Abstract
Pre-mRNA splicing is frequently coupled to transcription by RNA polymerase II (RNAPII). This coupling requires the C-terminal domain of the RNAPII largest subunit (CTD), although the underlying mechanism is poorly understood. Using a biochemical complementation assay, we previously identified an activity that stimulates CTD-dependent splicing in vitro. We purified this activity and found that it consists of a complex of two well-known splicing factors: U2AF65 and the Prp19 complex (PRP19C). We provide evidence that both U2AF65 and PRP19C are required for CTD-dependent splicing activation, that U2AF65 and PRP19C interact both in vitro and in vivo, and that this interaction is required for activation of splicing. Providing the link to the CTD, we show that U2AF65 binds directly to the phosphorylated CTD, and that this interaction results in increased recruitment of U2AF65 and PRP19C to the pre-mRNA. Our results not only provide a mechanism by which the CTD enhances splicing, but also describe unexpected interactions important for splicing and its coupling to transcription.
Keywords: Prp19 complex, RNA, U2AF65, splicing, transcription
Almost all mammalian RNA polymerase II (RNAPII) transcripts undergo three principal processing events before their export from the nucleus: capping, splicing, and polyadenylation. These events are frequently coupled to transcription by RNAPII in a manner that ensures the faithful and efficient execution of each step (Hirose and Manley 2000; Pandit et al. 2008; Perales and Bentley 2009). The repetitive C-terminal domain of the large subunit of RNAPII (CTD) has been shown to play a central role in coupling transcription to all three of the main processing reactions (Phatnani and Greenleaf 2006; Perales and Bentley 2009; Munoz et al. 2010). Deletion of most of the CTD can result in inefficient capping, splicing, and polyadenylation in vivo (McCracken et al. 1997b; Fong and Bentley 2001). Consistent with this, the stimulatory effect of the CTD on 3′ cleavage and splicing can be recapitulated in vitro (Hirose and Manley 1998; Hirose et al. 1999; Zeng and Berget 2000). In mammals, the CTD consists of 52 heptad repeats of the consensus sequence YSPTSPS. Multiple residues within the CTD heptad are phosphorylated throughout the transcription cycle (Egloff and Murphy 2008; Buratowski 2009). Phosphorylation of Ser 5 (S5) of the heptad is most prominent at the 5′ end of genes (Komarnitsky et al. 2000; Schroeder et al. 2000), while Ser 2 (S2) phosphorylation increases toward the 3′ end (Komarnitsky et al. 2000). CTD phosphorylation plays an important role in generating elongation-competent RNAPII (Sims et al. 2004), and is required for the stimulatory effect of the CTD on splicing (Hirose et al. 1999; Millhouse and Manley 2005).
A number of interactions linking pre-mRNA processing and the CTD have been documented. For capping, the functional connection with the CTD is straightforward; the guanylytransferase and methyltransferase enzymes necessary for capping both bind to the S5- phosphorylated CTD, which allosterically activates guanylytransferase activity (McCracken et al. 1997a; Yue et al. 1997; Ho and Shuman 1999). Connections between the polyadenylation machinery and the CTD have also been demonstrated. Human CstF50 was shown to interact physically with both the phosphorylated and unphosphorylated CTD, an interaction that appears important for efficient cleavage/polyadenylation in vivo (Fong and Bentley 2001). The yeast CFI subunit Pcf11 also interacts with the S2-phosphorylated CTD, the functional importance of which was suggested by a genetic interaction between a Pcf11 allele and an RBP1 CTD truncation allele (Licatalosi et al. 2002). Also, a CTD phosphatase, Ssu72, was shown recently to be important for transcription-coupled 3′ processing in vitro (Xiang et al. 2010).
The machinery that carries out pre-mRNA splicing is considerably more complex than those responsible for capping and polyadenlyation. The spliceosome, the protein–RNA assembly that catalyzes intron removal, contains at least 150 proteins and undergoes dynamic changes in conformation and protein composition during the series of events that begin with splice site recognition and end after the execution of the two catalytic steps (Jurica and Moore 2003; Smith et al. 2008; Wahl et al. 2009; Valadkhan and Jaladat 2010). In vitro, spliceosome assembly proceeds through the formation of a series of stable intermediate complexes, which are biochemically separable and amenable to proteomic analysis (Wahl et al. 2009). Among the earliest steps in spliceosome assembly is recognition of the 5′ and 3′ splice sites by the U1 snRNP and U2AF, respectively. U2AF is a dimer comprised of U2AF65 and U2AF35 (Zamore and Green 1989). U2AF65 binds to polypyrimidine-rich sequences found near the 3′ end of most introns and promotes stable U2 snRNP association with the pre-mRNA, an activity that requires its N-terminal arginine–serine-rich (RS) domain (Valcarcel et al. 1996). U2AF35 contacts a well-conserved AG dinucleotide at the 3′ end of the intron (e.g., Wu et al. 1999) and can interact with exon-bound SR proteins; both interactions can stabilize U2AF binding to suboptimal polypyrimidine tracts (Zuo and Maniatis 1996). Later steps in spliceosome assembly involve the activity of numerous additional factors, including the U4/U6.U5 tri-snRNP and the Prp19 complex, or PRP19C (Wahl et al. 2009). PRP19C was first discovered in yeast, where it was shown to be an essential splicing factor that does not tightly associate with snRNPs (Hogg et al. 2010). PRP19C, which consists of four polypeptides that form a salt-stable core (CDC5L, PRLG1, Prp19, and SPF27) and three more loosely associated polypeptides (HSP73, CTNNBL1, and AD002) (Grote et al. 2010), is found at the core of catalytically activated spliceosomes and plays a critical but poorly understood role in activation of the spliceosome (Chan et al. 2003; Bessonov et al. 2008; Song et al. 2010). Because PRP19C does not contain any proteins known to bind RNA, it is likely that PRP19C recruitment to the spliceosome occurs through protein–protein interactions with RNA-bound factors, although no such interaction has yet been described.
Most of what is known about the process of spliceosome assembly has come from the use of in vitro systems that are uncoupled from transcription, leaving the role of the transcriptional machinery in the process relatively poorly understood. However, a few physical interactions between splicing factors and the CTD have been documented. The yeast U1 snRNP component Prp40 was shown to bind to the phosphorylated CTD through multiple WW domains (Morris and Greenleaf 2000; Gasch et al. 2006). In humans, splicing factors that have been shown to bind directly to the CTD include CA150 (Carty et al. 2000), PSF, and p54/NRB (Emili et al. 2002). Of these, support for a functional significance to the CTD interaction has been provided only for PSF, which can be recruited to promoters by strong transcriptional activators to promote splicing in a CTD-dependent manner in vivo (Rosonina et al. 2005).
