Abstract
Removal of the assembly factor eukaryotic initiation factor 6 (eIF6) is critical for late cytoplasmic maturation of 60S ribosomal subunits. In mammalian cells, the current model posits that eIF6 release is triggered following phosphorylation of Ser 235 by activated protein kinase C. In contrast, genetic studies in yeast indicate a requirement for the ortholog of the SBDS (Shwachman-Bodian-Diamond syndrome) gene that is mutated in the inherited leukemia predisposition disorder Shwachman-Diamond syndrome (SDS). Here, by isolating late cytoplasmic 60S ribosomal subunits from Sbds-deleted mice, we show that SBDS and the GTPase elongation factor-like 1 (EFL1) directly catalyze eIF6 removal in mammalian cells by a mechanism that requires GTP binding and hydrolysis by EFL1 but not phosphorylation of eIF6 Ser 235. Functional analysis of disease-associated missense variants reveals that the essential role of SBDS is to tightly couple GTP hydrolysis by EFL1 on the ribosome to eIF6 release. Furthermore, complementary NMR spectroscopic studies suggest unanticipated mechanistic parallels between this late step in 60S maturation and aspects of bacterial ribosome disassembly. Our findings establish a direct role for SBDS and EFL1 in catalyzing the translational activation of ribosomes in all eukaryotes, and define SDS as a ribosomopathy caused by uncoupling GTP hydrolysis from eIF6 release.
Keywords: bone marrow failure syndromes, ribosome assembly, eIF6, human genetics, leukemia, ribosomopathy, NMR
Shwachman-Diamond syndrome (SDS; OMIM 260400) is an autosomal recessive disorder characterized by bone marrow dysfunction with a striking cumulative risk of progression to myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) (Donadieu et al. 2005) that is caused by mutations in the essential, highly conserved SBDS (Shwachman-Bodian-Diamond syndrome) gene (Boocock et al. 2003), whose specific function remains unknown. Genetic studies in Saccharomyces cerevisiae indicate that the SBDS ortholog Sdo1 and the GTPase elongation factor-like 1 (Efl1), a structural homolog of elongation factor G (EF-G), function in a pathway to release and recycle the essential nucleolar factor Tif6 (mammalian eukaryotic initiation factor 6 [eIF6]) from late cytoplasmic pre-60S ribosomal subunits (Menne et al. 2007). Tif6 is critical for the biogenesis and nuclear export of pre-60S subunits (Basu et al. 2001), and acts as a ribosomal anti-association factor by physically blocking intersubunit bridge formation (Gartmann et al. 2010). Therefore, dissociation of Tif6 from nascent 60S ribosomes is essential to allow the assembly of actively translating 80S subunits (Ceci et al. 2003).
Multiple gain-of-function TIF6 alleles suppress the pre-60S nuclear export defect and cytoplasmic mislocalization of Tif6 observed in cells deleted for SDO1 (sdo1Δ) (Menne et al. 2007) or EFL1 (efl1Δ) (Becam et al. 2001; Senger et al. 2001) by reducing the affinity of Tif6 for the ribosome, suggesting that the release of Tif6 may be directly catalyzed by the concerted action of Sdo1 and Efl1. However, direct biochemical evidence supporting this notion is lacking. Indeed, Efl1 alone was reported to dissociate 60S–Tif6 complexes in vitro in the presence of GTP (Senger et al. 2001). Although cosedimentation of human SBDS with free cytoplasmic 60S ribosomal subunits in sucrose gradients (Ganapathi et al. 2007) would be consistent with a conserved role for SBDS in 60S subunit maturation, the current model in mammalian cells posits that eIF6 removal is triggered following phosphorylation of Ser 235 by protein kinase C (PKC) and RACK1 (receptor for activated protein C) (Ceci et al. 2003). Furthermore, diverse alternate functions for SBDS in mammalian cells have been suggested, including mitotic spindle stabilization (Austin et al. 2008), chemotaxis (Wessels et al. 2006), Fas ligand-induced apoptosis (Rujkijyanont et al. 2008), cellular stress responses (Ball et al. 2009), and Rac2-mediated monocyte migration (Leung et al. 2010). Thus, despite the prior genetic studies in yeast, the mechanism of eIF6 release in mammalian cells is controversial, biochemical evidence supporting direct catalysis of eIF6 release by SBDS and EFL1 in eukaryotic cells is currently lacking, and the specific function of the SBDS protein, its mode of action, and the molecular mechanism of the cooperative interaction with EFL1 remain obscure.
To resolve these issues, we solved a high-resolution NMR structure of the human SBDS protein and biochemically reconstituted eIF6 release for the first time ex vivo using genetically stalled pre-60S subunits isolated from Sbds-deleted mice. We demonstrate that human SBDS and EFL1 cooperate to directly catalyze eIF6 removal from mammalian pre-60S subunits by a mechanism that requires GTP binding and hydrolysis by EFL1 but not eIF6 phosphorylation on Ser 235. We reveal that the essential role of the SBDS protein is to tightly couple the activation of GTP hydrolysis on the ribosome and eIF6 release, and identify a conserved residue (Lys151) mutated in SDS that is required for the cooperative interaction with EFL1. Furthermore, complementary NMR studies suggest unanticipated mechanistic parallels between this late step in 60S ribosome biogenesis and aspects of bacterial ribosome disassembly. By elucidating the conserved mechanism of eIF6 release in eukaryotes, this study provides compelling evidence that SDS is a ribosomopathy caused by uncoupling GTP hydrolysis by EFL1 from eIF6 release on the ribosome. Our data support the evaluation of strategies that promote eIF6 removal for SDS therapy.
Results
Histopathological abnormalities in Sbds-deleted liver
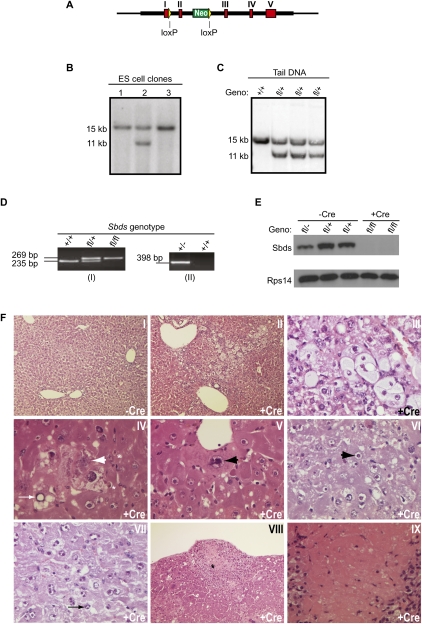
To test the hypothesis that SBDS and EFL1 cooperate to directly trigger the release of eIF6 from pre-60S ribosomal subunits, we generated a floxed Sbds allele (Sbdsfl) by gene targeting in mouse embryonic stem cells (Fig. 1A,B; Supplemental Fig. S1), designing the targeting construct such that Cre-mediated recombination of the loxP elements would remove Sbds exon 2. The resulting chimeras were crossed with wild-type C57Bl/6 mice to produce floxed heterozygous Sbdsfl/+ animals (Fig. 1C). Sbdsfl/+ mice were crossed with a germline deleter transgenic strain (Schwenk et al. 1995) to generate mice that were heterozygous for a deleted Sbds allele (Sbds-/+). Excision of exon 2 was confirmed by PCR of tail DNA (Fig. 1D).
Figure 1.
