Abstract
Proneural genes such as Ascl1 are known to promote cell cycle exit and neuronal differentiation when expressed in neural progenitor cells. The mechanisms by which proneural genes activate neurogenesis—and, in particular, the genes that they regulate—however, are mostly unknown. We performed a genome-wide characterization of the transcriptional targets of Ascl1 in the embryonic brain and in neural stem cell cultures by location analysis and expression profiling of embryos overexpressing or mutant for Ascl1. The wide range of molecular and cellular functions represented among these targets suggests that Ascl1 directly controls the specification of neural progenitors as well as the later steps of neuronal differentiation and neurite outgrowth. Surprisingly, Ascl1 also regulates the expression of a large number of genes involved in cell cycle progression, including canonical cell cycle regulators and oncogenic transcription factors. Mutational analysis in the embryonic brain and manipulation of Ascl1 activity in neural stem cell cultures revealed that Ascl1 is indeed required for normal proliferation of neural progenitors. This study identified a novel and unexpected activity of the proneural gene Ascl1, and revealed a direct molecular link between the phase of expansion of neural progenitors and the subsequent phases of cell cycle exit and neuronal differentiation.
Keywords: proneural factor, cell proliferation, genomic location analyses, target genes
The generation of new neurons in the developing brain is a complex process that requires neural progenitors to progress through a succession of distinct cellular states. Important advances have been made in imaging the development of neural lineages, including the cell divisions of progenitor cells and the differentiation and migration of newborn neurons (Kriegstein and Noctor 2004; Shioi et al. 2009). In contrast, there is still little known of the genetic programs that guide these different steps of neurogenesis.
Different types of progenitor cells have been identified in the embryonic telencephalon, the rostral-most division of the forebrain. Radial glial cells in the ventricular zone (VZ) have characteristic features of neural stem (NS) cells, as they self-renew by asymmetric division and have the potential to generate both neurons and glial cells (Gotz and Huttner 2005). Some radial glial cells directly give rise to post-mitotic neurons, while others generate intermediate progenitors that divide symmetrically before they produce neurons (Haubensak et al. 2004; Miyata et al. 2004; Noctor et al. 2004). Radial glial cells divide at the apical surface of the VZ, while intermediate progenitors, also called basal or non-surface-dividing progenitors, divide away from the apical surface in the subventricular zone (SVZ). The dorsal telencephalon, which generates the projection neurons of the cerebral cortex, contains a relatively small population of basal progenitors (at least in rodents) (see Smart et al. 2002), while the ventral telencephalon, which generates projection neurons of the basal ganglia as well as cortical and basal ganglia interneurons, has a larger SVZ (Smart 1976; Sheth and Bhide 1997). Telencephalic progenitors can be expanded in vitro as nonadherent “neurospheres” or as adherent NS cells, which can be maintained as proliferating and undifferentiated progenitors in the presence of mitogens or differentiated into neurons, oligodendrocytes, and astrocytes when the mitogens are withdrawn (Reynolds and Weiss 1992; Johe et al. 1996; Conti and Cattaneo 2010).
Neurogenesis in the embryonic brain is regulated primarily by proneural transcription factors (TFs), including Mash1/Ascl1 and Ngn1/2/Neurog1/2. Extensive genetic analysis in the mouse has shown that these factors promote cell cycle exit and neuronal differentiation in diverse progenitor populations (Bertrand et al. 2002; Ross et al. 2003). For example, overexpression of Ascl1 in NS cell cultures results in rapid differentiation of transduced cells into functional neurons (Berninger et al. 2007b; Geoffroy et al. 2009). Conversely, loss of Ascl1 results in a severe loss of basal ganglia neurons and cortical interneurons in the telencephalon (Casarosa et al. 1999; Horton et al. 1999; Marin et al. 2000; Yun et al. 2002). Proneural factors have been shown to regulate early steps of neurogenesis, including the neuronal commitment, subtype specification, and cell cycle exit of progenitors (Farah et al. 2000; Fode et al. 2000; Nieto et al. 2001; Nakada et al. 2004), as well as later steps such as the migration of post-mitotic neurons and the oriented growth of their axons (Seibt et al. 2003; Hand et al. 2005; Ge et al. 2006). In contrast to the understanding of the developmental functions of proneural factors, little is known of the molecular pathways that mediate their activities. Both Ascl1 and Neurog1/2 are known to activate Notch signaling by directly inducing expression of the ligands Delta and Jagged, thereby inhibiting neurogenesis in adjacent cells (Castro et al. 2006; Henke et al. 2009). In addition, Ascl1 confers a GABAergic neurotransmission phenotype to ventral telencephalic neurons at least in part through direct regulation of the homeobox genes Dlx1 and Dlx2 (Schuurmans et al. 2004; Poitras et al. 2007). A few additional targets of Ascl1, including the genes encoding the zinc finger protein Insm1 and the ubiquitin ligase Fbxw7, have been discovered by their association with conserved binding motifs for Ascl1 and its coregulators, Brn1 and Brn2 (Castro et al. 2006). Because of the paucity of information on their transcriptional targets, it is unclear whether Ascl1 and other proneural genes control most aspects of neurogenesis directly or via a downstream transcriptional cascade. For example, it is not known how proneural proteins promote cell division arrest, although induction of cyclin-dependent kinase (Cdk) inhibitors has been proposed (Farah et al. 2000). Several TFs and chromatin-modifying enzymes—including N-Myc, Bmi1, Tlx, and FoxM1—are known to maintain NS cell divisions in the embryonic or adult brain (Molofsky et al. 2003; Fasano et al. 2007; Schuller et al. 2007; Zhao et al. 2009; Qu et al. 2010). Whether proneural factors interact with such factors to inhibit cell proliferation and promote neurogenesis remains to be addressed.
The goal of this study was to characterize on a genomic scale the transcriptional targets of Ascl1 in order to better understand how this factor regulates neurogenesis and possibly discover new functions that have eluded genetic analysis. We combined genomic location and expression profiling analyses to identify Ascl1 targets in the embryonic telencephalon and cultured NS cells. This study showed that Ascl1 regulates a large number of target genes with diverse molecular functions and with potential involvement in a broad range of cellular processes, suggesting direct control of both early and late phases of neurogenesis. Unexpectedly, as this had not been identified by functional analysis, Ascl1 activates a large number of positive cell cycle regulators, and loss-of-function (LOF) analysis in the embryo and NS cell cultures confirmed that Ascl1 is indeed required for normal progenitor divisions. Our results therefore demonstrate that Ascl1 plays a major role in coordinating the program of neurogenesis by controlling the progression of neural progenitors through the successive phases of proliferation, cell cycle exit, and differentiation.
Results
Identification of the genetic program regulated by Ascl1 in the embryonic telencephalon
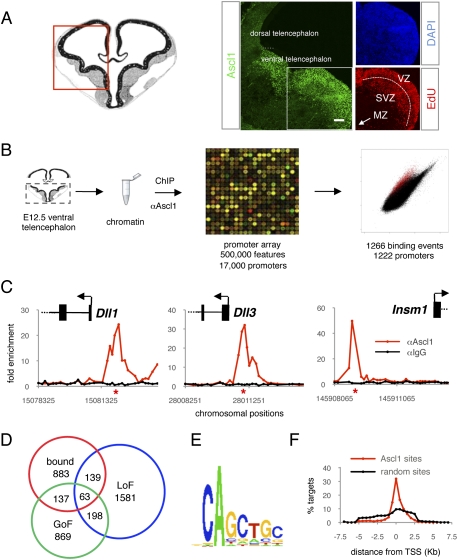
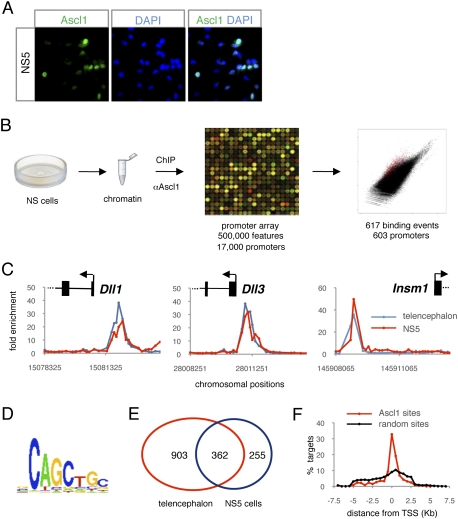
As a first step toward defining the genetic program activated by Ascl1 during telencephalon development, we performed an unbiased genome-wide study combining chromatin immunoprecipitation (ChIP) with promoter microarrays (ChIP–chip) to define the repertoire of Ascl1-binding sites in the embryonic telencephalon in vivo. We performed ChIP with an antibody against Ascl1 and chromatin prepared from the mouse ventral telencephalon at embryonic day 12.5 (E12.5), a stage when Ascl1 is highly expressed in this brain region (Fig. 1A). ChIP–chip experiments were carried out with oligonucleotide microarrays tiling ∼8 kb of proximal promoter regions associated with >17,000 genes (see the Materials and Methods). The result of this genome-wide location experiment revealed that Ascl1 is recruited to 1265 sites in 1222 promoters (with a false-positive rate of <10%), representing 7.2% of the promoters on the array (Fig. 1B; Supplemental Fig. S1; Supplemental Table S1). The majority of the sites (77%) were found within 1 kb of the transcription start sites of genes (Fig. 1F). All of the known Ascl1-binding sites tiled in the genomic array (Castro et al. 2006) were bound by Ascl1 in this study (Fig. 1C). Ascl1 binding to known sites outside of proximal promoters (i.e., in regulatory elements for Stk33, Fbxw7, and Dlx1/2) (Castro et al. 2006; Poitras et al. 2007) could not be detected in this study, as these were not tiled on the microarray.
Figure 1.