In order to study the functional connections between the CTD and pre-mRNA splicing, we previously constructed a fusion between the CTD and the SR protein SRSF1 (formerly ASF/SF2). This allowed recruitment of the CTD to splicing substrates harboring SRSF1-binding sites independent of transcription. Using this fusion protein, which we now call SRSF1-CTD, in in vitro splicing assays, we observed an increase in splicing kinetics in its presence when compared with SRSF1 alone (Millhouse and Manley 2005). In addition, we found that a HeLa nuclear fraction (NF20–40) was capable of activating splicing of one substrate, IgMA3, in HeLa S-100 in the presence of SRSF1-CTD but not SRSF1, suggesting that NF20–40 contains a factor capable of functionally interacting with the CTD. We purified and characterized the factor responsible for this activity and found that it consists of a complex containing both U2AF65 and PRP19C. U2AF65 and PRP19C interact directly in vitro and in an RNA-independent manner in vivo. Additionally, U2AF65 binds directly to the phosphorylated CTD, increasing U2AF association with the pre-mRNA and recruitment of PRP19C. U2AF65 thus bridges the transcriptional machinery and later stages of spliceosomal assembly through novel interactions with the RNAPII CTD and PRP19C.
Results
CTD-specific splicing activity copurifies with U2AF65 and PRP19C
We observed previously that a splicing substrate called IgMA3 (see Fig. 1A) is not spliced in S-100 complementation assays in the presence of 100 nM SRSF1 or SRSF1-CTD (structure of the fusion protein is shown in Fig. 1A). However, addition of an ammonium sulfate fraction derived from HeLa nuclear extract (NE) (NF20–40) specifically activated splicing in the presence of SRSF1-CTD, but not SRSF1 alone (Millhouse and Manley 2005). Tethering of the CTD to the pre-mRNA by the SRSF1 portion of the fusion protein was necessary, as addition of the CTD in trans failed to activate NF20–40-dependent splicing in the presence of SRSF1 (data not shown).
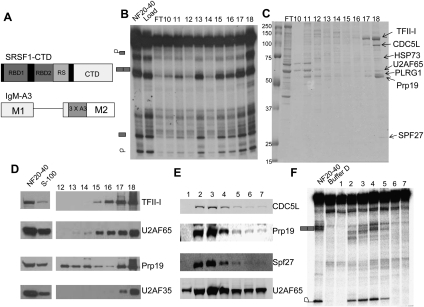
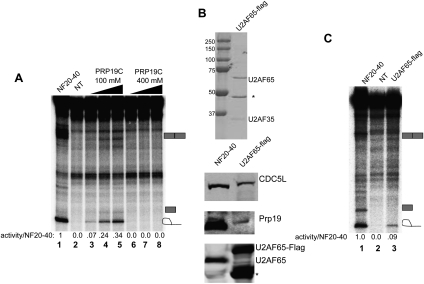
Figure 1.
U2AF65 and PRP19C cofractionate with CTD-dependent splicing activity. (A, top) Schematic diagrams of the SRSF1-CTD and IgMA3 constructs used in this study. IgMA3 contains three copies of a high-affinity binding site for SRSF1 (A3). (B) NF20–40 was fractionated by butyl-FF, Mono Q, followed by Mono S chromatography, and then Mono S fractions were assayed for CTD-dependent splicing. Splicing reactions were incubated in the presence of SRSF1-CTD, S-100, and the fraction indicated for 2 h, then RNA was isolated, separated by 6% denaturing PAGE, and then visualized by autoradiography. (C) Mono S fractions from B were separated by 10% SDS-PAGE gel and stained by colloidal Coomassie. Protein bands in fraction 18 were excised and identified by MALDI-TOF mass spectrometry. Protein identities are indicated at right. (D) NF20–40, S-100, and the indicated Mono S fractions from B were immunoblotted with the indicated antibodies. Comparisons of protein levels in NF20–40 and S-100 are provided at left. (E) NF20–40 was fractionated on a Mono Q column, and then gradient fractions were immunoblotted with the indicated antibodies. (F) Mono Q fractions from E were used in IgMA3 splicing assays in the presence of SRSF1-CTD and S-100 and then processed as in B.
To identify the factor(s) involved in the CTD-dependent splicing activity, we chromatographically purified the activity (purification scheme shown in Supplemental Fig. 1). We found that the NF20–40 activity bound to a butyl-FF column and eluted during a gradient between 400 and 0 mM ammonium sulfate (data not shown). The active fractions were applied to a Mono Q column, with the activity eluting early in the gradient, between 40 and 60 mM ammonium sulfate. The active Mono Q fractions were then loaded on a Mono S column and the activity was bound tightly, with the strongest peak of activity eluting at 800 mM ammonium sulfate (Fig. 1B, fractions 17 and 18). The Mono S fractions were separated by SDS-PAGE and stained with Coomassie blue (Fig. 1C). Protein bands in the most active fraction, 18, were excised and proteins were identified by mass spectrometry (Fig. 1C). Unexpectedly, we identified not only all four of the core components of PRP19C, as well as HSP73, but also the early-acting splicing factor U2AF65. The only species identified that were not associated with PRP19C or U2AF65 were multiple isoforms of the transcription factor TFII-I. TFII-I is known to regulate transcription of certain genes in response to mitogenic signals (Roy 2007), but has not been implicated in pre-mRNA splicing. The identities of U2AF65, Prp19, and TFII-I were confirmed by immunoblotting (Fig. 1D). Importantly, U2AF65 and Prp19 were present in all active Mono S fractions. TFII-I eluted later in the gradient than U2AF65 and Prp19 (Fig. 1D, fractions 15–18), and was barely detectable in the first active fraction (data not shown). Because U2AF65 usually exists as a dimer with U2AF35, we tested the Mono S fractions for the presence of U2AF35 by immunoblotting (Fig. 1D). U2AF35 was weakly detectable in fractions 17 and 18 but was undetectable in all earlier fractions, indicating that U2AF35 is dispensable for the CTD-dependent splicing activity.