Histopathological abnormalities in Sbds-deleted mouse livers. (A) Schematic of the targeted Sbds allele on mouse chromosome 5. Exons I–V are shown in red, loxP sites are shown in yellow, and the neomycin cassette is shown in green. (B) Representative filter hybridization analysis of neomycin-resistant embryonic stem cell clones. Genomic DNA was digested with BamH1 and hybridized to the 5′ probe (Supplemental Fig. S1). The 15-kb- and 11-kb bands represent wild-type Sbds+ (lanes 1,3) and targeted Sbdsfl (lane 2) alleles, respectively. (C) Representative filter hybridization for genotyping (Geno) of genomic tail DNA from mice carrying wild-type (+) and targeted (fl) Sbds alleles. DNA was digested with BamH1 and hybridized with the 5′ probe. (D) PCR genotyping of tail DNA to detect wild-type (+) and floxed (fl) (panel I) or exon 2 deleted (−) (panel II) Sbds alleles. The sizes of the PCR products are indicated. (E) Sbds protein is absent in Sbds-deleted mouse livers. The Sbds genotype and the absence (−Cre) or presence (+Cre) of the pMx1-cre transgene are indicated. Liver extracts were immunoblotted to visualize Sbds and Rps14. (F) Histopathological abnormalities in Sbds-deleted mouse livers. (Panels I–IX) H&E sections of livers from representative Sbdsfl/- mice after 4 wk of treatment with poly(I:C) in the absence (−Cre) or presence (+Cre) of the pMx1-cre transgene demonstrating normal zone 2 (50×) (panel I); disordered architecture in zone 2 between the portal triads and central veins (50×) (panel II); hydropic cytoplasmic swelling of hepatocytes in zone 2 (400×) (panel III); hepatocytes showing apparent cavitation of the nucleus (thin arrow), degenerative nuclear change (thick arrow), and apoptotic cells (asterisks) (400×) (panel IV); a swollen hepatocyte containing an enlarged abnormal nucleus (arrow) (400×) (panel V); an enlarged, ring-shaped nucleolus with an eosinophilic center (arrow) (400×) (panel VI); multiple enlarged eosinophilic nucleoli (arrow) (400×) (panel VII); the area of necrosis adjacent to the liver capsule with surrounding neutrophil infiltrate (asterisk) (100×) (panel VIII); and the necrotic area showing karyolysis of hepatocytes (400×) (panel IX). See also Supplemental Figure S1.
As homozygous exon 2 deletion was embryonic-lethal (data not shown), Sbdsfl/flTg:Mx1-cre mice were bred with Sbdsfl/-Tg:Mx1-cre mice to conditionally delete the Sbds gene. Following induction of Cre recombinase by administration of poly(I:C) for 2 wk, PCR for Sbds transcripts (data not shown) and immunoblotting for the Sbds protein indicated Sbds deletion of 100% in the livers of Sbdsfl/flTg:Mx1-cre and Sbdsfl/-Tg:Mx1-cre mice (Fig. 1E). No recombination was observed in control mice lacking the Tg:Mx1-cre transgene or in mice injected with vehicle alone (data not shown).
Prominent histological abnormalities were observed in the livers of Sbds-deleted mice (Fig. 1F, panels I–IX). Compared with undeleted controls (Fig. 1F, panel I), Sbds-deleted livers showed disordered architecture in zone 2 between the portal triads and central veins (Fig. 1F, panel II), with a range of degenerative hepatocyte appearances, including hydropic cytoplasmic swelling (Fig. 1F, panel III), nuclear cavitation, degenerative nuclear change, necrosis, and apoptosis (Fig. 1F, panels IV,V). Many zone 2 hepatocytes showed striking nucleolar abnormalities, including enlarged, ring-shaped nucleoli with eosinophilic centers (Fig. 1F, panel VI) and multiple enlarged eosinophilic nucleoli of varying sizes and shapes (Fig. 1F, panel VII). There were also scattered subcapsular areas of hepatocyte necrosis with an associated acute inflammatory reaction (Fig. 1F, panels VIII,IX).
Sbds deletion causes a ribosomal subunit-joining defect
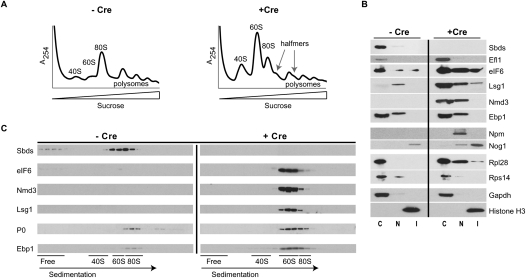
We postulated that the striking histological abnormalities in Sbds-deleted livers might be a consequence of a primary defect in 60S ribosomal subunit maturation. To test this hypothesis, we examined the abundance of cytoplasmic ribosomal subunits, monosomes, and polysomes in liver cell extracts by performing sucrose gradient centrifugation (Fig. 2A). Compared with undeleted controls, Sbds-deleted extracts showed accumulation of free cytoplasmic 40S and 60S subunits and halfmer ribosomes (representing 43S initiation complexes that are stalled at the AUG start codon awaiting binding of 60S subunits), with no significant difference in the ratio of 60S to 40S subunits (Supplemental Fig. S2). These data indicate that Sbds deletion causes a ribosomal subunit-joining defect in mammalian cells in vivo.
Figure 2.
60S subunit maturation defect in Sbds-deleted mouse livers. (A) Polysome profiles of liver cell extracts from undeleted (−Cre) or Sbds-deleted (+Cre) mice. Halfmer ribosomes are indicated with arrows. (B) Cytoplasmic accumulation of late pre-60S assembly factors in Sbds-deleted cells. Subcellular fractions from undeleted (−Cre) or Sbds-deleted (+Cre) liver cells were immunoblotted to visualize the indicated factors. Rps14 and Rpl28 reveal the 40S and 60S ribosomal subunits, respectively. Gapdh is a cytoplasmic marker, Npm is a nuclear marker, and histone H3 is a chromatin marker. (C) Cytoplasmic fraction; (N) soluble nuclear fraction; (I) insoluble nuclear fraction containing nucleoli and chromatin. (C) Cosedimentation of eIF6, Nmd3, and Lsg1 with pre-60S subunits in Sbds-deleted cells. Extracts from undeleted (−Cre) or Sbds-deleted (+Cre) liver cells as above were fractionated by sucrose gradient sedimentation and immunoblotted to visualize the indicated factors. P0 reveals the 60S ribosomal stalk, and the sedimentation positions of the 40S, 60S, and 80S ribosomal particles are indicated. See also Supplemental Figure S2.
Nucleolar eIF6 is not limiting for 60S subunit biogenesis in Sbds-deleted cells
The absence of a ribosomal subunit imbalance following Sbds deletion in mouse livers contrasts with the underaccumulation of 60S ribosomal subunits in sdo1Δ yeast cells where 60S subunit deficiency is secondary to the delayed preribosomal RNA (rRNA) processing and defective nuclear export of pre-60S subunits caused by impaired nucleolar recycling of Tif6 (Menne et al. 2007). We hypothesized that nuclear export of pre-60S subunits might be maintained in Sbds-deleted liver because eIF6 is not limiting in the nucleolus. To test this, liver cells were fractionated into cytoplasmic, soluble, and insoluble nuclear extracts and immunoblotted to detect eIF6. In Sbds-deleted liver cells, eIF6 accumulates in the cytoplasmic, nuclear, and nucleolar fractions (Fig. 2B). Thus, in contrast with sdo1Δ yeast cells where Tif6 is markedly redistributed from the nucleolus to the cytoplasm, the level of eIF6 is not limiting in the nucleolus of Sbds-deleted cells, thereby maintaining 60S subunit biogenesis and export.
Accumulation of stalled late cytoplasmic pre-60S ribosomes in Sbds-deleted cells
We hypothesized that the subunit-joining defect in Sbds-deleted cells might be a consequence of failure to release one or more late trans-acting ribosome assembly factors from the 60S intersubunit interface, and anticipated that such factors would accumulate in the cytoplasm in the absence of Sbds. Indeed, compared with undeleted controls, we observed significant cytoplasmic accumulation of the late pre-60S assembly factors eIF6, Nmd3, and Lsg1 in Sbds-deleted extracts relative to Gapdh (Fig. 2B). Additionally, we observed cytoplasmic accumulation of the GTPase Efl1 (NP_780526) but not the early nucleolar GTPase Nog1 or the nuclear protein nucleophosmin (Npm). Furthermore, Ebp1, the mammalian ortholog of the yeast 60S biogenesis factor Arx1, did not appear to accumulate or show redistribution upon Sbds deletion. The Sbds protein itself was clearly cytoplasmic, with no detectable signal in the nucleolar fraction (marked by Nog1).