Characterization of the transcriptional program regulated by Ascl1 in the ventral telencephalon. (A) Expression of Ascl1 (green) in E12.5 mouse ventral telencephalons detected by immunohistochemistry is confined to the proliferative zones (VZ and SVZ; their border is marked with a white dashed line). Labeling for EdU (red) 6 h after administration marks progenitor cells in G2/M phases of the cell cycle. The MZ, which contains post-mitotic neurons, is outside the enlarged area, as shown with the white arrow. All nuclei are labeled with DAPI (blue). The red rectangle in the left drawing indicates the area of the ventral telencephalon documented. The white square in the left panel indicates the area enlarged in the right panels. Bar, 150 μm. (B) DNA segments bound by Ascl1 in the ventral telencephalon were identified by ChIP with an Ascl1-specific antibody and hybridization to a promoter microarray (ChIP-on-chip). (C) Ascl1 binding to known target genes. The plots display ChIP enrichment ratios for Ascl1 (red) and control (black) samples for all probes in target promoters. Genes are shown to scale above the plots (exons represented as boxes and introns represented as lines) and genomic regions below the plots. Arrows indicate the transcription start site and direction of transcription. Red stars show the positions of previously validated Ascl1-binding sites. (D) Venn diagram showing the overlap between genes associated with Ascl1-binding events (red), genes deregulated in Ascl1 GOF experiments (green), and genes deregulated in Ascl1 LOF experiments (blue). The number of genes in each section of the diagram is indicated. (E) Motif enriched in Ascl1-bound segments from a de novo search using Trawler software. (F) Plot representing the fraction of Ascl1-binding events located at consecutive 0.5-kb intervals away from the transcription start site of target genes (red dots and line). A plot for a randomized sample of binding events is shown as control (black dots and line).
To identify DNA sequence motifs that are overrepresented in the promoters of Ascl1 telencephalic targets and might therefore be recognized by Ascl1, we used the Trawler algorithm (Ettwiller et al. 2007; Haudry et al. 2010). The only position weight matrix (PWM) found to be overrepresented in the data set corresponded to the consensus CAGCTGC (Fig. 1E), which is very similar to the consensus sequences reported previously for Ascl1 binding (CAGCTG) (Bertrand et al. 2002; Hu et al. 2004; Castro et al. 2006) and binding of its Drosophila orthologs Asense (Southall and Brand 2009) and Achaete/Scute (GCAGGTG) (Supplemental Fig. S2; Singson et al. 1994).
To assess the impact of Ascl1 promoter binding on the regulation of the corresponding genes, we used genome-wide expression profiling data sets that we had generated previously using embryonic ventral telencephalons that were either overexpressing or mutant for Ascl1 (Supplemental Tables S2, S3; Gohlke et al. 2008; C Parras, unpubl.). Intersection of the lists of Ascl1-bound genes and of genes deregulated in these Ascl1 LOF and gain-of-function (GOF) experiments identified 339 likely direct targets of Ascl1 (Fig. 1D). Of these target genes, 220 were positively regulated and 101 were negatively regulated by Ascl1, with 18 showing conflicting regulation (Supplemental Fig. S3).
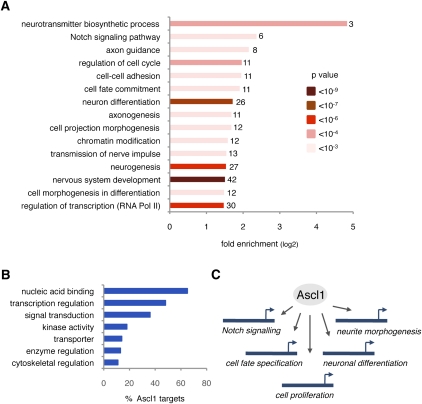
We used the unbiased identification of Ascl1 targets as a tool to examine the functions of Ascl1 in telencephalon development. A survey of the functional annotations of Ascl1 direct targets by Gene Ontology (http://www.geneontology.org) revealed a wide spectrum of biological activities (Fig. 2; Supplemental Table S4). Biological processes significantly enriched in this data set correspond to diverse phases of neurogenesis, including early steps of lateral inhibition (e.g., “Notch signaling pathway”), cell fate decisions (e.g., “cell fate commitment”) and control of cell proliferation (e.g., “regulation of cell cycle”), and later steps of neuronal differentiation (e.g., “neurotransmitter biosynthetic process”) and neurite outgrowth (e.g., “cell projection morphogenesis”) (Fig. 2A). Ascl1 targets also display a diverse range of molecular functions, including transcriptional activity (48% of Ascl1 targets), signal transduction (36%), transporter activity (14%), and cytoskeletal activity (11%) (Fig. 2B).
Figure 2.
Ascl1 directly regulates different steps in the neurogenic program. (A) Enrichment of Gene Ontology biological process terms in Ascl1 target genes. Number of target genes in each category is shown. (B) Enrichment of Gene Ontology molecular function terms (GO Slim) in Ascl1 target genes. (C) Ascl1 directly binds and regulates genes associated with all of the major steps of neurogenesis.
Systematic analysis of expression patterns using publicly available in situ hybridization data (http://www.genepaint.org) further showed that Ascl1 targets are expressed during different phases of neurogenesis in the ventral telencephalon (Supplemental Fig. S4; data not shown). A first set of targets is transcribed exclusively in the VZ, where radial glial stem cells are located (n = 112); genes in a second set are expressed only in the SVZ, where intermediate progenitors are found (n = 24), while genes in a third set are expressed mainly in the mantle zone (MZ) (n = 35), which contains post-mitotic and differentiating neurons. Genes in a fourth set present a more uniform or complex expression pattern (n = 65) or their expression is not detected in the embryonic telencephalon (n = 23), while expression of 80 targets has not been documented. Together, these results suggest that Ascl1 promotes neurogenesis by directly regulating a large number of genes that contribute to all phases of the neurogenic program (Fig. 2C).
Ascl1 promotes progenitor divisions in the embryonic telencephalon
Although Ascl1 expression has been shown to arrest progenitor divisions at the onset of neurogenesis, like other proneural proteins (Farah et al. 2000; Nakada et al. 2004), the genes that promote cell cycle exit downstream from Ascl1 are not known. We therefore scanned the list of Ascl1 targets for genes implicated previously in cell cycle regulation. As expected, we identified several genes that have been shown to inhibit cell cycle progression and/or promote cell cycle arrest in neural progenitors or other cell types, including Btg2 (Park et al. 2008), CcnG2 (Arachchige Don et al. 2006), Ebf3 (Zhao et al. 2006), Gadd45g (Ying et al. 2005; Huang et al. 2010), Hipk2 (Wei et al. 2007), and Prmt2 (Fig. 3; Supplemental Table S5; Yoshimoto et al. 2006). More surprising, we also found among Ascl1 targets a large number of positive cell cycle regulators (Fig. 3; Supplemental Table S5), suggesting that Ascl1 may have a previously unreported role of promoting cell proliferation. This group of targets included E2f1, the main transcriptional activator of genes promoting G1/S transition (Helin 1998); its coactivator, Ep400 (Tyteca et al. 2006); and one of their targets, Cdca7 (Goto et al. 2006). Other genes in this group included the TFs Tead1 and Tead2 and their coactivator, TAZ, which have been shown to promote the proliferation of neural progenitors (Cao et al. 2008) and are inhibited by the Hippo pathway (Ota and Sasaki 2008); the oncogene FoxM1, which promotes the G2/M transition in cerebellar granule cells and other cell types (Schuller et al. 2007); and the transcriptional inhibitors Id1, which promote cell proliferation and block differentiation by inhibiting the activity of the Retinoblastoma protein (Ruzinova and Benezra 2003). Binding of Ascl1 to these positive cell cycle regulators in ChIP–chip and ChIP-PCR experiments was very robust overall compared with Ascl1-binding events as a whole (Fig. 3; data not shown). Analysis of Ascl1-bound elements in a transcription assay in P19 cells identified eight elements with Ascl1-inducible enhancer activity, including six associated with cell cycle-promoting genes (E2f1, Gpc1, Id1, Taz, Tead1, and Tead2) and two with cell cycle arrest genes (Ccng2 and Gadd45g) (Supplemental Fig. S5). Most of these positive cell cycle regulators are activated by Ascl1, as shown by their down-regulation in Ascl1 mutant embryos and/or up-regulation in embryos overexpressing Ascl1 (Fig. 4E; Supplemental Table S5). These results suggest that Ascl1 actively promotes cell proliferation during telencephalic development, and that it acts by directly regulating a large number of genes that control cell cycle progression at different phases of the cycle (Fig. 3C).
Figure 3.
Illustration of Ascl1 target genes with a predicted role in the regulation of progenitor cell divisions. (A) Promoter regions and expression patterns of selected Ascl1 targets that are expected to promote (names in blue) or inhibit (names in green) cell proliferation. The plots display ChIP enrichment ratios for Ascl1 (red) and control (black) samples in promoter regions. Black arrows indicate the transcription start site and direction of transcription. Blue arrowheads indicate the position of primers used for ChIP-PCR validation. The expression patterns on sagittal sections of an E14.5 mouse telencephalon are from the public in situ hybridization database GenePaint (http://www.genepaint.org). Note that many Ascl1 targets are expressed throughout the telencephalon but are likely to be regulated by Ascl1 only in the ventral telencephalon. (B) Validation of Ascl1-bound segments by ChIP-PCR on immunoprecipitated material from an E12.5 ventral telencephalon. ORF1–3 are negative control regions described in the Supplemental Material. Mean ± SEM; quantification in triplicate from at least two immunoprecipitations. (C) Ascl1 target genes promote progression through multiple phases of the cell cycle. The genes listed include Ascl1 targets in both embryonic telencephalon and NS5 cells. Genes in black and red encode TFs and other types of proteins, respectively. Arrowheads indicate the main cell cycle checkpoints (G1, G2, and metaphase).
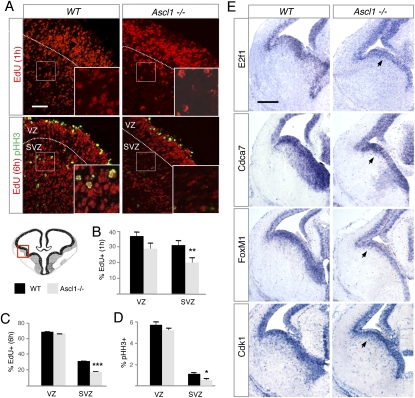
Figure 4.