We also examined NF20–40 Mono Q fractions for a potential correlation between the presence of PRP19C and U2AF65 and CTD-dependent splicing activity. Immunoblotting showed that PRP19C—specifically, CDC5L, Prp19, and SPF27—eluted early in the Mono Q gradient and coincided with U2AF65 in fractions 2–5 (Fig. 1E). U2AF65 eluted in a broader peak, beginning in the earliest gradient fraction and continuing through gradient fraction 7. Notably, splicing activity was restricted to fractions that contained both U2AF65 and PRP19C (Fig. 1F, fractions 2–5). Later fractions, which were also enriched in U2AF65, were inactive in the splicing assay.
U2AF65 interacts with the PRP19C in vitro and in vivo
Given that U2AF65 and PRP19C copurify in active fractions, we next wished to test the possibility that U2AF65 and PRP19C interact physically. To this end, we expressed and purified GST-tagged U2AF65 in Escherichia coli (Fig. 2A). Using GST-U2AF65 in a pull-down assay with NF20–40, a strong interaction with Prp19 was detected by Western blotting with anti-Prp19 antibodies (Fig. 2B, lane 6), while a somewhat weaker interaction was observed with S-100 (Fig. 2B, lane 3). The Prp19 antibody detected additional bands (most prominently in S-100) (Fig. 2B, lanes 1,2) that may correspond to previously described ubiquitinated forms of Prp19 (Lu and Legerski 2007). Consistent with that report, which showed that Prp19 ubiquitination disrupts its association with other complex members, these modified forms failed to cofractionate with other PRP19C subunits on Mono Q (data not shown), suggesting that the modification disrupts interactions between Prp19 and other complex subunits. The modified forms of Prp19 also failed to interact with U2AF65 (Fig. 2B), suggesting that only unmodified, complex-associated Prp19 interacts with GST-U2AF65. To identify the region(s) of U2AF65 required to interact with PRP19C, we constructed deletion mutants lacking the N-terminal RS domain or the C-terminal U2AF homology (UHM) domain—regions implicated in protein–protein interactions (Kielkopf et al. 2004; Shepard and Hertel 2009). The mutant protein lacking the UHM domain bound Prp19 with efficiency comparable with full-length U2AF65 (Fig. 2B, lanes 5,8). In contrast, truncation of the RS domain completely eliminated the interaction with Prp19 (Fig. 2B, lanes 4,7).
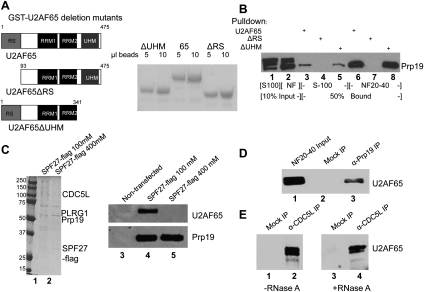
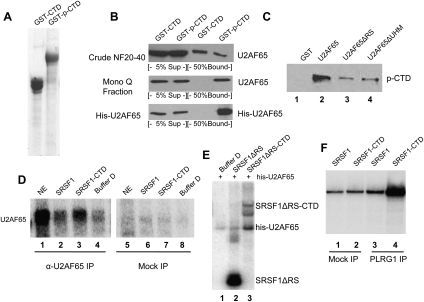
Figure 2.
U2AF65 interacts with PRP19C in an RNA-independent manner. (A, left) Schematic of U2AF65 and deletion constructs used for GST pull-downs. (Right) These constructs were expressed in E. coli and then purified by GSH sepharose beads. The beads were run on SDS-PAGE and Coomassie-stained. (B) GST pull-down was performed using GST-tagged U2AF65, U2AF65ΔRS, or U2AF65ΔUHM. After a 3-h incubation at 22°C with the indicated extract, beads were washed and then eluted with 15 mM glutathione. Eluted proteins were immunoblotted with an anti-Prp19 antibody. (C) A vector expressing Flag-tagged SPF27 was transfected into 293T cells, cells were harvested and used to make NE, and PRP19C was purified using anti-Flag agarose and washed with buffer D containing the indicated salt concentration. In parallel, beads were incubated with NE from nontransfected cells amM KCl. Beads were eluted with Flag peptide, separated by SDS-PAGE, and Coomassie stained (left) or immunoblotted with the indicated antibody (right). (D) Co-IPs were carried out in NF20–40 using anti-Prp19 antibodies or anti-GST antibodies (mock). Beads were washed with buffer D and then boiled and immunoblotted for U2AF65. (E) Co-IP was performed using anti-CDC5L antibodies or anti-GST antibodies as above, except NF20–40 was mock- or RNase A-treated beforehand for 30 min at 37°C. The effectiveness of RNase A digestion was confirmed by agarose gel electrophoresis followed by ethidium staining (not shown).
We next wished to determine whether U2AF65 and PRP19C interact in vivo. As a first experiment, we transiently transfected a plasmid expressing Flag-tagged SPF27 into 293T cells, prepared NE, and incubated the NE with anti-Flag M2 agarose beads. After incubation, aliquots of beads were washed with buffer containing 100 mM or 400 mM KCl. Bound proteins were eluted using Flag peptide and analyzed by Coomassie staining and immunoblotting. The resulting complexes contained polypeptides that correspond to all four core PRP19C subunits: CDC5L, PLRG1, Prp19, and SPF27 (see Fig. 2C, lanes 1,2; data not shown). The intensity of the Prp19 band was considerably stronger than the SPF27-Flag band, consistent with a 4:1 Prp19:SPF27 stoichiometry in PRP19C (Grote et al. 2010), and indicating that SPF27-Flag was efficiently incorporated into the endogenous PRP19C. Importantly, immunoblotting with anti U2AF65 antibodies indicated that washing with low-salt buffer resulted in copurification of U2AF65, while higher salt, which did not affect the amount of Prp19 that copurified with SPF27-Flag, eliminated the copurification of U2AF65 (Fig. 2C, lanes 4,5). Neither protein was detected in the eluate from beads incubated with NE from nontransfected cells (Fig. 2C, lanes 1–3). These data indicate that U2AF65 and PRP19C form a salt-labile complex in NE.
To examine whether endogenous U2AF65 and PRP19C interact, we performed immunoprecipitations (IPs) with NF20–40. Using anti-Prp19 antibodies for IP, we indeed detected coimmunoprecipitation (co-IP) of U2AF65 (Fig. 2D, lane 3). The same interaction was observed using an anti-CDC5L antibody, as well as antibodies against two additional PRP19C subunits: PLRG1 and SPF27 (Fig. 2E, lanes 2,4; data not shown). Importantly, co-IP of U2AF65 was maintained after extensive treatment of the extract with RNase A (Fig. 2E, cf. lanes 2 and 4), indicating that the interaction was not bridged by RNA, as might occur, for example, in spliceosomes.