In light of the ribosomal subunit-joining defect in the absence of Sbds, we hypothesized that the cytoplasmic accumulation of late pre-60S assembly factors was most likely due to their retention on late pre-60S particles. We therefore tested whether eIF6, Nmd3, and Lsg1 cofractionated with cytoplasmic pre-60S ribosomes by sucrose gradient sedimentation analysis and immunoblotting of liver cell extracts (Fig. 2C). The absorbance profile of the sucrose gradients at 254 nm revealed the distribution of 40S and 60S subunits, 80S monosomes, and polysomes, while antiserum to Rpl28 revealed the 60S subunits (also highlighted by antiserum to the stalk protein P0 and anti-Ebp1) and Rps14 antiserum detected the 40S subunits (data not shown). The assembly factors eIF6, Nmd3, and Lsg1 cosedimented with cytoplasmic pre-60S ribosomal subunits and did not accumulate significantly at the top of the sucrose gradient on sedimentation analysis of Sbds-deleted liver cell extracts. The Sbds protein cofractionated with pre-60S subunits in undeleted extracts, with a minor portion in the free fraction at the top of the gradient, but was undetectable in Sbds-deleted extracts, as expected. These data indicate that deletion of Sbds causes a ribosomal subunit-joining defect associated with the cytoplasmic accumulation of pre-60S particles carrying the ribosome assembly factors eIF6, Nmd3, and Lsg1.
As both Tif6 and Nmd3 bind to the intersubunit interface of the 60S subunit in yeast (Gartmann et al. 2010; Sengupta et al. 2010), we considered the most likely cause of the subunit-joining defect in Sbds-deleted cells to be due to a failure to release eIF6 and Nmd3 from the intersubunit interface of late pre-60S subunits.
Genome-wide arrays reveal extensive overlap in the genetic interactions between SDO1 and EFL1
We postulated that eIF6 rather than Nmd3 might be the direct target of SBDS and EFL1. To provide further support for this hypothesis, we turned to S. cerevisiae, as we had demonstrated previously that the fitness defect of yeast cells deleted for SDO1 (sdo1Δ), EFL1 (efl1Δ), or both (sdo1Δ efl1Δ) is rescued by a plasmid carrying the dominant TIF6-V192F suppressor allele (Menne et al. 2007). Strikingly, 11 spontaneous suppressor yeast strains that had bypassed the fitness defect of the sdo1Δ efl1Δ double-deletion mutant harbored point mutations in the TIF6 gene (N9Y, N9D, V15I, V15D, R61G, Q75R, L101I, L101V, A103S, A103T, and A103V).
We next screened for synthetic sick/lethal interactions in a genome-wide array of two-factor crosses using sdo1Δ and efl1Δ query strains (harboring a dominant gain-of-function TIF6 suppressor allele) and the set of ∼5000 viable deletion alleles. This analysis revealed an overlapping set of 36 identical genetic interactions shared between SDO1 and EFL1 (accounting for ∼50% of the SDO1 genetic interactions identified, P ∼ 0) (Supplemental Fig. S3; Supplemental Table S1). Taken together, these genetic data strongly support the hypothesis that Sdo1 and Efl1 function in a common pathway upstream of Tif6, raising the possibility that Tif6 is the direct target of Sdo1 and Efl1 in yeast, and that, by implication, eIF6 is the direct target of SBDS and EFL1 in mammalian cells.
Human SBDS and EFL1 are necessary and sufficient to catalyze eIF6 release
We set out to obtain direct evidence in mammalian cells that the release of eIF6 from pre-60S ribosomal subunits is directly catalyzed by the cooperative interaction between human SBDS and EFL1. We biochemically reconstituted an ex vivo eIF6 release assay by adding recombinant human SBDS and EFL1 to eIF6-loaded pre-60S subunits isolated from Sbds-deleted mouse liver. A schematic overview of the eIF6 release assay is shown in Figure 3A. Endogenous murine Sbds and Efl1 were not detected on the isolated pre-60S subunits by immunoblotting (data not shown). As shown in Figure 3B, dissociation of eIF6 was triggered by human SBDS and EFL1 and the nucleotide triphosphate GTP, but not the addition of SBDS, EFL1, or GTP alone. In the absence of added nucleotide triphosphate or the presence of GDP, SBDS and EFL1 failed to trigger eIF6 removal, and there was only marginal release in the presence of the nonhydrolysable GTP analog GDPNP (Fig. 3C). ATP was inactive in the assay (data not shown). Furthermore, two catalytically inactive EFL1 mutants (T33A and H96A) were defective for eIF6 dissociation (data not shown). Consistent with the hypothesis that eIF6 removal is specifically triggered by SBDS/EFL1, there was no detectable dissociation of Ebp1 or Nmd3 (Fig. 3B). Taken together, these data indicate that SBDS and EFL1 act in concert to directly catalyze eIF6 release from murine pre-60S particles by a mechanism that requires both GTP binding and GTP hydrolysis by EFL1. Furthermore, as EFL1 is highly conserved, we took advantage of a genetic complementation assay in yeast to determine whether GTP hydrolysis is critical for its function in vivo. Indeed, wild-type but not mutant EFL1 alleles carrying mutations in key conserved catalytic residues complemented the growth defect of efl1Δ yeast cells in vivo (Supplemental Fig. S4), indicating that GTP binding and hydrolysis are critical for EFL1 function both in vitro and in vivo.
Figure 3.
SBDS and EFL1 cooperate to directly catalyze eIF6 release. (A) Schematic of eIF6 release assay. Pre-60S subunits were isolated from Sbds-deleted liver cell extracts by sucrose gradient sedimentation, incubated with recombinant release factors, and pelleted through a 30% (w/v) sucrose cushion. Immunoblotting revealed the distribution of eIF6 in the supernatant (“free”) and pellet (“bound”). (B) SBDS and EFL1 are jointly required for GTP-dependent release of eIF6 from purified pre-60S subunits. Indicated combinations of recombinant human SBDS and EFL1 were incubated with pre-60S subunits in the presence of GTP. eIF6, Nmd3, and Ebp1 were visualized by immunoblotting. A minus sign (−) indicates no added factor. (C) Nucleotide dependence of eIF6 release by SBDS and EFL1. Recombinant human SBDS and EFL1 were incubated with pre-60S subunits in the presence of the indicated guanine nucleotides. eIF6 and Ebp1 were visualized by immunoblotting. (D) Ser 235 is not required for GTP-dependent eIF6 release by SBDS and EFL1 in vitro. (Left panel) Purified RRL 60S subunits (preloaded with eIF61-225) were incubated with the indicated combinations of recombinant human SBDS and EFL1 and GTP. (Right panel) RRL 60S subunits were incubated with SBDS and EFL1 and the indicated nucleotides. EDTA was used as a positive control for eIF6 release. eIF6 and Rpl28 were visualized by immunoblotting. See also Supplemental Figures S3 and S4 and Supplemental Tables S1 and S2.
Catalysis of eIF6 removal is independent of Ser 235 phosphorylation
A distinct mechanism of eIF6 release by RACK1 and PKC has been proposed in mammalian cells involving eIF6 phosphorylation on S235 (Ceci et al. 2003). To assess whether S235 phosphorylation is required for eIF6 removal in vitro, we prepared salt-washed 60S ribosomal subunits from rabbit reticulocyte lysate (RRL) and assayed the ability of SBDS and EFL1 to release prebound recombinant eIF6 protein truncated after residue N225 (eIF61-225) and therefore excluding S235. Indeed, GTP-dependent release of eIF61-225, but not ribosomal protein Rpl28, was promoted when eIF61-225-bound 60S subunits were incubated with both SBDS and EFL1 (Fig. 3D). As a positive control, EDTA promoted factor-independent eIF6 release. Thus, eIF6 that lacks residue S235 can be specifically released from mature 60S subunits by SBDS and EFL1 in vitro in the absence of exogenous 40S subunits, RACK1, or PKC. We therefore conclude that SBDS and EFL1 are necessary and sufficient for GTP-dependent release of eIF6 from 60S subunits in vitro.