Depletion of intermediate progenitors in the ventral telencephalon of Ascl1-null mutant embryos. (A) Immunolabeling of progenitor cells in E14.5 wild-type (WT) or Ascl1-null mutant (Ascl1−/−) embryos. Cells having incorporated EdU 1 h (S phase) or 6 h (M/early G1) after administration are labeled in red in the top and bottom panels, respectively. Cells expressing pHH3 (G2/M) are shown in green in the bottom panel. White dotted lines show the border between the VZ and SVZ, and small white rectangles show the areas enlarged in the insets. The red rectangle in the drawing below indicates the area of the ventral telencephalon illustrated. Bar, 50 μm. (B–D) Quantification of the fraction of cells having incorporated EdU 1 h (B) or 6 h (C) after administration or expressing pHH3 (D) in the VZ and SVZ of the ventral telencephalon in Ascl1-null mutant (gray bars) and wild-type (black bars) embryos. Mean ± SEM; (***) P < 0.001; (**) P < 0.01; (*) P < 0.05 with Student's t-test; n > 3000 cells for each condition; quantification from at least three sections from at least three embryos. (E) Expression of selected Ascl1 target genes with a predicted role in regulation of neural progenitor divisions, analyzed by in situ hybridization on frontal sections of ventral telencephalon from E14.5 wild-type and Ascl1-null mutant embryos. Arrowheads point to the domains of gene expression in the SVZ present in the wild-type and absent in the mutant embryo. Bar, 500 μm.
The regulation by Ascl1 of genes promoting cell proliferation might result in cell cycle defects in the absence of Ascl1. To address this possibility, we examined cell proliferation in the ventral telencephalon of Ascl1-null mutant embryos (Guillemot et al. 1993). Cells in S phase of the cell cycle, labeled with an antibody against the nucleoside analog EdU after a 1-h EdU pulse, were reduced by 32% in the SVZ of E14.5 Ascl1 mutant embryos when compared with wild-type embryos, but were not significantly reduced in the VZ (Fig. 4A,B). Cells in G2/M phase, labeled with an antibody against pHH3 or with the EdU antibody after a 6-h EdU pulse-chase, were also much less numerous in the mutant than the wild-type ventral telencephalic SVZ (reduced by 53% or 44%, respectively), while dividing stem cells in the VZ of Ascl1 mutant embryos were again unaffected (Fig. 4A–D). These results thus indicate that the null mutation of Ascl1 results in a loss of cycling intermediate progenitors in the SVZ of the ventral telencephalon. In agreement with this finding, the expression of Ascl1 targets that encode components of the E2F1 and FoxM1 pathways is strongly reduced in the SVZ of mutant embryos (Fig. 4E), although expression of other targets is reduced also in the VZ (e.g., Sox9 expression) (data not shown), suggesting additional roles of Ascl1 in VZ stem cells.
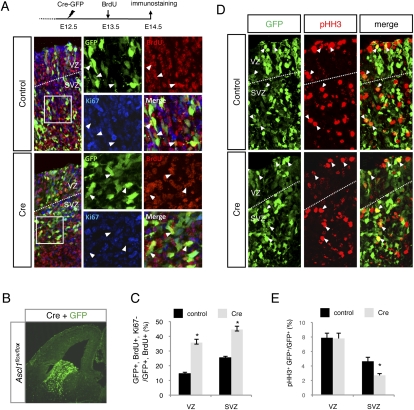
The absence of an overt proliferation defect in the VZ of the ventral telencephalon could be due to the activation of a compensatory pathway that sustains cell divisions in VZ stem cells in the absence of Ascl1. To address this possibility, we deleted Ascl1 acutely by electroporation of the Cre recombinase together with GFP in the ventral telencephalon of embryos homozygous for a conditional mutant allele of Ascl1 (Ascl1floxed) (Pacary et al. 2011). Embryos electroporated at E12.5 received an injection of BrdU at E13.5, and the phenotype of electroporated cells was analyzed at E14.5. Triple labeling for Ki67, BrdU, and GFP showed that the fraction of electroporated progenitors (GFP+) that had divided at E13.5 (BrdU+) and stopped cycling at E14.5 (Ki67−) was significantly increased in both the VZ and SVZ of Cre-electroporated embryos compared with control (GFP electroporated) embryos (by 146% and 76%, respectively) (Fig. 5A–C). The rate of cell cycle exit of GFP− nonelectroporated cells, however, was not increased, indicating that the cell cycle exit phenotype of Ascl1 mutant cells is cell-autonomous (data not shown). Since caspase 3 labeling did not show any increase in cell death (data not shown), we conclude that a large fraction of ventral telencephalic progenitors in both the VZ and SVZ exit the cell cycle prematurely when Ascl1 is acutely eliminated. Triple labeling of the same brains for GFP, BrdU, and the neuronal marker HuC/D indicated that Ascl1 mutant cells differentiate into neurons as efficiently as control cells (Supplemental Fig. S6). Double labeling for pHH3 and GFP also showed that acute loss of Ascl1 impaired progenitor divisions in the SVZ, although VZ cell divisions were unaffected (Fig. 5D,E).
Figure 5.
Reduced cell proliferation following acute deletion of Ascl1. (A) In utero electroporation of a control GFP vector (top panel) or a plasmid expressing Cre recombinase (bottom panel) together with GFP into the ventral telencephalon of E12.5 embryos carrying a conditional Ascl1-null mutant allele (Ascl1floxed), followed by BrdU administration at E13.5 and coimmunolabeling at E14.5 for GFP (green), BrdU (red), and Ki67 (blue). White dotted lines show the border between the VZ and SVZ, white arrowheads point to GFP+/BrdU+/Ki67− cells, and white rectangles show the areas enlarged in the insets. The design of the experiment is shown above the panels. Bar, 100 μm. (B) Illustration of an electroporated embryo labeled for GFP (green) showing extensive electroporation of the ventral telencephalon. (C) Quantification of the rate of cell cycle exit, corresponding to the fraction of electroporated cells cycling at E13.5 (GFP+/BrdU+) that are not dividing at E14.5 (GFP+/BrdU+/Ki67−), in the VZ and SVZ of control and Cre electroporated embryos. Mean ± SEM; (**) P < 0.01 with Student's t-test; n > 300 for each condition; quantification from at least three sections from at least three embryos from three different litters. (D) Same experiment as in A, except that electroporated brain sections were colabeled for pHH3 and GFP. White arrowheads point to double-labeled cells. Bar, 50 μm. (E) Quantification of the fraction of GFP+ coexpressing pHH3 in the VZ or SVZ of control (black bars) or Cre electroporated (gray bars) embryos. Mean ± SEM; (*) P < 0.05 with Student's t-test; n > 1400 cells for each condition; quantification from four sections from at least three embryos from three different litters.
Altogether, our genomic and genetic analyses showed that Ascl1 regulates a large number of cell cycle progression genes, and that this activity is required to maintain ventral telencephalic progenitors in cycle.
Ascl1 activates cell proliferation genes in NS cell cultures
ChIP–chip and the expression profiling experiments described above showed that Ascl1 activates two sets of target genes in the embryonic brain, with opposite roles in cell cycle control. Because the embryonic tissue used in these experiments contains a mixture of cells at different stages of the neurogenic process, it is unclear whether these two sets of target genes are regulated simultaneously or sequentially during neurogenesis. Although the distribution of their transcripts suggests that at least some of the Ascl1 targets that promote and inhibit the cell cycle are expressed sequentially (Fig. 3A), the low resolution of in situ hybridization data and the lack of information on the distribution of the protein products leave open the question of when, exactly, the different categories of Ascl1 target genes are active.
To focus our analysis on Ascl1 targets in cycling progenitors, we turned to a model of NS cells in culture. NS cell cultures contain homogenous populations of progenitors that divide indefinitely in the presence of EGF and FGF2 and differentiate into neurons and glial cells when these mitogens are withdrawn from the culture medium (Conti et al. 2005; Conti and Cattaneo 2010). The NS5 cell line dividing in the presence of mitogens expresses Ascl1, albeit in a heterogeneous manner due to temporal fluctuations in its expression levels (Fig. 6A; Supplemental Fig. S7). To identify Ascl1 target genes in these cycling cells, we characterized the genome-wide repertoire of Ascl1-bound promoters by performing ChIP–chip experiments with the same antibody against Ascl1 and the same promoter microarrays as before. This analysis showed that, in NS5 cells, Ascl1 is recruited to 617 sites corresponding to 603 promoters (with a false-positive rate of <10%) (Fig. 6B; Supplemental Fig. S8; Supplemental Table S6). A large proportion of these sites is also occupied by Ascl1 in the embryonic telencephalon (362 or 59%) (Fig. 6E), and Ascl1-binding profiles are very similar in the two cell types (Fig. 6C). Analysis of the promoters bound by Ascl1 in NS5 cells with Trawler produced a PWM almost identical to that obtained in the embryonic telencephalon (cf. Figs. 6D and 1E and Supplemental Fig. S2).
Figure 6.
Genome-wide location analysis of Ascl1 in NS5 cells. (A) Expression of Ascl1 (green) in proliferating NS5 cells analyzed by immunocytochemistry. The nuclei of NS5 cells are labeled with DAPI (blue). (B) ChIP-on-chip analysis of DNA segments bound by Ascl1 in proliferating NS5 cells. (C) Ascl1 binding to known target genes. The plots display ChIP enrichment ratios for NS5 (black) and embryonic telencephalon (red) samples for all probes in target promoters. Genes are shown to scale above the plots and genomic regions are shown below the plots. Arrows indicate the transcription start site and direction of transcription. (D) Motif enriched in Ascl1-bound segments in NS5 cells from a de novo search using Trawler software. (E) Venn diagram showing the overlap in Ascl1 promoter occupancy data between the embryonic telencephalon (red) and NS5 cells (blue). (F) Plot representing the fraction of Ascl1-binding events located at consecutive 0.5-kb intervals away from the transcription start site of target genes (red dots and line). A plot for a randomized sample of binding events is shown as control (black dots and line).