U2AF and PRP19C are required for CTD-dependent splicing activity
We next wished to determine whether U2AF65 and PRP19C are in fact required for CTD-dependent splicing activity. We first examined the effect of depleting U2AF from NF20–40. To do this, we passed the NF20–40 through a Poly-U sepharose column at a salt concentration of 1 M KCl, while a mock sample was prepared using an equal volume of glutathione sepharose beads. As expected (Zamore and Green 1991), passage through a Poly-U column resulted in efficient depletion of U2AF65 from the NF20–40 (Fig. 3A, top panel, lane 2). Because, under high-salt conditions, U2AF65 and PRP19C do not interact (see above), depletion of U2AF65 in this manner resulted in minimal codepletion of PRP19C, as judged by CDC5L levels (Fig. 3A, bottom panel). Importantly, depletion of U2AF resulted in an approximately fivefold reduction in CTD-dependent splicing activity compared with the mock-depleted NF20–40 (Fig. 3B, cf. lanes 2 and 3). We next tested purified U2AF65 for its ability to restore activity to the depleted NF20–40. While neither baculovirus-expressed and purified U2AF65 nor the U2AF heterodimer was capable of activating CTD-dependent splicing alone (Fig. 3B, lanes 6,7), we found that both were able to partially restore activity to depleted NF20–40 (Fig. 3B, lanes 4,5). To confirm these results, we also used an anti-U2AF65 antibody to immunodeplete U2AF from NF20–40 at 500 mM NaCl, which resulted in an ∼85% depletion of U2AF65 but no change in PRP19C levels, as judged by CDC5L (Supplemental Fig. 2A). This treatment resulted in a >60% decrease in activity, compared with mock-depleted NF20–40 (Supplemental Fig. 2B). On the basis of these experiments, we conclude that U2AF65 is necessary but not sufficient for CTD-dependent splicing activity.
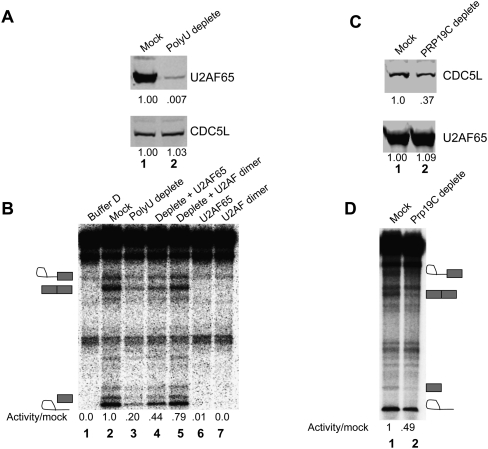
Figure 3.
U2AF and PRP19C are required for CTD-dependent splicing activity. (A) NF20–40 was depleted of U2AF65 at 1 M KCl using Poly-U sepharose beads, followed by immunoblotting against U2AF65 and CDC5L. Band intensities quantified using Li-cor software are indicated below. (B, lanes 2 and 3) Mock and depleted extracts from A were used in the CTD-dependent splicing assay. One-hundred nanomolar U2AF65 and U2AF produced in baculovirus-infected insect cells were tested for their ability to complement the depleted NF20–40 (lanes 4,5) or activate splicing when added alone (lanes 6,7). Relative intensities of spliced product were quantified using ImageQuant software and are indicated below. (C) PRP19C was depleted from an active Mono Q fraction at 500 mM NaCl using an anti-CDC5L antibody or was mock-depleted using an anti-Flag antibody. Mock and depleted samples were immunoblotted for U2AF65 and CDC5L. Relative CDC5L levels were normalized to U2AF65 and are indicated below. (D) CTD-dependent splicing assays were performed using the mock- and PRP19C-depleted samples from C. Relative amounts of spliced product are indicated below.
We next wished to determine whether the PRP19C is necessary for CTD-dependent splicing activity. To do this, we used anti-CDC5L antibodies to immunodeplete PRP19C, in this case from an active Mono Q fraction. This resulted in an ∼60% depletion of PRP19C, as judged by CDC5L levels (Fig. 3C, top panel, cf. lanes 1 and 2). Importantly, this caused a >50% reduction in activity when compared with a mock-depleted control (Fig. 3D, cf. lanes 1 and 2). As PRP19C depletion was performed at 500 mM NaCl, levels of U2AF65 were not affected (Fig. 3C, bottom panel, lanes 1,2). Although additional rounds of incubation with antibodies led to near complete depletion of PRP19C, this treatment resulted in loss of activity in both the mock and depleted fractions (data not shown). Nonetheless, the reduction of activity that we observed upon partial depletion of PRP19C indicates that PRP19C contributes to the CTD-dependent splicing activity of NF20–40.
U2AF65 and PRP19C association is required for CTD-dependent splicing
U2AF65 and PRP19C are both present in S-100, which nonetheless is unable to substitute for NF20–40 to activate CTD-dependent splicing. To investigate the basis for this, we next examined the behavior of S-100-derived U2AF65 and PRP19C on a Mono Q column. The elution profile of each factor was strikingly different than observed with NF20–40, with PRP19C (represented by CDC5L) eluting in a narrow peak early in the Mono Q gradient (Fig. 4A, fraction 6) and U2AF65 eluting much later (Fig. 4A, fractions 10–14). Consistent with the inability of S-100 to activate CTD-dependent splicing, none of the S-100-derived Mono Q fractions were active, even when concentrated PRP19C and U2AF65-containing fractions were mixed together (data not shown).
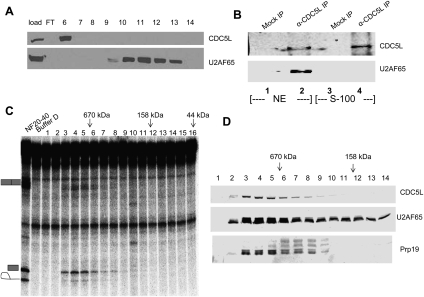
Figure 4.