To identify specific eIF6 phosphopeptides in mammalian cells, we subjected eIF6-enriched pre-60S particles isolated from Sbds-deleted mice to analysis by mass spectrometry. Although we identified eIF6 phosphopeptides containing phosphates at residues S174, S175, T165, and S166 and unambiguously identified multiple tryptic peptides containing S235, we found no evidence for in vivo phosphorylation of this residue (Supplemental Table S2). Thus, phosphorylation of eIF6 on S235 is neither present in vivo nor required for eIF6 release ex vivo (Fig. 3B,C) or in vitro (Fig. 3D). These data do not support a requirement for S235 phosphorylation for eIF6 release in mammalian cells.
High-resolution solution structure of human SBDS and the impact of disease mutations
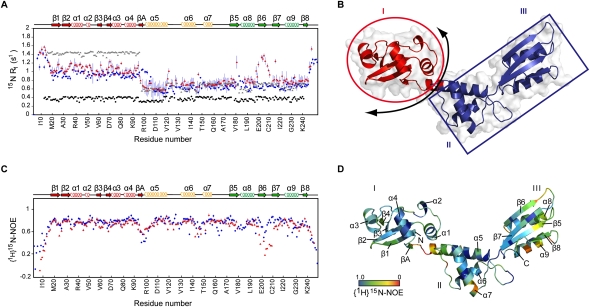
We next set out to determine the specific role of the SBDS protein in the mechanism of eIF6 release. As a prerequisite for this, we solved a high-resolution solution NMR structure of the human SBDS protein (Fig. 4A–E; Supplemental Table S3). The secondary structure elements and overall fold of the human and archaeal SBDS orthologs (AF0491) (Shammas et al. 2005) are identical, despite only 22% amino acid sequence identity and 42% amino acid similarity (Supplemental Fig. S3A–D), strongly supporting an evolutionarily conserved function. The human SBDS protein has a three-domain architecture: domain I, also called the FYSH domain (residues S2–S96); domain II (residues D97–A170); and domain III (residues H171–E250). The N (residues S2–V15) and C (residues N238–E250) termini are unstructured. The N-terminal domain consists of a twisted five-stranded anti-parallel β sheet with four α helices positioned on one face of the sheet. The central domain consists of a three-helical, right-handed twisted bundle, while the C-terminal domain has a typical ferredoxin-like fold. Importantly, helix α5 (residues D97–D117) is well defined in our structure, in contrast with a recent study in which 11 of these residues (D97–M107) were unassigned and therefore were assumed to be unstructured (de Oliveira et al. 2010).
Figure 4.
Solution structure of human SBDS and impact of SDS-associated mutations. (A) Ribbon representation of the lowest-energy human SBDS NMR structure surrounded by the solvent-accessible surface (radius probe 1.4 Å), prepared using the program PyMOL (http://www.pymol.org). Domain I is colored red, domain II is colored yellow, domain III is colored green, and loops are colored gray. The indicated SDS-associated mutations modify surface epitopes (blue spheres) or protein stability (pink spheres). (B–D) Overlay of the backbone atoms of the 20 lowest-energy structures from domain I (A16–T89) (A), domain II (D97–K164) (B), and domain III (H171–L237) (C). The 20 conformers were overlaid using Clusterpose (Diamond 1995). (E) Representation of the electrostatic surface potential of the human SBDS protein, calculated by the program APBS (Baker et al. 2001) and colored using a linear color ramp from −15 kT (red) to +15 kT (blue). The SDS-associated mutation K151N is indicated. (F) Overlays of the 1H,15N HSQC spectra for wild-type (blue) and three SDS-associated mutants (C84R, R126T, and K151N) (red). Arrows indicate peaks visible in the wild-type spectrum but not in the C84R mutant. See also Supplemental Figures S5 and S6 and Supplemental Tables S3–S5.
We directly assessed the impact of SDS-associated mutations using 15N,1H heteronuclear single-quantum correlation (HSQC) spectroscopy. Chemical shift perturbation caused by a mutation can be interpreted in terms of local or global changes in protein conformation, as 15N,1H HSQC spectra provide a fingerprint of the local environment of all amide resonances. The 29 amino acids targeted by missense mutations in SDS patients (Supplemental Table S4) predominantly map to domains I and II (Fig. 4A). Overlays of the 15N,1H HSQC spectra for wild-type SBDS and three representative SDS-associated disease mutants (C84R, R126T, and K151N) are shown in Figure 4F, and similar overlays for an additional 25 SBDS mutants are displayed in Supplemental Figure S2, A–D. Based on these data, we subdivided SDS-associated missense mutations into two categories: Class A mutations affect SBDS protein stability or fold, and class B mutations alter surface epitopes without affecting the SBDS protein stability or fold (Supplemental Table S5).
Conserved global domain motion of the SBDS proteins
To provide potential insight into its mechanism of action, we next characterized the dynamic properties of human and yeast SBDS. Two sets of relaxation experiments were recorded to distinguish between the overall mobility of individual domains (longitudinal relaxation rate 15N R1) and the fast time scale (picosecond) internal mobility of backbone amide residues ({1H}15N-heteronuclear NOE). The 15N R1 values show significant variation along the protein sequence (Fig. 5A,B), indicating that the tumbling of the SBDS protein is anisotropic. 15N R1 measurements for domain I yielded values significantly lower than those predicted by hydrodynamic calculations for an independently mobile domain, but were higher than the values anticipated for a completely rigid module. These data indicate that domain I tumbles largely, but not completely, independently of domains II and III (Figure 5B).
Figure 5.
Conserved interdomain motion of the SBDS protein. (A) Quasi-independent mobility of domain I relative to domains II and III. The experimentally determined longitudinal relaxation rate 15N R1 (sec−1) for human (blue circles) and yeast (red triangles) SBDS is plotted against human SBDS residue number. Black circles represent 15N R1 (sec−1) values calculated for a simulated fully rigid SBDS model, and gray triangles represent 15N R1 (sec−1) values calculated for a simulated model in which the motion of domain I is completely independent of domains II and III. (B) Schematic representation of the quasi-independent mobility of domain I (red) relative to domains II and III (blue). Arrow indicates the hinge region at the N terminus of helix α5. (C) {1H}15N-heteronuclear NOE values for human (blue circles) and yeast (red triangles) SBDS proteins plotted against human SBDS residue number. Residues for which no data are shown correspond to prolines or overlapped cross-peaks. (D) {1H}15N-heteronuclear NOE values mapped onto a ribbon representation of the human SBDS structure. Residues are colored from most (red) to least (blue) flexible.
Interestingly, the transition point or hinge between regions of the SBDS molecule with differing global motion is centered within the N terminus of helix α5 (residue T102), which has a high degree of internal flexibility (Fig. 5C,D), suggesting that it may convert rapidly between a fully folded and partially unfolded conformation. Strikingly, comparison of the backbone relaxation measurements between humans and yeast reveals that the dynamic properties of the SBDS proteins are highly conserved (Fig. 5A,C), although the transition point (residue Q94) within the yeast protein is shifted eight residues toward the N terminus. These data indicate that domain I of SBDS can sample a variety of positions and potentially propagate conformational change. We considered the dynamic mobility of SBDS to be reminiscent of the interdomain motion of bacterial ribosome-recycling factor (RRF) that is critical for the cooperative interaction with EF-G in the disassembly of post-termination ribosomes (Yoshida et al. 2003; Gao et al. 2007; Savelsbergh et al. 2009), raising the possibility that the key role of SBDS might be to tightly couple GTP-dependent conformational change in EFL1 (an EF-G homolog) to the release of eIF6.