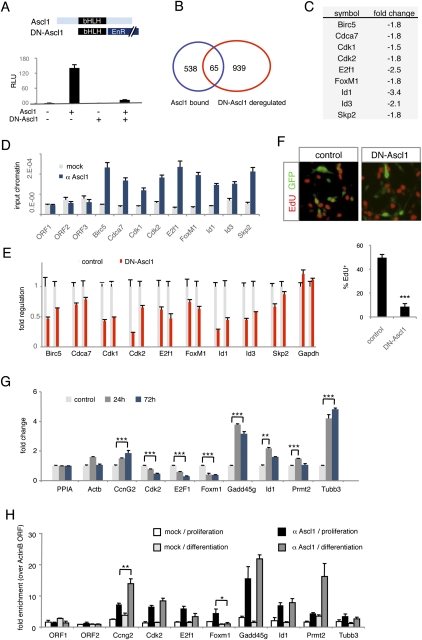
To identify the promoters that are actively regulated by Ascl1 in cultured NS cells, we transfected NS5 cells with a construct containing the basic helix–loop–helix (bHLH) domain of Ascl1 fused to the repressor domain of the Drosophila engrailed gene, which exerts a dominant-negative effect over wild-type Ascl1 (DN-Ascl1) (Fig. 7A). Expression profiling of DN-Ascl1-expressing NS cells isolated by fluorescence-activated cell sorting (FACS) 24 h after transfection identified 1004 significantly regulated genes, of which 518 (52%) were up-regulated and 486 (48%) were down-regulated (Supplemental Table S7). Of the 603 Ascl1-bound genes identified by location analysis in NS5 cells, 65 (11%) were regulated in this experiment (Fig. 7B; Supplemental Table S7). Among these direct targets, we found nine genes with well-described roles in cell cycle control, including four targets already identified in the embryonic telencephalon (Cdca7, E2f1, FoxM1, and Id1) (Fig. 7C; Supplemental Table S8). Three additional proliferation targets in NS cells (Birc5, Cdk2, and Id3) were also bound by Ascl1 in the embryonic telencephalon, although their expression was not significantly regulated in Ascl1 GOF or LOF experiments in vivo. Importantly, all cell cycle targets of Ascl1 in NS5 cells are known to promote cell proliferation. In contrast, the cell cycle arrest genes regulated by Ascl1 in the telencephalon were induced only when overexpression of Ascl1 drove NS5 cells out of the cell cycle (Supplemental Fig. S9; data not shown). Therefore, Ascl1 expressed in dividing progenitors regulates cell cycle progression genes and not cell cycle exit genes.
Figure 7.
A cell cycle regulatory module activated by Ascl1 in NS5 cells. (A) Ascl1-EnR acts as a dominant-negative version of Ascl1 in NS5 cells. The drawings depict wild-type Ascl1 and the Ascl1-EnR fusion protein containing the bHLH domain of Ascl1 (in black) and the Drosophila engrailed repressor domain (EnR, in dark blue). The graph shows the result of a transcriptional assay in which Ascl1-EnR blocks the transcriptional activity of wild-type Ascl1 in NS5 cells, measured by activation of a reporter construct expressing luciferase under the control of a multimerized Ascl1-binding site (E-box) and a minimal β-globin promoter. Mean ± SEM; n = 4. (B) Venn diagram showing the overlap between genes associated with an Ascl1-bound promoter (blue) and genes deregulated in an expression profiling analysis of DN-Ascl1-transfected NS5 cells (red). (C) Cell cycle regulators among Ascl1 target genes (bound by Ascl1 and deregulated by DN-Ascl1) and their respective fold changes in expression profiling analysis of DN-Ascl1-transfected cells compared with control vector-transfected cells. (D) Validation of Ascl1-bound segments by ChIP-PCR on immunoprecipitated material from NS5 cells. Mean ± SEM; quantification in triplicate from at least two immunoprecipitations. (E) Changes in expression levels of cell cycle target genes 24 h after transfection of NS5 cells with a control (black) or DN-Ascl1-expressing vector (red), quantified by real-time PCR. Mean ± SEM; n = 3. (F) Proliferation of NS5 cells expressing a control vector or DN-Ascl1 measured by the rate of EdU incorporation among transfected cells following exposure for a period of 9 h, 24 h after transfection. The pictures above the graph show labeling for EdU (red) and GFP (green) to mark cells transfected with the control vector (left) or Ascl1-EnR (right). Mean ± SEM; (***) P < 0.001 with Student's t-test; n = 6. (G) Change in expression levels of Ascl1 cell cycle targets in NS5 cells 24 and 72 h after induction of neuronal differentiation compared with proliferating NS5 cells (control) measured by real-time PCR. Mean ± SEM; n = 3. (H) Changes in Ascl1 binding to cell cycle targets in NS5 cells 48 h after induction of neuronal differentiation compared with proliferating NS5 cells, measured by ChIP-PCR. Mean ± SEM; (**) P < 0.01 with Student's t-test; n = 6.
To directly examine the role of Ascl1 in NS cell proliferation, we exposed DN-Ascl1-transfected and control GFP-transfected NS5 cells to EdU 24 h after transfection and measured the fraction of GFP+-transfected cells having incorporated EdU and that had thus progressed through S phase. Expression of DN-Ascl1 resulted in an 83% reduction in the fraction of EdU+ NS5 cells (Fig. 7F). To confirm this result, we performed a partial knockdown of Ascl1 in NS5 cells by adenoviral administration of an Ascl1 shRNA or a control shRNA, which resulted in a 38% reduction of EdU incorporation 24 h later (Supplemental Fig. S10). Ascl1 is therefore an important regulator of cell cycle progression in cultured NS cells.
We next investigated the mechanisms that promote the switch of Ascl1 targets between proliferating and differentiating cells. First, we differentiated NS5 cells into neurons using an efficient procedure described in Spiliotopoulos et al. (2009), and we examined how expression of Ascl1 and its targets changed as NS5 cells differentiate. Ascl1 remained expressed at similar levels in differentiating NS5 cells as in proliferating cells (Supplemental Fig. S11), but expression of numerous Ascl1 targets changed significantly, including a down-regulation of cell cycle progression genes (including Cdk2, E2f1, and FoxM1) and an up-regulation of the cell cycle arrest genes Ccng2 and Gadd45g and the neuronal gene Tubb3 (Fig. 7G). We next examined whether Ascl1 binding to its target genes was affected by NS5 cell differentiation. Ascl1 binding was indeed reduced at the Foxm1 locus and increased at the Ccng2 locus (Fig. 7H). However, binding to other targets did not change significantly (e.g., Cdk2, E2f1, Gadd45g, and Tubb3) (Fig. 7H), suggesting that other mechanisms that change binding site occupancy contribute to the changes of Ascl1 target gene expression between proliferating and differentiating cells.
We next considered the possibility that Ascl1 interacts with different TFs on promoters active in proliferating cells versus promoters active in differentiating cells. To address this possibility, we searched for DNA sequence motifs that are differentially represented in the vicinity of Ascl1-binding events in the promoters of Ascl1 targets promoting cell cycle progression (18 genes) (Supplemental Table S5) and the promoters of targets promoting cell cycle exit or expressed in differentiating neurons (18 genes) (Supplemental Fig. S12) using the algorithm Cisfinder (Sharov and Ko 2009). As expected, the PWM GCAGCTG, corresponding to the consensus Ascl1-binding motif, was enriched in the two data sets, but another PWM, CTGGGAAC—matching closely the consensus binding sequence GTGGGAA for the DNA-binding nuclear adaptor protein from the Notch signaling pathway, CBF1/RBPj (Tun et al. 1994)—was overrepresented only in the promoters of cell cycle progression genes (Supplemental Fig. S12). This finding therefore suggests a model whereby coregulation of the cell cycle progression targets of Ascl1 by Notch signaling results in their expression in proliferating progenitors where Notch signaling is active, and their repression in differentiating neurons where Notch signaling is inhibited and the CBF1/NotchICD-activating complex is replaced by a CBF1/corepressor complex (Kao et al. 1998).
Discussion
In this study, we characterized the genetic program regulated by Ascl1 in the embryonic brain by combining location analysis of Ascl1-bound sites in the embryonic telencephalon and NS cell cultures with expression profiling of genes deregulated by overexpression or deletion of Ascl1. This is the first genome-scale analysis of the transcriptional program activated by a mammalian proneural factor. We chose to perform this study by ChIP–chip (hybridization of immunoprecipitated DNA to promoter oligonucleotide arrays) rather than ChIP-seq (high-throughput sequencing of the immunoprecipitated DNA) to focus on the identity of target genes and the discovery of novel biological functions of Ascl1 rather than its global regulatory activity. An ongoing location analysis by ChIP-seq has shown that Ascl1 is recruited at many genomic sites located at large distances from transcription start sites and is therefore difficult to associate unambiguously with target genes (DS Castro, B Martynoga, and F Guillemot, unpubl.).
This study led to the identification of a novel role for Ascl1 in the proliferation of neural progenitors. We discuss below the nature of the genetic program of neurogenesis that we uncovered, and particularly of the subprogram promoting cell divisions, and we consider the mechanisms that may promote the switch of Ascl1 activity from promoting cell proliferation to triggering cell cycle arrest and differentiation.