Association of U2AF65 and PRP19C is necessary for CTD-dependent IgMA3 splicing. (A) S-100 was separated on a Mono Q column under conditions identical to those used in Figure 1E. Immunoblotting for U2AF65 and CDC5L was performed using S-100 (load) and Mono Q fractions. (B) Co-IP was performed using an anti-CDC5L antibody in NE and S-100, followed by anti-U2AF65 immunoblotting. (C) NF20–40 was fractionated on a Superdex 200 column. Fractions were directly assayed for CTD-dependent splicing. Elution locations of gel filtration standards are indicated at the top. (D) Immunoblot of Superdex 200 fractions for U2AF65, CDC5L, and Prp19.
The failure of U2AF65 and PRP19C to cofractionate when derived from S-100 suggests that the two complexes are not physically associated when present in the cytoplasmic extract. To test this, we performed parallel IPs in NE and S-100 using the anti-CDC5L antibody. In NE, as with the NF20-40, CDC5L coimmunoprecipitated U2AF65, while, in S-100, co-IP did not occur (Fig. 4B, cf. lanes 2 and 4). On the basis of co-IP and chromatographic data, we conclude that endogenous U2AF65 and PRP19C are physically associated in NE but not S-100. Possible explanations for this behavior are discussed below. In any event, these data support the idea that a physical interaction between U2AF65 and PRP19C in NF20–40 is required for CTD-dependent splicing of IgMA3.
The above data suggest that CTD-dependent splicing activity should reside in a complex containing U2AF65 and PRP19C. To estimate the molecular weight of the CTD-dependent splicing activity, we separated NF20–40 on a Superdex 200 gel filtration column, then performed splicing assays using the resulting fractions. Importantly, the activity eluted at an apparent mass of >670 kDa (Fig. 4C, fractions 3–9), consistent with the expected molecular mass of a putative U2AF65–PRP19C complex. The elution of the activity in fractions 3–9 coincided perfectly with fractions in which PRP19C and U2AF65 were both present, as judged by immunoblotting (Fig. 4D), while a number of lower-molecular-weight fractions containing U2AF65 alone (Fig. 4D, fractions 10–14) exhibited no splicing activity (Fig. 4C, fractions 10–14). Taken together, these results strongly suggest that CTD-dependent splicing activity resides in a complex containing U2AF65 and PRP19C.
CTD-dependent splicing activity can be reconstituted by U2AF65/PRP19C purified from mammalian cells
We next tested the ability of purified U2AF/PRP19C to reconstitute CTD-dependent splicing activity. To isolate a PRP19C–U2AF complex, we transfected a plasmid encoding Flag-tagged SPF27 into 293T cells, then used anti-Flag beads to purify PRP19C from NE made from these cells by washing the beads with buffer containing either 100 mM or 400 mM KCl. As shown above (Fig. 2C), washing the beads with 100 mM KCl resulted in copurification of U2AF65, while washing with 400 mM KCl removed U2AF65. Importantly, when used in CTD-dependent splicing assays, the low-salt PRP19C activated splicing in a concentration-dependent manner, while the stringently purified PRP19C exhibited no more activity than the mock eluate (Fig. 5A, cf. lanes 3–5 and 6–8). As a complementary approach, we prepared the U2AF65/PRP19C complex from 293T cells transfected with a plasmid encoding U2AF65-Flag, using 100 mM KCl to wash the beads (Fig. 5B). U2AF65 purified under these conditions was associated with PRP19C, as Western blotting revealed that both CDC5L and Prp19 were present in the U2AF65 preparation (Fig. 5B, bottom panel). In contrast to the more stringently purified baculovirus preparations of U2AF/U2AF65 analyzed above (see Fig. 3B), the PRP19C-containing U2AF65 preparation was able to activate splicing when used in the CTD-dependent splicing assay (Fig. 5C, lane 3). Together, these results indicate that a U2AF65–PRP19C complex can activate CTD-dependent splicing.
Figure 5.
U2AF65 affinity-purified from mammalian cells can reconstitute CTD-dependent splicing. (A) NF20–40 (lane 1), anti-Flag eluate prepared from nontransfected cells (lane 2), and PRP19C preparations from Figure 2C were used in the CTD-dependent splicing assay. For each, 40, 80, or 120 nM was used in splicing reactions. (B) Anti-Flag eluate prepared from 293T cells transfected with U2AF65-Flag was separated by SDS-PAGE and Coomassie-stained (top panel) or immunoblotted (bottom panel) for the indicated proteins. The band marked with an asterisk (*) in the U2AF65 preparation is an N-terminally proteolyzed U2AF65 species. (C) NF20–40 (lane 1), anti-Flag eluate prepared from nontransfected cells (lane 2), and 50 nM U2AF65-Flag from B were used in the CTD-dependent splicing assay.
The RNAPII CTD binds U2AF65 directly
We next wished to elucidate the role of the CTD in U2AF65–PRP19C splicing stimulatory activity. Based on its function in other processes, a likely possibility is that one or more U2AF65–PRP19C subunits interact directly with the CTD, and this interaction helps recruit the complex to the RNA substrate. Given that U2AF65 has been shown to associate tightly with elongating RNAPII in vitro (Ujvari and Luse 2004), it was a logical candidate to participate in such an interaction. We therefore tested whether U2AF65 can interact with the CTD, first by performing pull-down assays with GST-CTD that was either unphosphorylated or phosphorylated (p-CTD) in and repurified from NE (Fig. 6A; Hirose and Manley 1998. In vitro phosphorylation using NE results in a mixture of S2 and S5 phosphorylation (e.g., Xiang et al. 2010). When we used NF20–40 as input for the pull-down, we found that U2AF65 interacted with both CTD and p-CTD (Fig. 6B, top panel). However, after partial purification by Mono Q chromatography, U2AF65 bound only to p-CTD, suggesting that the interaction with the unphosphorylated CTD observed with NF20–40 was mediated by other factors present in the fraction (Fig. 6B, middle panel). Interestingly, we also detected pull-down of PRP19C (Prp19) from NF20–40 by p-CTD (Supplemental Fig. 3A). This interaction may be bridged by U2AF65, as it was not observed with S-100, where the two complexes fail to interact, but in which U2AF65 bound strongly to p-CTD (Supplemental Fig. 3B).
Figure 6.