SBDS stimulates 60S-dependent GTP hydrolysis by EFL1
To address this hypothesis, we first asked whether SBDS might function as an adaptor that recruits EFL1 to the pre-60S subunit. However, we found that EFL1 and SBDS can bind independently to 60S ribosomal subunits in vitro, with no evidence of significant binding cooperativity in salt titration experiments (Fig. 6A). We next considered whether SBDS might activate GTP hydrolysis by EFL1. We first confirmed biochemically that recombinant human EFL1 does indeed have GTPase activity by incubating the protein with [γ-32P]GTP and measuring the release of inorganic phosphate (Pi). The low intrinsic GTPase activity of EFL1 was not significantly enhanced by SBDS alone (Fig. 6B, lanes 1,2). However, as the sarcin–ricin loop (SRL) is critical for the activation of translational GTPases on the ribosome (Voorhees et al. 2010), we next asked whether the GTPase activity of EFL1 was stimulated by RRL 60S subunits. Although 60S subunits alone had no significant intrinsic GTPase activity, they robustly activated GTP hydrolysis by EFL1 (Fig. 6B, lanes 3,5). As a control, we tested the ability of 60S subunits to stimulate the GTPase activity of a catalytically inert EFL1 variant carrying a missense mutation in a highly conserved catalytic residue (H96A, corresponding to yeast H106A). In contrast to wild-type EFL1, we detected no significant 60S-dependent stimulation of the GTPase activity of the EFL1-H96A mutant (data not shown). We conclude that the 60S subunit specifically activates the GTPase activity of EFL1.
Figure 6.
SBDS tightly couples activation of EFL1 GTP hydrolysis to eIF6 release. (A) SBDS and EFL1 bind independently to 60S subunits. SBDS and EFL1 were bound to RRL 60S subunits over the indicated range of KCl concentrations and pelleted through 30% (w/v) sucrose cushions. Bound SBDS, EFL1, and Rpl28 were visualized by immunoblotting. (B) SBDS stimulates 60S-dependent GTP hydrolysis by EFL1. 60S-dependent GTPase activity of human wild-type EFL1 or a catalytically inactive mutant (H96A) was measured in the presence of wild-type or variant (R126T and K151N) SBDS. The experiment was repeated three times and the average values with SD are presented. (Inset) Dose response curve for 60S-dependent EFL1 catalytic activity as a function of GTP concentration in the presence (black line) or absence (red line) of human SBDS. Measurements were performed in duplicate. (C) SDS-associated SBDS variants are defective in eIF6 release. Pre-60S subunits were incubated with GTP, EFL1, and either wild-type or variant (R126T or K151N) SBDS proteins. Following sucrose pelleting, “bound” and “free” fractions were immunoblotted to visualize eIF6 and Ebp1. See also Supplemental Figure S7.
However, despite 60S-dependent activation of its GTPase activity, EFL1 alone is insufficient to promote the release of eIF6 in the absence of SBDS in vitro (Fig. 3B,D) or in vivo (Fig. 2A–C). We therefore asked whether SBDS significantly enhances the 60S-dependent GTPase activity of EFL1. Indeed, we found that SBDS significantly stimulated 60S-dependent GTP hydrolysis by EFL1 (P < 0.01) (Fig. 6B, lanes 5,7). In contrast, Pi release by the catalytically inert EFL1 mutant H96A was not enhanced on incubation with 60S subunits and SBDS (Fig. 6B, lane 6), indicating that SBDS specifically stimulates Pi release from EFL1 and not from another contaminating ribosomal GTPase. In an experiment in which 60S-dependent GTP hydrolysis by EFL1 was measured as a function of GTP concentration, SBDS induced a twofold increase in Vmax for the reaction (Fig. 6B, inset). We conclude that SBDS significantly stimulates the 60S-dependent GTPase activity of EFL1 under multiple turnover conditions.
SBDS tightly couples GTP hydrolysis by EFL1 on the ribosome to eIF6 release
We postulated that the critical role of SBDS might be to tightly couple the activation of EFL1 GTP hydrolysis on the ribosome with eIF6 release, and that specific SDS-associated missense mutants might abrogate this activity. To test this hypothesis, we examined the function of two class B missense SBDS mutants (R126T and K151N) in three independent assays: pre-60S subunit binding, stimulation of EFL1 60S-dependent GTPase activity, and ability to drive eIF6 release from pre-60S ribosomal subunits.
The SbdsR126T disease-associated allele is a functional hypomorph in vivo and the R126T mutant protein is expressed at wild-type levels in fibroblasts homozygous for the SbdsR126T allele (Ball et al. 2009), indicating that it is functionally defective. Residue R126 lies on the surface of the α5–α6 loop at the interface between domains II and III (Fig. 4A). The 15N,1H HSQC spectrum of the R126T mutant displayed only local chemical shift perturbation of residues within loops α5–α6 and α7–β6 (Fig. 4F), indicating that it is has the same fold as wild-type SBDS. Although the R126T mutant bound efficiently to pre-60S subunits (Supplemental Fig. S7), it did not significantly enhance the 60S-dependent GTPase activity of EFL1 compared with wild-type SBDS (Fig. 6B, lanes 5,7,8). Furthermore, R126T was strongly defective compared with wild-type SBDS in triggering eIF6 release in concert with EFL1 and GTP (Fig. 6C). These data are consistent with the hypothesis that the primary defect in SDS is impaired release of eIF6 from pre-60S ribosomes.
Residue K151 is conserved between yeast and human SBDS and localizes to a basic patch on helix α7 on the surface of domain II (Fig. 4A; Supplemental Fig. S5). The 15N,1H HSQC spectrum of the K151N mutant displayed no significant chemical shift perturbation compared with wild-type SBDS (Fig. 4F). Although K151N bound efficiently to pre-60S ribosomal subunits (Supplemental Fig. S7), it was strongly defective compared with wild-type SBDS in triggering eIF6 dissociation with EFL1 and GTP (Fig. 6C). However, in contrast to R126T, the K151N mutant significantly stimulated the 60S-dependent GTPase activity of EFL1 (P < 0.01), and the degree of stimulation was not significantly different compared with wild type (Fig. 6B, lane 9). Thus, the K151N mutant is defective in eIF6 release despite significantly stimulating 60S-dependent GTP hydrolysis by EFL1.
Finally, we took advantage of the conservation of residue K151 in yeast SDO1 to examine the effect of the K151N mutation on SDO1 function in vivo by genetic complementation. As the sole source of SDO1, the sdo1-K151N allele failed to complement the fitness defect of sdo1Δ yeast cells despite appropriate expression (data not shown), indicating that residue K151 is critical for SDO1 function in vivo. Thus, we conclude that, although SBDS can significantly stimulate the 60S-dependent GTPase activity of EFL1 in multiple turnover conditions, its essential role is to tightly couple the activation of EFL1 GTP hydrolysis on the ribosome to eIF6 release.
Discussion
Our study describes the establishment of an ex vivo system that reconstitutes a late cytoplasmic step in the maturation of pre-60S ribosomal subunits. This system allows us to conclude that (1) human SBDS is an essential cofactor for the EFL1 GTPase, and together they cooperate to directly catalyze the release of eIF6 from mammalian pre-60S ribosomal subunits; (2) the mechanism of eIF6 release requires GTP binding and hydrolysis by EFL1, but is independent of eIF6 phosphorylation on Ser 235; (3) SBDS stimulates 60S-dependent GTP hydrolysis by EFL1, but its essential role is to tightly couple the activation of EFL1 GTP hydrolysis on the ribosome to eIF6 release; and (4) a conserved lysine (K151) that is mutated in SDS and maps to the central three-helical bundle of the SBDS protein is required for cooperativity with EFL1. Taken together with the comprehensive high-resolution NMR structural and dynamic analysis of both human and yeast SBDS proteins reported here, our findings provide key insights into the molecular mechanism of a fundamental and pivotal step that is required for the translational activation of ribosomes in all eukaryotes (summarized in the model shown in Fig. 7). This study is the first to link the etiology of the liver pathology associated with the clinical syndrome of SDS to defective 60S subunit maturation, and provides compelling evidence that SDS is a ribosomopathy that is caused by uncoupling GTP hydrolysis by EFL1 on the ribosome from eIF6 release.