The genetic program of neurogenesis in the ventral telencephalon
Ascl1 and other proneural genes have the remarkable capacity to activate an entire program of neurogenesis when overexpressed in neural progenitors or some nonneural cells (Farah et al. 2000; Nakada et al. 2004; Berninger et al. 2007b; Geoffroy et al. 2009; Vierbuchen et al. 2010). Whether proneural genes act by inducing TF cascades or regulating effector genes that function at multiple stages during neurogenesis was not known. This study provides evidence that Ascl1 does indeed induce many TFs, but also many other types of protein. Ascl1 regulates a large array of downstream TFs, some of which were implicated previously in the specification or differentiation of neural cells; e.g., Sox4 (Bergsland et al. 2006), Dlx2 (Anderson et al. 1997; Petryniak et al. 2007), Ebf3 (Zhao et al. 2006), Gli3 (Kuschel et al. 2003), and Nf1b (Steele-Perkins et al. 2005). These results greatly expand the number of TFs known to act downstream from Ascl1 in the ventral telencephalon (Castro et al. 2006; Poitras et al. 2007; Farkas et al. 2008) or other cell lineages (Cau et al. 1997, 2002; Hu et al. 2004). These downstream TFs must amplify and diversify the initial input from Ascl1 expression and, assuming they regulate each other, may confer stability and robustness to the neurogenic program (Blais and Dynlacht 2005; Blais et al. 2005; Orkin et al. 2008).
However, most Ascl1 targets are not involved in transcriptional regulation. Their molecular functions range from signal transduction (e.g., Cxcr4 and Rgs16) to cell adhesion (Itgb1 and Cdh8) and cytoskeleton regulation (Kif1a and Pak3), and the cellular activities they regulate include axonogenesis and neurite morphogenesis (Efna1 and Plxna2), neurotransmitter biosynthesis (Gad2 and Grip1), and cell cycle control (see below). These results indicate that Ascl1 directly controls both early and late stages of neurogenesis. This finding may hold true for other proneural proteins such as Neurogenin2, which has been shown to promote neuronal migration, a late step in neurogenesis, via direct regulation of a small Rho GTPase and other effector genes (Ge et al. 2006; Heng et al. 2008). The direct transcriptional activation of a large part of the neurogenic program is a unique feature of proneural proteins, which may be largely responsible for the neuronal reprogramming activity of Ascl1 in astrocytes (Berninger et al. 2007a) and fibroblasts (Vierbuchen et al. 2010).
Ascl1 sequentially activates cell cycle progression and cell cycle arrest genes
The most unexpected finding from this study is that Ascl1 activates a large number of positive cell cycle regulators in the embryonic telencephalon and NS cells. Ascl1 target genes include several canonical cell cycle regulators that are essential for G1/S transition (e.g., E2f1, Cdk1, Cdk2, and Skp2) or entry into mitosis (e.g., Cdk1 and Cdc25b, a target identified in a previous study) (Castro et al. 2006). Other Ascl1 targets have additional roles besides cell cycle control. FoxM1 has an important role in progression of normal and tumor cells through M phase and also promotes angiogenesis through activation of vascular endothelial growth factor (VEGF) expression (Schuller et al. 2007; Fu et al. 2008; Zhang et al. 2008). Cyr61/Ccn1, a secreted heparin-binding protein, also promotes both cell proliferation and angiogenesis (Chen and Du 2007). Ascl1 therefore promotes cell cycle progression at both G1/S and G2/M transitions, and may also coordinate cell cycle regulation with other aspects of the biology of neural progenitors. Ascl1 shares some of its targets with c-Myc and N-Myc—including Birc5, Cdca7, Ccnd2, Foxm1, and the Notch ligand Dll3 (Bouchard et al. 1999; Goto et al. 2006; Kidder et al. 2008; Wierstra and Alves 2008; Zhao et al. 2009)—suggesting that its role in mitotic progenitors may involve cooperation with Myc proteins. In the absence of Ascl1, Myc proteins may be able to sustain the division of certain progenitors (e.g., in the telencephalic VZ).
Direct targets of Ascl1 in the embryonic telencephalon also include genes involved in cell cycle arrest—including Fbxw7, Gadd45g, Ccng2, Hipk2, and Prmt2—thus demonstrating that Ascl1 regulates two sets of targets with opposite roles in cell cycle control. Interestingly, several of the cell cycle arrest targets have distinct spatial and/or temporal expression patterns from those of cell cycle progression targets in both the embryonic brain and NS5 cells, suggesting that they are regulated during a later phase of neurogenesis. Two findings support this assertion. First, cell cycle arrest targets are strongly up-regulated when NS5 cells are induced to differentiate by overexpression of Ascl1, and they are not affected by overexpression of DN-Ascl1 (Supplemental Fig. S5). Second, their transcripts are found mostly outside of the VZ in the embryonic telencephalon (Fig. 3). Gadd45g and Fbxw7 transcripts are localized to the SVZ (Castro et al. 2006) and may be expressed by progenitor cells that are going through their last division or are already post-mitotic. Ccng2 and Prmt2 transcripts are present mostly in the MZ, where these genes may prevent cell cycle re-entry of post-mitotic neurons.
Together, our results suggest that Ascl1 has a role in both cell cycle promotion and cell cycle termination, and that the transition between these two functions involves the coordinated down-regulation and induction of multiple target genes. It is unusual that the same TF promotes in the same lineage both the expansion of progenitors and their subsequent division arrest and differentiation. Interestingly, Asense, the Ascl1 ortholog in Drosophila, has been shown recently to promote neuroblast self-renewal (Southall and Brand 2009), while earlier studies had suggested a role of Asense in inhibiting cell divisions of neuroblast daughter cells (Wallace et al. 2000), suggesting that this dual activity is an evolutionarily conserved feature of the Ascl1 gene family. A selective advantage may be that it allows for a rapid and efficient switch from a proliferating progenitor state to a post-mitotic neuronal state and ensures that the regulatory programs that underpin these two states are mutually exclusive.
The processes that select which set of target genes Ascl1 regulates in progenitors versus differentiating neurons are not currently known. A differential response of cell cycle progression genes and cell cycle arrest genes to different levels of Ascl1 (e.g., Gaudet and Mango 2002) is ruled out by our observation that Ascl1 protein levels do not increase substantially when NS cells differentiate. Other mechanisms, however, can be envisaged. The differential binding of Ascl1 to the Ccng2 and Foxm1 promoters in proliferating versus differentiating NS5 cells suggests that changes in Ascl1 structure (e.g., post-translational modifications such as phosphorylation) (Hand et al. 2005; Ma et al. 2008) and/or Ascl1 regulation (e.g., a switch from oscillatory to stable expression) (Kageyama et al. 2008) control Ascl1–promoter interactions and possibly other aspects of target gene regulation (e.g., Chen and Greene 2004; Mengel et al. 2010). Other pathways coregulating Ascl1 targets may also contribute to the change in the Ascl1 transcriptional program as neural progenitors stop proliferating and begin to differentiate. The enrichment of a consensus motif for CBF1/RBPj binding in promoters of cell cycle progression targets raises the intriguing possibility that coregulation by Ascl1 and the Notch pathway results in expression of these genes in proliferating progenitors where Notch is signaling and their repression in differentiating neurons where the Notch pathway is inactive. Elucidating the mechanisms involved in the selection of Ascl1 targets will shed light on the pathways that control the switch from proliferation to differentiation during neural development.
Ascl1 promotes cell divisions in the embryonic brain and NS cell cultures
Prompted by the discovery of positive cell cycle regulators among Ascl1 targets, we re-examined the phenotype of Ascl1 mutant progenitors and found that mutation of Ascl1 results in a loss of dividing intermediate progenitors in the ventral telencephalon. Inhibition of Ascl1 activity in cultured NS cells also reduced their rate of divisions. Ascl1 was well known to terminate progenitor divisions (Farah et al. 2000; Nakada et al. 2004), but there was little evidence so far that it also sustains cell proliferation (Yi et al. 2008). Promoting progenitor divisions, however, may be a general property of Ascl1, since deletion of Ascl1 in the adult brain also results in a severe loss of dividing stem cells and progenitors in the SVZ and the dentate gyrus (M Lebel-Potter, C Parras, C Hunt, and F Guillemot, unpubl.).
Some Ascl1-expressing neural lineages contain highly proliferative intermediate progenitors, as in the ventral telencephalon and the adult SVZ, but other lineages have intermediate progenitors that divide only briefly (as in the embryonic cortex and adult dentate gyrus) or have no intermediate progenitors at all (in the midbrain and spinal cord). Interestingly, in these latter situations, Ascl1 expression is quickly followed by that of Neurog2, which, at least in the cortex, switches off Ascl1 expression (Helms et al. 2005; Britz et al. 2006; Kele et al. 2006; Ozen et al. 2007). This suggests that Neurog2 does not share the proproliferative property of Ascl1, and that, on the contrary, it may be involved in suppressing the Ascl1-dependent generation and/or expansion of intermediate progenitors.
The discovery of the role of Ascl1 in neural progenitor divisions may have implications beyond brain development, particularly in the cancer field. High levels of Ascl1 expression have been observed in a wide range of neuroendocrine tumors, including small cell lung carcinomas (Ball 2004), prostate tumors (Vias et al. 2008), medullary thyroid cancers (Chen et al. 2005), and gastroenteropancreatic tumors (Shida et al. 2008). A direct role of Ascl1 in neurodendocrine tumor formation is supported by experiments of overexpression of Ascl1 together with SV40 Large T Antigen in lung epithelium, which produce massive neuroendocrine tumors (Linnoila et al. 2000), and Ascl1 knockdown experiments, which inhibit proliferation of lung cancer cells both in vitro and in vivo (Osada et al. 2005; Jiang et al. 2009). Ascl1 is also overexpressed in several types of gliomas, including grade II and grade III astrocytomas and a subset of glioblastoma multiforme (Somasundaram et al. 2005; Phillips et al. 2006; Verhaak et al. 2010). The demonstration that Ascl1 promotes the proliferation of neural progenitors through regulation of multiple cell cycle control genes—including a major oncogene in gliomas, E2f1 (Alonso et al. 2008)—raises the intriguing possibility that deregulation of Ascl1 is causally linked to the generation of these highly malignant and common forms of brain tumors.