The CTD binds directly to U2AF65 to recruit U2AF65–PRP19C to pre-mRNA. (A) GST-CTD was expressed in E. coli, purified, and phosphorylated in vitro. Five-microliter aliquots were subjected to SDS-PAGE to ensure equal recovery. (B) GST pull-downs were performed using unphosphorylated or NE-phosphorylated GST-CTD. Pull-downs were performed using NF20–40 (top panel), U2AF65 partially purified by Mono Q chromatography (middle panel), and His-tagged U2AF65 purified from baculovirus-infected insect cells (bottom panel). Input and bound samples were immunoblotted for U2AF65. (C) GST pull-down was performed using GST-U2AF65 constructs used in Figure 2A and p-CTD cleaved from GST by thrombin. After elution, the bound p-CTD was detected by immunoblotting using the anti-CTD antibody 8WG16. (D) Splicing reactions were assembled with 32P-UTP-labeled IgMA3 in the presence of NE, S-100 and SRSF1, SRSF1-CTD, or buffer D. After 10 min, reactions were exposed to UV light, followed by RNase treatment. Reactions were then immunoprecipitated using an anti-U2AF65 or anti-HA antibody (mock), and cross-linked proteins were then separated by 10% SDS-PAGE and visualized by autoradiography. (E) UV cross-linking reactions using IgMA3 were performed in the presence of purified U2AF65, SRSF1ΔRS-CTD, or both. After cross-linking and RNase treatment, proteins were separated by 8% SDS-PAGE and visualized by autoradiography. (F) Splicing reactions were performed in the presence of S-100, NF20–40, and SRSF1 or SRSF1-CTD. After a 30-min incubation at 30°C, reactions were immunoprecipitated with anti-GST antibodies (mock) or anti-PLRG1 antibodies. After IP, RNA was extracted and separated by denaturing 6% PAGE and visualized by autoradiography.
We next wished to provide evidence that the CTD–U2AF65 interaction was direct. To this end, we used purified His-tagged U2AF65 produced in baculovirus-infected cells in the GST-pull-down assay. Importantly, a strong, p-CTD-specific interaction was detected (Fig. 6B, bottom panel), indicating that U2AF65 interacts directly with the p-CTD. In addition, the U2AF65–CTD interaction was robust enough to persist after washing with up to 500 mM NaCl (Supplemental Fig. 4A) and was resistant to RNase A (Supplemental Fig. 4B). In order to identify the regions of U2AF65 that contribute to p-CTD binding, we used GST-U2AF65 and the RS and UHM domain truncations described above in pull-down assays with the purified p-CTD that had been cleaved from GST using thrombin. We found that only full-length U2AF65 bound strongly to the p-CTD, while each of the truncation mutants displayed significantly reduced p-CTD binding (Fig. 6C, lanes 2–4). This suggests that U2AF65 interacts with the p-CTD in a manner that requires both ends of the protein for optimal binding.
The RNAPII CTD–U2AF65 recruits U2AF and PRP19C to pre-mRNA
The interaction of U2AF65 with the p-CTD has the potential to underlie the CTD dependence of U2AF–PRP19C stimulatory activity. The IgMA3 splicing substrate contains a weak polypyrimidine tract to which U2AF65 is recruited inefficiently in the absence of an exonic splicing enhancer in the second exon (Graveley et al. 2001). A model for how SRSF1-CTD stimulates IgMA3 splicing is that substrate-bound SRSF1-CTD recruits U2AF65 to the pre-mRNA more efficiently than SRSF1 alone as a result of the p-CTD–U2AF65 interaction. To test this possibility, we performed UV cross-linking with 32P-IgMA3 RNA and S-100 plus NF20–40 in the presence of SRSF1 or SRSF1-CTD, followed by IP with anti-U2AF65 antibodies. Significantly, we detected an increase in U2AF65 cross-linking in the presence of SRSF1-CTD compared with SRSF1 (Fig. 6D, cf. lanes 2,3). To extend this result, we performed the cross-linking assay with purified baculovirus U2AF65 together with SRSF1 derivatives lacking the SRSF1 RS domain (SRSF1ΔRS and SRSF1ΔRS-CTD) (Millhouse and Manley 2005). As above, enhanced cross-linking was observed with SRSF1ΔRS-CTD relative to SRSF1ΔRS (Fig. 6E, lanes 2,3). Together, these results provide strong evidence that the CTD–U2AF65 interaction is sufficient to facilitate U2AF recruitment to the pre-mRNA.
We next asked whether the increased recruitment of U2AF65 to the IgMA3 pre-mRNA by SRSF1-CTD leads to increased recruitment of PRP19C. To test this, we again set up splicing reactions with 32P-IgMA3 RNA in the presence of NF20–40 and S-100, as well as SRSF1 or SRSF1-CTD. After a 30-min incubation, reaction mixtures were immunoprecipitated using a mock antibody (anti-GST) or anti-PLRG1 antibodies, and immunoprecipitated RNA was purified and resolved by denaturing PAGE. Strikingly, we found that PLRG1 strongly associated with pre-mRNA in the presence of SRSF1-CTD but not SRSF1, indicating that Prp19C recruitment to the pre-mRNA was significantly enhanced by the CTD (Fig. 6F, cf. lanes 3 and 4). We conclude that an interaction between U2AF65 and p-CTD promotes U2AF65 binding to the IgMA3 substrate, and this leads to enhanced Prp19C recruitment, reflecting the interaction between U2AF65 and Prp19C.
Discussion
Here we used a biochemical complementation assay described previously to purify and characterize an activity capable of activating CTD-dependent splicing in vitro. This resulted in the unexpected discovery that two well-studied splicing factors not previously known to associate (U2AF65 and PRP19C) interact functionally and physically to activate splicing. This was first indicated by our observation that both factors copurify in active fractions after extensive chromatography. We also showed that U2AF65 and PRP19C interact in an RNA-independent manner, and that the two indeed exist in a salt-sensitive complex in vivo. Importantly, several experimental approaches confirmed that the U2AF65/PRP19C complex is responsible for CTD-dependent splicing activity. The role played by the CTD was suggested by our discovery that U2AF65 binds directly to the p-CTD, and we showed that this enhanced both U2AF65 binding and PRP19C recruitment to the pre-mRNA. Below, we discuss the implications of these findings with respect to both the link between splicing and transcription and splicing per se.
How might U2AF65 and PRP19C cooperate to activate CTD-dependent splicing? Our data suggest that the association of the two factors in a complex, and not simply their concentration, is important. For example, the concentration of each is considerably higher in S-100 than in some active fractions (CJ David and JL Manley, unpubl.), but S-100 is incapable of activating CTD-dependent splicing. Also, addition of purified U2AF65 and PRP19C in combination did not result in a significant increase in activity. U2AF65/PRP19C-containing fractions activated CTD-dependent splicing only when the two factors were associated, and gel filtration indicated that CTD-dependent splicing activity resides in a high-molecular-weight (HMW) complex of a size consistent with a PRP19C–U2AF65 complex. What determines whether the two splicing factors interact is not clear, but may reflect the presence or absence of a bridging factor and/or differential protein modifications.