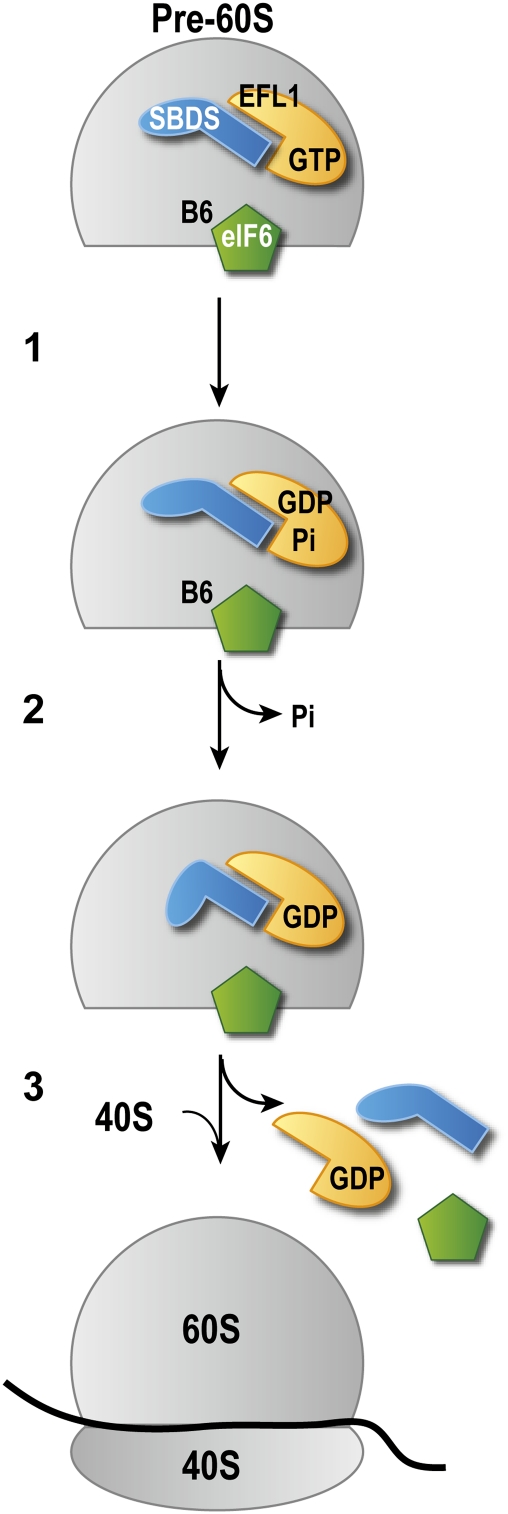
Figure 7.
Model of eIF6 release by SBDS and EFL1. (1) SBDS stimulates 60S-dependent GTP hydrolysis by EFL1, generating EFL1.GDP.Pi. (2) Following release of inorganic Pi, EFL1 adopts its GDP-bound conformation and domain I of SBDS is rotated relative to domains II and III, directly or indirectly disrupting the intersubunit bridge B6. (3) Binding of eIF6 is destabilized, release of eIF6 is triggered, and EFL1.GDP and SBDS dissociate from the ribosome. Release of eIF6 allows the formation of actively translating 80S ribosomes. The putative conformations of EFL1 are not indicated.
Molecular mechanism of eIF6 release
Based on our genetic, structural, and biochemical data, we propose an evolutionarily conserved model in which SBDS tightly couples the energy of EFL1 GTP hydrolysis and Pi release to eIF6 dissociation through a conformational coupling mechanism (Fig. 7), mimicking aspects of the cooperative interaction between RRF and EF-G in ribosome disassembly (Savelsbergh et al. 2009). As the cellular concentration of GTP is higher than that of GDP, EFL1 likely associates in its GTP-bound form with the GTPase-associated center of the pre-60S subunit (Graindorge et al. 2005; Voorhees et al. 2010), probably in close physical proximity to SBDS (C Wong and N Basse, unpubl.), with eIF6 localized in the vicinity of the dynamic intersubunit bridge B6 (Gartmann et al. 2010). As GTP binding and hydrolysis precede the dissociation of eIF6 from the pre-60S subunit (Fig. 3A–D), it is likely that EFL1 undergoes GTP-dependent conformational change on the ribosome, analogous to its structural homolog, EF-G. The flexible attachment of domain I of SBDS to domains II and III (Fig. 5A–D) raises the possibility that interdomain motion may be important for its function in eIF6 dissociation. We therefore suggest that, following GTP hydrolysis and Pi release (Fig. 6B), conformational change in EFL1 induces a domain rotation in SBDS that in turn drives a structural rearrangement of the ribosome, displacing the B6 bridge, destabilizing the interaction of eIF6 with the pre-60S subunit, and triggering eIF6 dissociation. However, further experiments will be required to fully elucidate the structural basis for the cooperative interaction between SBDS and EFL1. As SBDS and EFL1 are not observed on mammalian polysomes, they likely dissociate following eIF6 release, but the precise timing for this event is unknown. Furthermore, SBDS and EFL1 can directly evict eIF6 from mature 60S subunits in vitro, suggesting that, at least in mammals, this eIF6 recycling mechanism may function to promote translation by enhancing the pool of available free 60S subunits.
In yeast, the release of eIF6 appears to be upstream and a prerequisite for the removal of Nmd3 from pre-60S particles by the GTPase Lsg1 (Lo et al. 2010). Consistent with these data, we observed accumulation of cytoplasmic pre-60S particles carrying Lsg1, Nmd3, and eIF6 in the absence of Sbds (Fig. 2C). In the presence of Lsg1, it was therefore unexpected that Nmd3 persisted on the pre-60S particles following release of eIF6, suggesting that an additional factor or modification of Lsg1 may be required to drive Nmd3 release. Our ability to reconstitute the final steps of 60S maturation ex vivo provides an important platform to further dissect the mechanism of Nmd3 release.
SDS as a ribosomopathy
Our findings provide compelling evidence that autosomal recessive SDS is a ribosomopathy in which the primary defect is the uncoupling of GTP hydrolysis by EFL1 on the ribosome from eIF6 release during late cytoplasmic maturation of pre-60S ribosomal subunits. Autosomal dominant Diamond-Blackfan anemia is associated with mutations in an increasing number of genes encoding ribosomal proteins (RPs) of the small and large ribosomal subunits (Boria et al. 2010), and the 5q− syndrome, an acquired subtype of MDS, is linked to haploinsufficiency of the RPS14 gene (Ebert et al. 2008; Barlow et al. 2010a). Thus, the primary defect underlying SDS is distinct from the other ribosomopathies associated with hematological disease. We suggest that the diverse alternate functions proposed for SBDS in mammalian cells (Wessels et al. 2006; Austin et al. 2008; Rujkijyanont et al. 2008; Ball et al. 2009; Leung et al. 2010) are downstream secondary consequences of the primary defect in 60S ribosomal subunit maturation.
How do we explain the SDS phenotype? It seems likely to reflect, at least in part, the sensitivities of specific cell types to cellular stress responses during development that result from defective ribosome biogenesis. Indeed, recent in vivo data have established the RP–Mdm2 interaction as a p53 stress signaling pathway that is activated by aberrant ribosome biogenesis and is essential to protect against oncogenic c-MYC-induced tumorigenesis (Macias et al. 2010). Furthermore, the p53 dependence of the pathologies associated with defective ribosome biogenesis is supported by genetic data in a number of animal models (Barlow et al. 2010b).
The dramatic disruption of nucleolar architecture in Sbds-deleted livers is consistent with the hypothesis that nucleolar stress may contribute to the observed hepatocyte defects following complete deletion of Sbds. However, as SDS is a consequence of partial, not complete, loss of SBDS function, further work will be required to clarify the importance of nucleolar stress responses to the clinical SDS phenotype. Nevertheless, as abnormal liver function tests and hepatomegaly are recognized features of SDS at an early age (Toiviainen-Salo et al. 2009), this study directly links the etiology of a pathological abnormality associated with the clinical syndrome of SDS to defective 60S subunit maturation and a ribosomal subunit-joining defect.
Our findings demonstrate the feasibility and power of biochemically recapitulating late maturation of mammalian ribosomal subunits ex vivo to elucidate the underlying molecular mechanisms. Finally, this study strongly supports the hypothesis that small molecules that mimic the effects of TIF6 suppressor mutations in yeast may have utility in the treatment of SDS and possibly other forms of bone marrow failure.
