. Data were processed with HKL2000 software (Otwinowski and Minor 1997). The crystal belongs to the P312 space group, and there is one HJURP–CENP-A–H4 trimer per asymmetric unit. The structure was solved by molecular replacement using Molrep of the CCP4 program suite (Collaborative Computational Project, Number 4 1994), with the CENP-A–H4 heterodimer structure (Protein Data Bank [PDB] ID: 3NQJ) as the search model (Sekulic et al. 2010). The resultant electron density map was of high quality (Supplemental Fig. S1), allowing unambiguous building of the HJURP model. Model building and refinement were carried out using Coot (Emsley and Cowtan 2004) and PHENIX (Adams et al. 2010), and TLS refinement was applied to improve the electron density map. The refined model contained residues 14–74 of HJURP, residues 59–140 of CENP-A, and residues 23–96 of histone H4. Structure figures were prepared using PyMol (http://www.pymol.org), and the coordinates were deposited in PDB under accession code 3R45.
Molecular weight determination by MALS
The molecular masses in solution were determined by MALS using the DAWN HELEOSTM II 18-angle static light-scattering system (Wyatt Technology), connected to an Agilent HPLC, hooked up with a WTC SEC column (Wyatt Technology). The system was first equilibrated using a buffer containing 20 mM Tris-Cl (pH 8.0), 1 M NaCl, and 1 mM DTT for 12 h. The equilibrated system was then calibrated with BSA at a concentration of 2 mg/mL. The HJURP–CENP-A–H4 complex was prepared as described before, and the CENP-A–H4 tetramer was reconstituted and purified according to a published protocol (Tanaka et al. 2004). Protein samples at three different concentrations (HJURP–CENP-A–H4: 1 mg/mL, 2 mg/mL, and 4 mg/mL; CENP-A–H4: 1 mg/mL, 2 mg/mL, and 5 mg/mL) were subjected to MALS analyses at a flow rate of 0.5 mL/min at 16°C. The molecular mass was calculated using ASTRA5.3.4.14 software (Wyatt Technology).
GST pull-down experiments
CENP-A mutations were generated in the pCDFDUET (CENPA + H4) plasmid by PCR. Wild-type and mutant CENP-A complexes were purified on a HisTrap FF column (GE Healthcare). GST-tagged HJURP CBD was produced in E. coli using a pGEX-6P-1 vector. Histone H3–H4 complexes were reconstituted using bacterially expressed human histone H3, H3(Q68S), or H3CATD, and H4 using an established protocol (Luger et al. 1999). In vitro binding assays were performed as described previously (Foltz et al. 2009). Briefly, a portion of GST or GST-HJURP CBD-bound glutathione-sepharose beads (∼5.0 μg of protein each) were incubated with recombinant wild-type or mutant CENP-A–H4 complexes in the binding buffer (100 mM NaH2PO4 at pH 7.4, 300 mM NaCl, 0.025% NP-40, 10% glycerol, 1 mM DTT) for 30 min at room temperature. GST pull-down experiments of histone H3 complexes were carried out similarly, with the exception that 0.05% of NP-40 was used. Protein-bound glutathione resins were washed six times with the binding buffer, and the bound samples were analyzed by Coomassie blue-stained SDS-PAGE on a 6%–25% polyacrylamide gradient gel.
DNA binding
A biotin-labeled 208-bp α-satellite DNA fragment (0.2 μg) was incubated with 0.5 μg of reconstituted wild-type CENP-A–H4 complex in the absence or presence of increasing amounts of GST-HJURP CBD or GST for 1 h at room temperature in the binding buffer containing 10 mM HEPES-KOH (pH 7.6), 0.1 mM EDTA, and 100 mM NaCl. DNA binding was analyzed by 5% native PAGE in 0.5× TBE, transferred, cross-linked onto a Hybond-Nx nylon membrane, and detected with a streptavidin-alkaline phosphatase conjugate and CDP-Star chemiluminescent reagent (Roche Applied Science) as described in Li et al. (2003).
Acknowledgments
We thank SSRF beamline scientists for technical support during data collection. The work was supported by grants from the National Basic Research Program of China (2009CB825501 and 2010CB944903 to R.M.X., 2011CB966300 to G.L., 2011CB966302 to Y.S., and 2010CB912103 to Y.X.), the Natural Science Foundation of China (90919029 and 3098801 to R.M.X., and 91019007 to G.L.), Chinese Academy of Sciences (CAS), and the Novo Nordisk-CAS foundation.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.2045111.
Supplemental material is available for this article.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Karpen GH 2008. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 9: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K 2010. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell 37: 834–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Cleveland DW 2011. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell 144: 471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL Jr, Cleveland DW 2004. Structural determinants for generating centromeric chromatin. Nature 430: 578–582 [DOI] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL 2007. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell 26: 853–865 [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y 2010. Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci 107: 20317–20322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137: 485–497 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK 2006. Structural basis for the histone chaperone activity of Asf1. Cell 127: 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR III, Cleveland DW 2006. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 8: 458–469 [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR III, Bassett EA, Wood S, Black BE, Cleveland DW 2009. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell 137: 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Rice JC 2004. Regulation of heterochromatin by histone methylation and small RNAs. Curr Opin Cell Biol 16: 230–238 [DOI] [PubMed] [Google Scholar]
- Henikoff S, Dalal Y 2005. Centromeric chromatin: what makes it unique? Curr Opin Genet Dev 15: 177–184 [DOI] [PubMed] [Google Scholar]
- Henikoff S, Furuyama T 2010. Epigenetic inheritance of centromeres. Cold Spring Harb Symp Quant Biol. doi: 10.1101/sqb.2010.75.001 [DOI] [PubMed] [Google Scholar]
- Li G, Tolstonog GV, Sabasch M, Traub P 2003. Type III intermediate filament proteins interact with four-way junction DNA and facilitate its cleavage by the junction-resolving enzyme T7 endonuclease I. DNA Cell Biol 22: 261–291 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ 1999. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol 304: 3–19 [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C 2007. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell 129: 1153–1164 [DOI] [PubMed] [Google Scholar]
- Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T 2007. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature 446: 338–341 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC 2009. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell 137: 1173–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N, Bassett EA, Rogers DJ, Black BE 2010. The structure of (CENP-A–H4)(2) reveals physical features that mark centromeres. Nature 467: 347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A 2010. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci 107: 1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE 2007. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci 104: 10571–10576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Tawaramoto-Sasanuma M, Kawaguchi S, Ohta T, Yoda K, Kurumizaka H, Yokoyama S 2004. Expression and purification of recombinant human histones. Methods 33: 3–11 [DOI] [PubMed] [Google Scholar]