Earlier work on the IgM substrate may provide some clues as to why a preformed complex containing U2AF65 and PRP19C is necessary to activate IgMA3 splicing. Green and colleagues (Kan and Green 1999; Shen et al. 2004) showed that the second exon of the IgM substrate contains an inhibitory element that is bound by the splicing inhibitory protein polypyrimidine tract-binding protein (PTB). Kan and Green (1999) showed that splicing inhibition by PTB does not prevent early steps in spliceosome assembly, but rather results in an ATP-dependent complex that is similar in electrophoretic mobility and composition to spliceosomal A complex, but that is unable to progress to later stages in spliceosome assembly (Kan and Green 1999). A similar stalled A-like complex also forms on an in vitro splicing substrate containing the PTB-inhibited c-src N1 exon (Sharma et al. 2008). Proteomic analysis comparing complexes formed in the presence or absence of PTB inhibition showed that PRP19C recruitment is impaired when PTB functions to inhibit splicing (Sharma et al. 2008). PTB inhibition is likely relevant to our S-100 complementation assay, as the PTB-binding sites described by Shen et al. (2004) are intact in the IgMA3 substrate, and UV cross-linking reveals PTB as the major cross-link in splicing reactions using S-100 (CJ David and JL Manley, unpubl.).
Based on the above, we propose the following model for CTD-dependent activation of IgMA3 splicing. This substrate is unable to recruit U2AF efficiently due to its weak polypyrimidine tract (Graveley et al. 2001). Instead, a U2AF65–PRP19C complex is recruited to the pre-mRNA by RNA-bound SRSF1-CTD through the direct interaction between the CTD and U2AF65. U2AF35, which is normally required to mediate interactions between enhancer-bound SR proteins and U2AF65 (Graveley et al. 2001), appears to be dispensable, as U2AF35 was not detectable in some active fractions and was not required to reconstitute activity. It is likely that the CTD–U2AF65 interaction makes U2AF35 unnecessary for recruitment of U2AF65 to the IgMA3 polypyrimidine tract. The p-CTD–U2AF65 interaction also results in recruitment of PRP19C, most likely through its interaction with U2AF65. The combination of SRSF1-CTD and the U2AF65–PRP19C complex thus overcomes two important barriers to the assembly of active IgMA3 spliceosomes: first, the recruitment of U2AF65 to the suboptimal IgM polypyrimidine- tract, and second, recruitment of PRP19C to PTB-inhibited early spliceosomes.
Aside from a possible role in IgMA3 splicing, the U2AF65–PRP19C interaction likely plays a broader role in spliceosome assembly. As mentioned above, PRP19C is a stable component of catalytically activated spliceosomes (Bessonov et al. 2008), but the interactions that underlie its recruitment to the spliceosome are currently poorly understood. Studies in yeast originally suggested that PRP19C associates with the spliceosome at a late stage in its assembly, after the recruitment of the U4/U6.U5 tri-snRNP (Tarn et al. 1993). However, more recent data have shown that human PRP19C is a part of the first ATP-dependent spliceosomal complex (A complex) and its recruitment is independent of the tri-snRNP (Behzadnia et al. 2007). PRP19C association may occur even earlier than A complex, as it can associate with pre-mRNA in the absence of ATP (Jurica et al. 2002). These data indicate that PRP19C interacts with a factor that associates with early spliceosomes, resulting in its initial recruitment to the pre-mRNA. The interaction with U2AF65 that we documented here is a strong candidate for such an interaction. Additionally, the fact that the U2AF65 RS domain is required to interact with PRP19C raises the possibility that RS domains on other spliceosomal proteins may be able to perform a similar function. Interestingly, a U2AF–PRP19C connection appears to be conserved in yeast, as the yeast U2AF65 homolog Mud2 was shown to interact physically with a subunit of yeast PRP19C, Clf1 (Chung et al. 1999). Clf1 has a human homolog, Crn, which has been shown to associate with spliceosomes (Chung et al. 2002) but has not been found to associate with human PRP19C. Additional work will be necessary to identify the factor that bridges U2AF65 and PRP19C in mammalian cells.
Our results may also have implications for regulation of alternative splicing (AS). Since it appears likely that increased PRP19C recruitment can counteract PTB inhibition, PRP19C recruitment may serve as a regulatory point in the control of AS. Consistent with this idea, it was shown recently that the AS regulatory protein hnRNP M can interact directly with PRP19C, and this interaction appears to be important for regulation of AS by this protein (Lleres et al. 2010). Additionally, our results point to the possibility that splicing of different introns might be affected differently by modulation of PRP19C levels, which occurs during processes such as neuronal differentiation (Urano et al. 2006).
In addition to the association between U2AF and PRP19C, we also identified a novel interaction between U2AF65 and the CTD. This raises the possibility that U2AF65 plays an important role in the physical coupling of transcription and splicing. The fact that U2AF65 binds the CTD is consistent with previous studies. For example, U2AF65 has been reported to associate very tightly with RNAPII transcription elongation complexes (TECs) in vitro (Ujvari and Luse 2004). Importantly, these experiments showed that U2AF65 in the TEC was positioned to interact with RNA immediately upon its extrusion from the polymerase. Such positioning appears consistent with CTD binding, as the base of the CTD is located near the RNA channel (Cramer et al. 2001). Additionally, U2AF65 has been shown by chromatin IP to associate with transcriptionally active genes, including transcribed regions well upstream of the first 3′ splice site, consistent with RNAPII-mediated recruitment of U2AF65 to intron-containing genes (Listerman et al. 2006). Similar experiments with yeast have shown that Mud2 is cotranscriptionally recruited to active genes, raising the possibility that the interaction we identified here is evolutionarily conserved (Gornemann et al. 2005; Lacadie et al. 2006).