Materials and methods
Protein expression and purification
Recombinant human eIF6 (NP_852133) and SBDS (NP_057122) were expressed and purified from Escherichia coli, and human EFL1 (NP_078856) was expressed and purified from S. cerevisiae. Recombinant proteins were purified by Ni-NTA affinity and gel filtration chromatography. See the Supplemental Material for detailed protocols.
NMR spectroscopy
NMR spectra were assigned and the solution structure was determined using standard techniques (Wüthrich 1986; Bax 1994).
Mouse gene targeting
The mouse genomic Sbds locus was PCR-amplified from mouse 129 DNA. The targeting vector pMCTV1 was generated such that exposure to Cre recombinase results in deletion of exon 2 and the selection cassette. All animal experiments were undertaken with the approval of the UK Home Office.
Sucrose density gradients
Frozen liver (typically 30 mg) was homogenized with a pestle in detergent lysis buffer A (10 mM Tris-HCl at pH 7.4, 10 mM NaCl, 1.5 mM MgCl2, 0.5% [v/v] Triton X-100, 0.5% [w/v] deoxycholate, 1% [v/v] Tween 20, 100 mg/mL cycloheximide) with complete EDTA-free protease inhibitors (Roche) and 0.5 U/mL RNase inhibitor (Promega) and incubated for 10 min on ice. Lysates were cleared in a microfuge. Equal amounts (typically 10–20 A254 U) were applied to a 10%–50% (w/v) sucrose gradient in 30 mL of buffer B (10 mM Tris-HCl at pH 7.4, 75 mM KCl, 1.5 mM MgCl2) and centrifuged (Beckmann SW32 rotor) at 32,000 rpm for 2 h and 46 min at 4°C. For ribosome profiles, liver lysates were prepared in buffer A (without MgCl2 or cycloheximide) in the presence of 5 mM EDTA; MgCl2 was removed from the sucrose gradient buffer B.
Samples were unloaded using a Brandel gradient fractionator, the polysome profiles were detected using a UV monitor (UV-1, Pharmacia) at A254, and 1-mL fractions were collected. The electronic outputs of the UV-1 monitor and fraction collector were fed into a Labjack U3-LV data acquisition device with a LJTick-InAmp preamplifier. Proteins were precipitated from sucrose gradient fractions with 10% (v/v) trichloroacetic acid (TCA), separated on SDS-PAGE gels and transferred to PVDF membranes for immunoblotting.
Preparation of liver extracts and immunoblotting
Whole-liver extracts were prepared by homogenizing 10 mg of liver in RIPA buffer C (20 mM HEPES at pH 7.4, 20 mM β-glycerophosphate, 10 mM NaF, 0.5 mM EDTA, 0.5 mM EGTA, 0.2 M NaCl, 1% [v/v] Nonidet P-40, 0.5% [w/v] sodium deoxycholate, 0.1% [w/v] SDS) with complete EDTA-free protease inhibitors (Roche) using a pellet pestle (Sigma) and leaving for 10 min on ice with occasional vortexing. Samples were cleared in a microfuge and normalized for protein concentration using a BCA protein assay kit (Pierce). Samples were fractionated using NE-PER nuclear and cytoplasmic extraction reagents (Pierce) according to the manufacturer's instructions. All fractions were normalized against the protein concentration of the cytoplasmic fractions.
eIF6 release assay
The reaction mixture (100 μL) contained 50 μL of sucrose gradient-purified 60S subunits (∼0.5–1 pmol); 5 pmol of SBDS; 5 pmol of EFL1; 100 μM purified GTP, GDP, or GDPNP (Sigma) in buffer B with complete EDTA-free protease inhibitors (Roche); and 0.5 U/mL RNasin (Promega). GDP and GTP were purified as in Wilden et al. (2006).
Alternatively, 5 pmol of 60S subunits was purified from RRL (Pisarev et al. 2007) and incubated with an excess of recombinant eIF61-225 for 20 min on ice, and free eIF6 was removed using a 100-kDa Microcon centrifugal filter unit (Sigma). The eIF6-loaded RRL was then used as a substrate in the eIF6 release assay. Reaction mixtures were incubated for 10 min at 37°C, layered onto a 150-μL 30% (w/v) sucrose cushion in buffer B, and centrifuged (Beckmann TLA120.1 rotor) at 120,000 rpm for 30 min at 4°C. Two-hundred microliters of supernatant (“free”) was removed, and the remaining 50 μL was resuspended (“bound”) in 150 μL of buffer B. Samples were TCA-precipitated for immunoblotting.
RRL 60S-binding assay
The reaction mixture (100 μL) contained 5 pmol of 60S subunits purified from RRL (Pisarev et al. 2007) with 25 pmol each of SBDS and EFL1 in buffer B. Additional KCl was added where specified. Samples were subsequently processed as above for eIF6 release.
60S-dependent GTP hydrolysis by EFL1
The reaction mixture (50 μL) contained 5 nmol of [γ-32P]GTP (6 Ci/μmol; Perkin-Elmer), 5 pmol of RRL 60S subunits, 6.3 pmol of SBDS, and 40 pmol of EFL1 in buffer D (50 mM Tris at pH 7.4, 2 mM MgCl2, 150 mM NaCl, 2 mM DTT). The reaction mixture was incubated for 30 min at 37°C. An aliquot (40 μL) of the reaction mix was added to 360 μL of chilled charcoal slurry (5% [v/v] Norit in 50 mM NaH2PO4, at pH 3.0) and centrifuged at 13,000 rpm for 10 min. The amount of radioactivity in 100 μL of supernatant was quantified by liquid scintillation counting.
Yeast strains and plasmids
S. cerevisiae strains used in this study are listed in Supplemental Table S6 and primers are listed in Supplemental Table S7. Deletion strains were generated by homologous recombination using appropriate PCR products to transform strain Y5538. The coding sequence for wild-type EFL1 was PCR-amplified from yeast genomic DNA and cloned into pYCT/C2 to generate plasmid pEFL1. Plasmids pEFL1-T33A, pEFL1-D102A, pEFL1-H106A, and pEFL1-H106I, pEFL1-D159A, pEFL1-W240E were generated by QuickChange site-directed mutagenesis (Stratagene).
Suppressor clone isolation
Suppressor clone isolation was performed as described (Menne et al. 2007).
Synthetic genetic array analysis and random sporulation assays
SGA and random sporulation analysis was performed as described (Tong and Boone 2006; Baryshnikova et al. 2010; Costanzo et al. 2010). MATα efl1Δ∷NatMX4 TIF6-R61G and MATα sdo1Δ∷NatMX4 TIF6-R61G query strains harboring SGA markers and haploid-specific reporters were crossed to an array containing the set of ∼5000 viable deletion mutant strains. Nourseothricin was obtained from Werner BioAgents.
Peptide separation, mass spectrometry, and database analysis
For peptide separation, mass spectrometry, and database analysis used in this study, see the Supplemental Material.
Antibodies
For antibodies used in this study, see the Supplemental Material.
Statistical analysis
Student's t-test was used to determine significant differences. P < 0.01 was considered significant.
Accession numbers
Coordinates for the human SBDS structure have been deposited in the Protein Data Bank (ID 2L9N), and the 1H, 15N, and 13C chemical shifts have been deposited in the BioMagResBank database (accession code 17479).
Acknowledgments
We thank the European NMR Large-Scale Facility (Utrecht, Holland) and E. Reynaud (University College Dublin) for Lsg1 antibodies; M. Tchórzewski (Maria Curie-Skodowska University) for P0 antisera; J. Sale for critical comments on the manuscript; A. Newman for strain BCY123; R. Mittal for assistance with GTPase assays; Mark Allen, Mark Bycroft, and Trevor Rutherford for critical advice and assistance with NMR spectroscopy; M. Schwarz (St. Mary's Hospital, Manchester) for communicating unpublished data; the LMB animal facility staff; and L. Easton and P. Lukavsky for RRL ribosomes. Leukemia and Lymphoma Research, the Association for International Cancer Research, the Sylvia Aitkin Trust, the Medical Research Council, the MDS Foundation, the Leukemia and Lymphoma Society of America, Shwachman-Diamond Support UK, Ted's Gang, the Tesni Parry Memorial Fund, and the Cambridge NIHR Biomedical Research Center supported this work. M.J.A. is supported by Cancer Research UK. J.D. and C.B.-C. are supported by the Institut de Veille Sanitaire, Inserm, the Association Laurette Fugain, and the Association Sportive de Saint Quentin Fallavier. C.B. is supported by the Canadian Institutes of Health Research (CIHR) (MOP-102629), the National Institutes of Health (NIH) (1R01HG005853-01), and the Ontario Research Fund (ORF-GL2) (GL2-01-22).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.623011.