Materials and methods
ChIP
Basal ganglia from E12.5 mouse embryos or cultured NS5 cells were fixed in 1% formaldehyde for 15 min and lysed in 1% SDS, 10 mM EDTA, and 50 mM Tris. Sonication was performed using a Bioruptor (Diagenode) at high power settings for 45 cycles (30 sec on/30 sec off). Immunoprecipitations with mouse anti-Ascl1 (Pharmingen) or control LEAF-purified mouse IgG1 (Cambridge Biosciences) antibodies were done as described (Castro et al. 2006) using 80 μg of chromatin per assay. DNA sequences were quantified by real-time PCR (primers are listed in the Supplemental Material) by using a 7500 Real-Time PCR System (Applied Biosystems) and a SYBR green-based kit for quantitative PCR (iQ Supermix, Bio-Rad). Quantities of immunoprecipitated DNA were calculated by comparison with a standard curve generated by serial dilutions of input DNA. The data were plotted as means of at least two independent ChIP assays and three independent amplifications; error bars represent standard error of the mean. For hybridization onto microarrays, each sample was obtained from pooling the product of two immunoprecipitations followed by amplification using a WGA kit (Sigma), as described previously (O'Geen et al. 2006).
ChIP-on-chip array hybridization and data extraction
Amplified ChIP material and chromatin input DNA were labeled by incorporation of Cy3 or Cy5-labeled nucleotides (Bio-Rad) using the Bioprime array CGH genomic labeling system (Invitrogen) and hybridized to Agilent mouse promoter 244K arrays according to the manufacturer's protocols. Slides were scanned in an Agilent scanner and data were extracted using an Agilent Feature Extractor. Data were normalized by Intra-array Median normalization followed by Intra-array Lowess normalization, and were analyzed with the neighbor error model in order to calculate the probability scores associated with Ascl1 binding to each array oligonucleotide {p[Xbar] and p(X}, all methods implemented in ChIP-Analytics software (Agilent). Binding events were identified by sliding a window encompassing three probes across genomic regions. A binding event was called if p[Xbar] < 0.001 and either the central probe has p(X) < 0.001 and at least one neighboring probe has p(X) < 0.1, or at least one neighbor has p(X) < 0.005. Consecutive bound probes with maximum 1000-base-pair (bp) gaps were collapsed into segments. Binding events from two data sets were considered overlapping if corresponding segments overlapped by at least 200 nucleotides (nt).
Expression profiling of Ascl1 GOF and LOF
Microarray data from Ascl1 GOF have been described previously (Gohlke et al. 2008). To generate the Ascl1 LOF data set, basal ganglia from litters of E13.5 Ascl1-null mutant embryos (Guillemot et al. 1993) were processed for RNA Tryzol extraction as described (Schuurmans et al. 2004). Three replicates of Ascl1 mutant and control embryos were analyzed, each sample with tissue pooled from at least three embryos. Expression profiling was done using Affymetrix MOE430 2.0 arrays. Raw image files were processed using Affymetrix GCOS and the Microarray Suite (MAS) 5.0 algorithm and normalization using GCRMA software (Irizarry et al. 2003). Statistical analysis was performed using ANOVA with a statistical significance threshold determined at α ≤ 0.05 with a P-value correction using the Benjamini-Hochberg method and a kernel-based false-discovery rate (FDR) method. The fold cutoff for deregulated genes was set to 1.25.
Motif search and Gene Ontology analyses
To find statistically overrepresented motifs associated with Ascl1-binding events in the genome-wide set of Ascl1 target genes, the highest-confidence probe (lowest p[Xbar]) was selected for each segment. Selected probes were extended by 100 nt in both directions, and the repeat masked sequence for the corresponding region was obtained. The background sets necessary to assess motif overrepresentation were generated by computing continuous regions spanning the full promoters after extending each probe to a maximum of 1.5 kb in both directions and merging overlapping regions. Two random sets (covering 10 times more sequence than the corresponding peak set) of background regions were selected for NS5 and telencephalon samples. Trawler was run combining peak and background sets (minimum motif length = 7; minimum number of occurrence = 15). Gene Ontology analysis was performed using GOToolBox (http://genome.crg.es/GOToolBox), using the list of tiled promoters as the reference data set.
In utero electroporation
In utero electroporation of Ascl1 floxed embryos was performed as described (Nguyen et al. 2006). DNA (pClG2 and pClG2-Cre) was prepared in endotoxin-free conditions (Qiagen), mixed at 1 μg/μL with 0.05% Fast Green (Sigma), and injected through the uterine wall into the telencephalic vesicle using pulled borosilicate needles (Harvard Apparatus) and a Femtojet microinjector (Eppendorf). Five electrical pulses were applied at 30 V (50-msec duration) across the uterine wall at 1-sec intervals using 5-mm platinum electrodes (Tweezertrode 45-0489, BTX, Harvard Apparatus) connected to an electroporator (ECM830, BTX). The day following the surgery, pregnant mice were injected with 100 mg/kg BrdU and sacrificed the next day.
Immunohistochemistry and in situ hybridization
Embryonic brains were fixed in 4% PFA overnight and then placed in 20% sucrose/PBS overnight. Brains were then embedded in OCT Compound and frozen before sectioning using a cryostat (Leica). Ten-micron cryostat sections were processed for in situ hybridization or immunostaining as described in the Supplemental Material. EdU incorporation was assessed using the Click-iT EdU system (Invitrogen) according to the manufacturer's instructions previous to immunostaining (1 mg of EdU per animal administered by intraperitoneal injection). Ascl1 staining was performed using anti-Ascl1 antibody from Pharmingen. All images were acquired with a laser-scanning confocal microscope (Radiance 2100, Bio-Rad), or bright field (Axioplan 2 imaging, Zeiss). The border between the VZ and SVZ was defined by the position of abventricular pHH3+ cells in the same sections or adjacent sections. Cell counts were performed using MetaMorph software (Molecular Devices). A statistical analysis was performed using an unpaired two-tailed Student's t-test between control and experimental condition (StatView software, version 5).
Culture and gene expression studies in NS cells
NS5 cells were cultured as described previously (Conti et al. 2005) and were nucleofected using an AAD-1001 nucleofector device with mouse NS cell nucleofector solution (Amaxa), program A-033, and 5 μg of DNA per 5 million cells. Proliferation assays were done with a 9-h pulse of EdU 24 h after nucleofection using the Click-iT EdU system (Invitrogen) according to the manufacturer's instructions. TaqMan gene expression assays (Applied Biosystems) were performed on cDNA produced with High-Capacity RNA-to-cDNA Master Mix (Applied Biosystems) after Trizol RNA extraction. For expression profiling, cells nucleofected with DN-Ascl1 (the bHLH domain, residues 115–173, in-frame with the EngR domain, was cloned upstream of an IRES and an NLS-tagged GFP in the pCAGGS expression vector) or empty plasmid were FACS-sorted for GFP expression 24 h after nucleofection and were profiled using MouseRef8 version 2 beadarrays (Illumina), three replicates used per condition. Luciferase assays were performed as described previously (Castro et al. 2006) after transfection with Lipofectamine Plus (Invitrogen) for NS5 cells or Lipofectamine 2000 (Invitrogen) for P19 cells.
NS5 cells were induced to differentiate into neurons according to the protocol described by Spiliotopoulis et al. (2009). Nearly confluent cultures were trypsinized and plated into laminin-coated flasks in D1 medium (NS cell basal medium with 1% B27, 0.5% N2, 5 ng/mL FGF2). After 3 d in these conditions, the majority of cells expressed the neuronal markers MAP2 and Tuj1, with very few GFAP-positive glial cells (data not shown), as described previously (Spiliotopoulos et al. 2009).
To generate Ascl1-GFP NS cells, ventral telencephalons from E14.5 Ascl1-GFP transgenic mice [NIH GENSAT Tg(Ascl1-EGFP)AU176Gsat/Mmnc] (Gong et al. 2003) were dissected, mechanically dissociated, and cultured according to Conti et al. (2005) to generate a homogeneous NS cell culture. Cells used in this study were subjected to 20–30 passages post-derivation, and Ascl1 promoter-driven GFP was confirmed by fluorescent microscopy. Ascl1-GFP NS cells were trypsinized and subjected to FACS on a FACS ARIA II (Becton Dickinson). The 5% of cells with the brightest and weakest levels of GFP fluorescence were sorted to 90%–95% purity. Dead cells were excluded by their incorporation of 7-Aminoactinomysin D (7-AAD; Calbiochem). Cells were sorted into fresh medium and cultured under normal NS cell conditions. After recovery and expansion for 7 d, cells were retrypsinized and subjected to exactly the same FACS protocol, maintaining all machine settings and analysis gates.
Acknowledgments
We thank Mike Hubank, Doug King, and UCL Genomics for the hybridization of the promoter arrays. We are grateful to Austin Smith for providing NS5 cells, David Rowitch for providing the FoxM1 in situ probe, and Martin Lévesque for discussion on Ascl1 expression in cancers. We thank James Briscoe and members of the Guillemot laboratory for suggestions on the manuscript. D.S.C. was supported by a salary from the Wellcome Trust (082347/Z/07/Z), and B.M. and M.L.P. were supported by career development fellowships from the Medical Research Council (UK). This work was supported by a project grant from the Wellcome Trust (082347/Z/07/Z) and institutional funds from the Medical Research Council to F.G. (U117570528).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.627811.
Supplemental material is available for this article.