Supplemental material is available for this article.
References
- Austin KM, Gupta ML, Coats SA, Tulpule A, Mostoslavsky G, Balazs AB, Mulligan RC, Daley G, Pellman D, Shimamura A 2008. Mitotic spindle destabilization and genomic instability in Shwachman-Diamond syndrome. J Clin Invest 118: 1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball HL, Zhang B, Riches JJ, Gandhi R, Li J, Rommens JM, Myers JS 2009. Shwachman-Bodian Diamond syndrome is a multi-functional protein implicated in cellular stress responses. Hum Mol Genet 18: 3684–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JL, Drynan LF, Hewett DR, Holmes LR, Lorenzo-Abalde S, Lane AL, Jolin HE, Pannell R, Middleton AJ, Wong SH, et al. 2010a. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q− syndrome. Nat Med 16: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JL, Drynan LF, Trim NL, Erber WN, Warren AJ, McKenzie AN 2010b. New insights into 5q− syndrome as a ribosomopathy. Cell Cycle 9: 4286–4293 [DOI] [PubMed] [Google Scholar]
- Baryshnikova A, Costanzo M, Kim Y, Ding H, Koh J, Toufighi K, Youn JY, Ou J, San Luis BJ, Bandyopadhyay S, et al. 2010. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Methods 7: 1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu U, Si K, Warner JR, Maitra U 2001. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol Cell Biol 21: 1453–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax A 1994. Multidimensional nuclear-magnetic-resonance methods for protein studies. Curr Opin Struct Biol 4: 738–744 [Google Scholar]
- Becam AM, Nasr F, Racki WJ, Zagulski M, Herbert CJ 2001. Ria1p (Ynl163c), a protein similar to elongation factors 2, is involved in the biogenesis of the 60S subunit of the ribosome in Saccharomyces cerevisiae. Mol Genet Genomics 266: 454–462 [DOI] [PubMed] [Google Scholar]
- Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM 2003. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet 33: 97–101 [DOI] [PubMed] [Google Scholar]
- Boria I, Garelli E, Gazda HT, Aspesi A, Quarello P, Pavesi E, Ferrante D, Meerpohl JJ, Kartal M, Da Costa L, et al. 2010. The ribosomal basis of Diamond-Blackfan anemia: mutation and database update. Hum Mutat 31: 1269–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, Marchisio PC, Biffo S 2003. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426: 579–584 [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. 2010. The genetic landscape of a cell. Science 327: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira JF, Sforca ML, Blumenschein TM, Goldfeder MB, Guimaraes BG, Oliveira CC, Zanchin NI, Zeri AC 2010. Structure, dynamics, and RNA interaction analysis of the human SBDS protein. J Mol Biol 396: 1053–1069 [DOI] [PubMed] [Google Scholar]
- Diamond R 1995. Coordinate-based cluster analysis. Acta Crystallogr D Biol Crystallogr 51: 127–135 [DOI] [PubMed] [Google Scholar]
- Donadieu J, Leblanc T, Bader Meunier B, Barkaoui M, Fenneteau O, Bertrand Y, Maier-Redelsperger M, Micheau M, Stephan JL, Phillipe N, et al. 2005. Analysis of risk factors for myelodysplasias, leukemias and death from infection among patients with congenital neutropenia. Experience of the French Severe Chronic Neutropenia Study Group. Haematologica 90: 45–53 [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, et al. 2008. Identification of RPS14 as a 5q− syndrome gene by RNA interference screen. Nature 451: 335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi KA, Austin KM, Lee CS, Dias A, Malsch MM, Reed R, Shimamura A 2007. The human Shwachman-Diamond syndrome protein, SBDS, associates with ribosomal RNA. Blood 110: 1458–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, Zavialov AV, Ehrenberg M, Frank J 2007. Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits. J Mol Biol 374: 1345–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartmann M, Blau M, Armache JP, Mielke T, Topf M, Beckmann R 2010. Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. J Biol Chem 285: 14848–14851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graindorge JS, Rousselle JC, Senger B, Lenormand P, Namane A, Lacroute F, Fasiolo F 2005. Deletion of EFL1 results in heterogeneity of the 60 S GTPase-associated rRNA conformation. J Mol Biol 352: 355–369 [DOI] [PubMed] [Google Scholar]
- Leung R, Cuddy K, Wang Y, Rommens J, Glogauer M 2010. Sbds is required for Rac2-mediated monocyte migration and signaling downstream of RANK during osteoclastogenesis. Blood 117: 2044–2053 [DOI] [PubMed] [Google Scholar]
- Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW 2010. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell 39: 196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias E, Jin A, Deisenroth C, Bhat K, Mao H, Lindstrom MS, Zhang Y 2010. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell 18: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menne TF, Goyenechea B, Sanchez-Puig N, Wong CC, Tonkin LM, Ancliff PJ, Brost RL, Costanzo M, Boone C, Warren AJ 2007. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat Genet 39: 486–495 [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Unbehaun A, Hellen CU, Pestova TV 2007. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol 430: 147–177 [DOI] [PubMed] [Google Scholar]
- Rujkijyanont P, Watanabe K, Ambekar C, Wang H, Schimmer A, Beyene J, Dror Y 2008. SBDS-deficient cells undergo accelerated apoptosis through the Fas-pathway. Haematologica 93: 363–371 [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Rodnina MV, Wintermeyer W 2009. Distinct functions of elongation factor G in ribosome recycling and translocation. RNA 15: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 23: 5080–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B, Lafontaine DL, Graindorge JS, Gadal O, Camasses A, Sanni A, Garnier JM, Breitenbach M, Hurt E, Fasiolo F 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol Cell 8: 1363–1373 [DOI] [PubMed] [Google Scholar]
- Sengupta J, Bussiere C, Pallesen J, West M, Johnson AW, Frank J 2010. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J Cell Biol 189: 1079–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shammas C, Menne TF, Hilcenko C, Michell SR, Goyenechea B, Boocock GR, Durie PR, Rommens JM, Warren AJ 2005. Structural and mutational analysis of the SBDS protein family. Insight into the leukemia-associated Shwachman-Diamond syndrome. J Biol Chem 280: 19221–19229 [DOI] [PubMed] [Google Scholar]
- Toiviainen-Salo S, Durie PR, Numminen K, Heikkil P, Marttinen E, Savilahti E, Makitie O 2009. The natural history of Shwachman-Diamond syndrome-associated liver disease from childhood to adulthood. J Pediatr 155: 807–811.e2 doi: 10.1016/j.jpeds.2009.06.047 [DOI] [PubMed] [Google Scholar]
- Tong AH, Boone C 2006. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol 313: 171–192 [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V 2010. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330: 835–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels D, Srikantha T, Yi S, Kuhl S, Aravind L, Soll DR 2006. The Shwachman-Bodian-Diamond syndrome gene encodes an RNA-binding protein that localizes to the pseudopod of Dictyostelium amoebae during chemotaxis. J Cell Sci 119: 370–379 [DOI] [PubMed] [Google Scholar]
- Wilden B, Savelsbergh A, Rodnina MV, Wintermeyer W 2006. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc Natl Acad Sci 103: 13670–13675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich K 1986. NMR of proteins and nucleic acid. John Wiley & Sons, Inc, New York [Google Scholar]
- Yoshida T, Oka S, Uchiyama S, Nakano H, Kawasaki T, Ohkubo T, Kobayashi Y 2003. Characteristic domain motion in the ribosome recycling factor revealed by 15N NMR relaxation experiments and molecular dynamics simulations. Biochemistry 42: 4101–4107 [DOI] [PubMed] [Google Scholar]