References
- Alonso MM, Alemany R, Fueyo J, Gomez-Manzano C 2008. E2F1 in gliomas: a paradigm of oncogene addiction. Cancer Lett 263: 157–163 [DOI] [PubMed] [Google Scholar]
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL 1997. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron 19: 27–37 [DOI] [PubMed] [Google Scholar]
- Arachchige Don AS, Dallapiazza RF, Bennin DA, Brake T, Cowan CE, Horne MC 2006. Cyclin G2 is a centrosome-associated nucleocytoplasmic shuttling protein that influences microtubule stability and induces a p53-dependent cell cycle arrest. Exp Cell Res 312: 4181–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball DW 2004. Achaete-scute homolog-1 and Notch in lung neuroendocrine development and cancer. Cancer Lett 204: 159–169 [DOI] [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J 2006. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev 20: 3475–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninger B, Costa MR, Koch U, Schroeder T, Sutor B, Grothe B, Gotz M 2007a. Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. J Neurosci 27: 8654–8664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninger B, Guillemot F, Gotz M 2007b. Directing neurotransmitter identity of neurones derived from expanded adult neural stem cells. Eur J Neurosci 25: 2581–2590 [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F 2002. Proneural genes and the specification of neural cell types. Nat Rev Neurosci 3: 517–530 [DOI] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD 2005. Constructing transcriptional regulatory networks. Genes Dev 19: 1499–1511 [DOI] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD 2005. An initial blueprint for myogenic differentiation. Genes Dev 19: 553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M 1999. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J 18: 5321–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britz O, Mattar P, Nguyen L, Langevin LM, Zimmer C, Alam S, Guillemot F, Schuurmans C 2006. A role for proneural genes in the maturation of cortical progenitor cells. Cerebral Cortex 16: i138–i151 doi: 10.1093/cercor/bhj168 [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH 2008. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev 22: 3320–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarosa S, Fode C, Guillemot F 1999. Mash1 regulates neurogenesis in the ventral telencephalon. Development 126: 525–534 [DOI] [PubMed] [Google Scholar]
- Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley J, Nguyen L, Gossler A, Gottgens B, et al. 2006. Proneural bHLH and Brn proteins co-regulate a neurogenic programme through cooperative binding to a conserved DNA motif. Dev Cell 11: 831–844 [DOI] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Fode C, Guillemot F 1997. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development 124: 1611–1621 [DOI] [PubMed] [Google Scholar]
- Cau E, Casarosa S, Guillemot F 2002. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development 129: 1871–1880 [DOI] [PubMed] [Google Scholar]
- Chen Y, Du XY 2007. Functional properties and intracellular signaling of CCN1/Cyr61. J Cell Biochem 100: 1337–1345 [DOI] [PubMed] [Google Scholar]
- Chen LF, Greene WC 2004. Shaping the nuclear action of NF-κB. Nat Rev Mol Cell Biol 5: 392–401 [DOI] [PubMed] [Google Scholar]
- Chen H, Kunnimalaiyaan M, Van Gompel JJ 2005. Medullary thyroid cancer: the functions of raf-1 and human achaete-scute homologue-1. Thyroid 15: 511–521 [DOI] [PubMed] [Google Scholar]
- Conti L, Cattaneo E 2010. Neural stem cell systems: physiological players or in vitro entities? Nat Rev Neurosci 11: 176–187 [DOI] [PubMed] [Google Scholar]
- Conti L, Pollard SM, Gorba T, Reitano E, Toselli M, Biella G, Sun Y, Sanzone S, Ying QL, Cattaneo E, et al. 2005. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol 3: e283 doi: 10.1371/journal.pbio.0030283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwiller L, Paten B, Ramialison M, Birney E, Wittbrodt J 2007. Trawler: de novo regulatory motif discovery pipeline for chromatin immunoprecipitation. Nat Methods 4: 563–565 [DOI] [PubMed] [Google Scholar]
- Farah MH, Olson JM, Sucic HB, Hume RI, Tapscott SJ, Turner DL 2000. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development 127: 693–702 [DOI] [PubMed] [Google Scholar]
- Farkas LM, Haffner C, Giger T, Khaitovich P, Nowick K, Birchmeier C, Paabo S, Huttner WB 2008. Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron 60: 40–55 [DOI] [PubMed] [Google Scholar]
- Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S 2007. shRNA knockdown of Bmi-1 reveals a critical role for p21–Rb pathway in NSC self-renewal during development. Cell Stem Cell 1: 87–99 [DOI] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F 2000. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev 14: 67–80 [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Malureanu L, Huang J, Wang W, Li H, van Deursen JM, Tindall DJ, Chen J 2008. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol 10: 1076–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, Mango SE 2002. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science 295: 821–825 [DOI] [PubMed] [Google Scholar]
- Ge W, He F, Kim KJ, Blanchi B, Coskun V, Nguyen L, Wu X, Zhao J, Heng JI, Martinowich K, et al. 2006. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc Natl Acad Sci 103: 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy CG, Critchley JA, Castro DS, Ramelli S, Barraclough C, Descombes P, Guillemot F, Raineteau O 2009. Engineering of dominant active basic helix–loop–helix proteins that are resistant to negative regulation by postnatal central nervous system antineurogenic cues. Stem Cells 27: 847–856 [DOI] [PubMed] [Google Scholar]
- Gohlke JM, Armant O, Parham FM, Smith MV, Zimmer C, Castro DS, Nguyen L, Parker JS, Gradwohl G, Portier CJ, et al. 2008. Characterization of the proneural gene regulatory network during mouse telencephalon development. BMC Biol 6: 15 doi: 10.1186/1741-7007-6-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. 2003. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425: 917–925 [DOI] [PubMed] [Google Scholar]
- Goto Y, Hayashi R, Muramatsu T, Ogawa H, Eguchi I, Oshida Y, Ohtani K, Yoshida K 2006. JPO1/CDCA7, a novel transcription factor E2F1-induced protein, possesses intrinsic transcriptional regulator activity. Biochim Biophys Acta 1759: 60–68 [DOI] [PubMed] [Google Scholar]
- Gotz M, Huttner WB 2005. The cell biology of neurogenesis. Nat Rev Mol Cell Biol 6: 777–788 [DOI] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL 1993. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell 75: 463–476 [DOI] [PubMed] [Google Scholar]
- Hand R, Bortone D, Mattar P, Nguyen L, Heng I-TK, Guerrier S, Boutt E, Peters E, Barnes AP, Parras C, et al. 2005. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron 48: 45–62 [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB 2004. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci 101: 3196–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry Y, Ramialison M, Paten B, Wittbrodt J, Ettwiller L 2010. Using Trawler_standalone to discover overrepresented motifs in DNA and RNA sequences derived from various experiments including chromatin immunoprecipitation. Nat Protoc 5: 323–334 [DOI] [PubMed] [Google Scholar]
- Helin K 1998. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev 8: 28–35 [DOI] [PubMed] [Google Scholar]
- Helms AW, Battiste J, Henke RM, Nakada Y, Simplicio N, Guillemot F, Johnson JE 2005. Sequential roles for Mash1 and Ngn2 in the generation of dorsal spinal cord interneurons. Development 132: 2709–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JI-T, Nguyen L, Castro DS, Zimmer C, Wildner H, Armant O, Skowronska-Krawczyk D, Bedogni F, Matter JM, Hevner RF, et al. 2008. Neurogenin2 controls cortical neuron migration through regulation of Rnd2. Nature 455: 114–118 [DOI] [PubMed] [Google Scholar]
- Henke RM, Meredith DM, Borromeo MD, Savage TK, Johnson JE 2009. Ascl1 and Neurog2 form novel complexes and regulate Delta-like3 (Dll3) expression in the neural tube. Dev Biol 328: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton S, Meredith A, Richardson JA, Johnson JE 1999. Correct coordination of neuronal differentiation events in ventral forebrain requires the bHLH factor MASH1. Mol Cell Neurosci 14: 355–369 [DOI] [PubMed] [Google Scholar]
- Hu Y, Wang T, Stormo GD, Gordon JI 2004. RNA interference of achaete-scute homolog 1 in mouse prostate neuroendocrine cells reveals its gene targets and DNA binding sites. Proc Natl Acad Sci 101: 5559–5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Kubish GM, Redmond TM, Turner DL, Thompson RC, Murphy GG, Uhler MD 2010. Direct transcriptional induction of Gadd45γ by Ascl1 during neuronal differentiation. Mol Cell Neurosci 44: 282–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jiang T, Collins BJ, Jin N, Watkins DN, Brock MV, Matsui W, Nelkin BD, Ball DW 2009. Achaete-scute complex homologue 1 regulates tumor-initiating capacity in human small cell lung cancer. Cancer Res 69: 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johe KK, Hazel TG, Muller T, Dugich-Djordjevic MM, McKay RD 1996. Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes Dev 10: 3129–3140 [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I 2008. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat Neurosci 11: 1247–1251 [DOI] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T 1998. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev 12: 2269–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kele J, Simplicio N, Ferri AL, Mira H, Guillemot F, Arenas E, Ang SL 2006. Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development 133: 495–505 [DOI] [PubMed] [Google Scholar]
- Kidder BL, Yang J, Palmer S 2008. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS ONE 3: e3932 doi: 10.1371/journal.pone.0003932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein AR, Noctor SC 2004. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci 27: 392–399 [DOI] [PubMed] [Google Scholar]
- Kuschel S, Ruther U, Theil T 2003. A disrupted balance between Bmp/Wnt and Fgf signaling underlies the ventralization of the Gli3 mutant telencephalon. Dev Biol 260: 484–495 [DOI] [PubMed] [Google Scholar]
- Linnoila RI, Zhao B, DeMayo JL, Nelkin BD, Baylin SB, DeMayo FJ, Ball DW 2000. Constitutive achaete-scute homologue-1 promotes airway dysplasia and lung neuroendocrine tumors in transgenic mice. Cancer Res 60: 4005–4009 [PubMed] [Google Scholar]
- Ma YC, Song MR, Park JP, Henry Ho HY, Hu L, Kurtev MV, Zieg J, Ma Q, Pfaff SL, Greenberg ME 2008. Regulation of motor neuron specification by phosphorylation of neurogenin 2. Neuron 58: 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL 2000. Origin and molecular specification of striatal interneurons. J Neurosci 20: 6063–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengel B, Hunziker A, Pedersen L, Trusina A, Jensen MH, Krishna S 2010. Modeling oscillatory control in NF-κB, p53 and Wnt signaling. Curr Opin Genet Dev 20: 656–664 [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M 2004. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development 131: 3133–3145 [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ 2003. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425: 962–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada Y, Hunsaker TL, Henke RM, Johnson JE 2004. Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus neuronal cell-type specification. Development 131: 1319–1330 [DOI] [PubMed] [Google Scholar]
- Nguyen L, Besson A, Heng JI, Schuurmans C, Teboul L, Parras C, Philpott A, Roberts JM, Guillemot F 2006. p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev 20: 1511–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M, Schuurmans C, Britz O, Guillemot F 2001. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron 29: 401–413 [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR 2004. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci 7: 136–144 [DOI] [PubMed] [Google Scholar]
- O'Geen H, Nicolet CM, Blahnik K, Green R, Farnham PJ 2006. Comparison of sample preparation methods for ChIP–chip assays. Biotechniques 41: 577–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Wang J, Kim J, Chu J, Rao S, Theunissen TW, Shen X, Levasseur DN 2008. The transcriptional network controlling pluripotency in ES cells. Cold Spring Harb Symp Quant Biol 73: 195–202 [DOI] [PubMed] [Google Scholar]
- Osada H, Tatematsu Y, Yatabe Y, Horio Y, Takahashi T 2005. ASH1 gene is a specific therapeutic target for lung cancers with neuroendocrine features. Cancer Res 65: 10680–10685 [DOI] [PubMed] [Google Scholar]
- Ota M, Sasaki H 2008. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development 135: 4059–4069 [DOI] [PubMed] [Google Scholar]
- Ozen I, Galichet C, Watts C, Parras C, Guillemot F, Raineteau O 2007. Proliferating neuronal progenitors in the postnatal hippocampus transiently express the proneural gene Ngn2. Eur J Neurosci 25: 2591–2603 [DOI] [PubMed] [Google Scholar]
- Pacary E, Heng J, Azzarelli R, Riou P, Castro D, Lebel-Potter M, Parras C, Bell DM, Ridley AJ, Parsons M, et al. 2011. Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signalling. Neuron 69: 1069–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Kim JY, Oh SP, Kang SY, Kim BW, Wang HJ, Song KY, Kim HC, Lim IK 2008. TIS21 negatively regulates hepatocarcinogenesis by disruption of cyclin B1–Forkhead box M1 regulation loop. Hepatology 47: 1533–1543 [DOI] [PubMed] [Google Scholar]
- Petryniak MA, Potter GB, Rowitch DH, Rubenstein JL 2007. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron 55: 417–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. 2006. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9: 157–173 [DOI] [PubMed] [Google Scholar]
- Poitras L, Ghanem N, Hatch G, Ekker M 2007. The proneural determinant MASH1 regulates forebrain Dlx1/2 expression through the I12b intergenic enhancer. Development 134: 1755–1765 [DOI] [PubMed] [Google Scholar]
- Qu Q, Sun G, Li W, Yang S, Ye P, Zhao C, Yu RT, Gage FH, Evans RM, Shi Y 2010. Orphan nuclear receptor TLX activates Wnt/β-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat Cell Biol 12: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S 1992. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255: 1707–1710 [DOI] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD 2003. Basic helix–loop–helix factors in cortical development. Neuron 39: 13–25 [DOI] [PubMed] [Google Scholar]
- Ruzinova MB, Benezra R 2003. Id proteins in development, cell cycle and cancer. Trends Cell Biol 13: 410–418 [DOI] [PubMed] [Google Scholar]
- Schuller U, Zhao Q, Godinho SA, Heine VM, Medema RH, Pellman D, Rowitch DH 2007. Forkhead transcription factor FoxM1 regulates mitotic entry and prevents spindle defects in cerebellar granule neuron precursors. Mol Cell Biol 27: 8259–8270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Seibt J, Brown C, Tang H, Cunningham JM, et al. 2004. Sequential phases of neocortical fate specification involve Neurogenin-dependent and –independent pathways. EMBO J 23: 2892–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibt J, Schuurmans C, Gradwhol G, Dehay C, Vanderhaeghen P, Guillemot F, Polleux F 2003. Neurogenin2 specifies the connectivity of thalamic neurons by controlling axon responsiveness to intermediate target cues. Neuron 39: 439–452 [DOI] [PubMed] [Google Scholar]
- Sharov AA, Ko MS 2009. Exhaustive search for over-represented DNA sequence motifs with CisFinder. DNA Res 16: 261–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth AN, Bhide PG 1997. Concurrent cellular output from two proliferative populations in the early embryonic mouse corpus striatum. J Comp Neurol 383: 220–230 [PubMed] [Google Scholar]
- Shida T, Furuya M, Kishimoto T, Nikaido T, Tanizawa T, Koda K, Oda K, Takano S, Kimura F, Shimizu H, et al. 2008. The expression of NeuroD and mASH1 in the gastroenteropancreatic neuroendocrine tumors. Mod Pathol 21: 1363–1370 [DOI] [PubMed] [Google Scholar]
- Shioi G, Konno D, Shitamukai A, Matsuzaki F 2009. Structural basis for self-renewal of neural progenitors in cortical neurogenesis. Cereb Cortex 19: i55–i61 doi: 10.1093/cercor/bhp042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singson A, Leviten MW, Bang AG, Hua XH, Posakony JW 1994. Direct downstream targets of proneural activators in the imaginal disc include genes involved in lateral inhibitory signaling. Genes Dev 8: 2058–2071 [DOI] [PubMed] [Google Scholar]
- Smart IH 1976. A pilot study of cell production by the ganglionic eminences of the developing mouse brain. J Anat 121: 71–84 [PMC free article] [PubMed] [Google Scholar]
- Smart IH, Dehay C, Giroud P, Berland M, Kennedy H 2002. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb Cortex 12: 37–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasundaram K, Reddy SP, Vinnakota K, Britto R, Subbarayan M, Nambiar S, Hebbar A, Samuel C, Shetty M, Sreepathi HK, et al. 2005. Upregulation of ASCL1 and inhibition of Notch signaling pathway characterize progressive astrocytoma. Oncogene 24: 7073–7083 [DOI] [PubMed] [Google Scholar]
- Southall TD, Brand AH 2009. Neural stem cell transcriptional networks highlight genes essential for nervous system development. EMBO J 28: 3799–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotopoulos D, Goffredo D, Conti L, Di Febo F, Biella G, Toselli M, Cattaneo E 2009. An optimized experimental strategy for efficient conversion of embryonic stem (ES)-derived mouse neural stem (NS) cells into a nearly homogeneous mature neuronal population. Neurobiol Dis 34: 320–331 [DOI] [PubMed] [Google Scholar]
- Steele-Perkins G, Plachez C, Butz KG, Yang G, Bachurski CJ, Kinsman SL, Litwack ED, Richards LJ, Gronostajski RM 2005. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol 25: 685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M 1994. Recognition sequence of a highly conserved DNA binding protein RBP-Jκ. Nucleic Acids Res 22: 965–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyteca S, Vandromme M, Legube G, Chevillard-Briet M, Trouche D 2006. Tip60 and p400 are both required for UV-induced apoptosis but play antagonistic roles in cell cycle progression. EMBO J 25: 1680–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vias M, Massie CE, East P, Scott H, Warren A, Zhou Z, Nikitin AY, Neal DE, Mills IG 2008. Pro-neural transcription factors as cancer markers. BMC Med Genomics 1: 17 doi: 10.1186/1755-8794-1-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M 2010. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463: 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace K, Liu TH, Vaessin H 2000. The pan-neural bHLH proteins DEADPAN and ASENSE regulate mitotic activity and cdk inhibitor dacapo expression in the Drosophila larval optic lobes. Genesis 26: 77–85 [DOI] [PubMed] [Google Scholar]
- Wei G, Ku S, Ma GK, Saito S, Tang AA, Zhang J, Mao JH, Appella E, Balmain A, Huang EJ 2007. HIPK2 represses β-catenin-mediated transcription, epidermal stem cell expansion, and skin tumorigenesis. Proc Natl Acad Sci 104: 13040–13045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierstra I, Alves J 2008. Cyclin E/Cdk2, P/CAF, and E1A regulate the transactivation of the c-myc promoter by FOXM1. Biochem Biophys Res Commun 368: 107–115 [DOI] [PubMed] [Google Scholar]
- Yi SH, Jo AY, Park CH, Koh HC, Park RH, Suh-Kim H, Shin I, Lee YS, Kim J, Lee SH 2008. Mash1 and neurogenin 2 enhance survival and differentiation of neural precursor cells after transplantation to rat brains via distinct modes of action. Mol Ther 16: 1873–1882 [DOI] [PubMed] [Google Scholar]
- Ying J, Srivastava G, Hsieh WS, Gao Z, Murray P, Liao SK, Ambinder R, Tao Q 2005. The stress-responsive gene GADD45G is a functional tumor suppressor, with its response to environmental stresses frequently disrupted epigenetically in multiple tumors. Clin Cancer Res 11: 6442–6449 [DOI] [PubMed] [Google Scholar]
- Yoshimoto T, Boehm M, Olive M, Crook MF, San H, Langenickel T, Nabel EG 2006. The arginine methyltransferase PRMT2 binds RB and regulates E2F function. Exp Cell Res 312: 2040–2053 [DOI] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabe de Angelis M, Weinmaster G, Rubenstein JL 2002. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development 129: 5029–5040 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang N, Dai B, Liu M, Sawaya R, Xie K, Huang S 2008. FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res 68: 8733–8742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LY, Niu Y, Santiago A, Liu J, Albert SH, Robertson KD, Liao D 2006. An EBF3-mediated transcriptional program that induces cell cycle arrest and apoptosis. Cancer Res 66: 9445–9452 [DOI] [PubMed] [Google Scholar]
- Zhao X, D'Arca D, Lim WK, Brahmachary M, Carro MS, Ludwig T, Cardo CC, Guillemot F, Aldape K, Califano A, et al. 2009. The N-Myc–DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev Cell 17: 210–221 [DOI] [PMC free article] [PubMed] [Google Scholar]