Our results, coupled with previous studies, provide an attractive model to explain how the efficiency of splicing in vivo can be enhanced by coupling it to transcription. Specifically, since U1 snRNP has been shown previously to associate with RNAPII (Das et al. 2007), the interaction between U2AF65 and elongating RNAPII via direct CTD binding means that both major factors that recognize the 5′ and 3′ splice sites are likely recruited to nascent transcripts. We envision a scenario (Fig. 7) in which RNAPII-associated U1 snRNP and SR proteins first recognize a 5′ splice site in the nascent RNA, resulting in tethering of the exon to RNAPII (Dye et al. 2006; Das et al. 2007). In the meantime, CTD-bound U2AF65 is positioned to interact with the 3′ splice site as soon as it is synthesized. Such an arrangement would result in rapid splice site recognition, the immediate juxtaposition of the two ends of the intron, and PRP19C recruitment through interaction with U2AF65, thereby facilitating efficient formation of mature spliceosomal complexes on nascent transcripts. In any event, our results have defined key molecular interactions that underlie the coupling of RNAPII transcription and splicing, and the role of the CTD in this process.
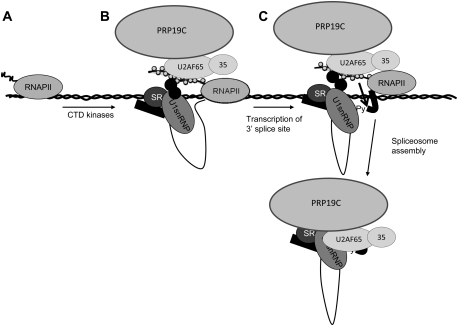
Figure 7.
Model for activation of CTD-dependent splicing by a U2AF–PRP19C complex. (A) At promoters, RNAPII is present in preinitiation complexes, but the CTD is unphosphorylated and unable to recruit splicing factors. (B) Transcription initiation results in CTD phosphorylation by multiple kinases, resulting in the association of splicing factors (including SR proteins), U1 snRNP through unknown interactions, and the U2AF–PRP19C complex via a direct interaction with U2AF65. RNAPII-associated U1 and SR proteins recognize a transcribed exon, resulting in its tethering to the RNAPII elongation complex through multiple interactions. (C) Transcription of the 3′ splice site results in a transition from protein–protein interactions between U2AF65 and the p-CTD to protein–RNA interactions, resulting in efficient recognition of the 3′ splice site. This facilitates rapid transition to a mature spliceosomal complex promoted by U2AF65-associated PRP19C.
Materials and methods
In vitro splicing
In vitro splicing assays were performed using S-100, appropriate nuclear fractions, and SRSF1 or SRSF1-CTD as described (Millhouse and Manley 2005). ImageQuant software was used to quantify the amount of spliced product (mRNA + intron) in each lane, with the equivalent regions in buffer D-containing reactions set as background.
Chromatography
NE for chromatography was prepared as described (Dignam et al. 1983). NF20–40 was prepared as described (Millhouse and Manley 2005). Chromatography was performed in buffer D (20 mM HEPES at pH 7.9, 5% glycerol, 0.2 mM EDTA, 100 mM KCl, 0.5 mM DTT, 1 mM PMSF), except for the Mono Q, which was run with buffer containing 20 mM tris-HCl (pH 7.9) instead of HEPES. For chromatography, 120 mg of NF20–40 was loaded on a 40-mL butyl-FF Hitrap column (GE Healthcare) and eluted with an 800–0 mM ammonium sulfate gradient in buffer D. Active fractions were pooled, concentrated by ammonium sulfate precipitation, desalted, and then loaded on an 8-mL Mono Q column (GE Healthcare). The Mono Q column was developed with a 0–250 mM ammonium sulfate gradient. Mono Q fractions were pooled, concentrated by ammonium sulfate precipitation, dialyzed in a microdialyzer (Gibco), and then assayed. Active fractions were loaded on a 1-mL Mono S column, which was developed with a 0–250 mM ammonium sulfate gradient, followed by a step to 800 mM ammonium sulfate. Mono S fractions were dialyzed as above, prior to CTD-dependent splicing assay. Excised proteins were identified using MALDI-TOF mass spectrometry essentially as described (Shevchenko et al. 2006).
Plasmids and recombinant proteins
SRSF1-CTD and SRSF1, SRSF1ΔRS-CTD, SRSF1ΔRS, and IgMA3 splicing substrate were prepared as described (Millhouse and Manley 2005). Baculovirus producing His-U2AF65 and His-U2AF65/35, a gift from Brenton Graveley, was used to infect Hi5 cells and was purified as described (Graveley et al. 2001). To produce U2AF65 and Prp19C in mammalian cells, U2AF65 and SPF27 were cloned into p3xFlag-CMV-14 and transfected into 293T cells. The following day, cells were split 1:5, then, 48 h after transfection, cells were collected and used to make NE. Purification of complexes was performed overnight using M2-agarose beads (Sigma), washed three times with buffer D containing the specified concentration of KCl, and then eluted with 0.2 mg/mL 3xFlag peptide (Sigma). GST-U2AF65 and deletion mutants were cloned into pGEX-6P1, induced in Rosetta cells overnight at 16°C, and purified using glutathione sepharose (GE Healthcare).
Antibodies and immunoblotting
The following antibodies were used: U2AF65 (Sigma, MC3), PLRG1 (Novus, NBP1-06556), SPF27 (Novus, NB110-40681), anti-CTD 8WG16 (laboratory stock), TFII-I (a gift of Ananda Roy, Tufts University), U2AF35 (a gift from Tom Maniatis, Columbia University Medical Center), GST (Molecular Probes, A5800), and anti-Flag M2 (Sigma, F1084). A Prp19 antibody from Bethyl Laboratories (A300-102A) was used in Figures 1E, 2B, and 5B, and a Prp19 antibody provided by R. Luhrmann was used in Figures 1D and 2D. A rabbit anti-CDC5L antibody from Abcam (ab31779-100) was used for the co-IP experiments in Figure 2A, while a mouse anti-CDC5L antibody from Santa Cruz Biotechnology (sc-81220) was used for immunodepletion. Immunoblotting was performed with Li-cor secondary antibodies, and was quantified using Li-cor software.
GST pull-down
For GST pull-downs using GST-tagged U2AF65 or truncated derivatives, 5 μg of immobilized protein was used for pull-down in 250 μg of S-100/NF20–40. Proteins were incubated for 3 h at 22°C, and then washed with buffer D. Proteins were eluted with 15 mM glutathione prior to SDS-PAGE and immunoblot analysis. GST-CTD was prepared and phosphorylated as described (Hirose and Manley 1998). For pull-downs, 5 μg of each CTD fusion protein was used for each 250 μg of extract or column fraction, incubated for 3 h at 22°C, and then washed three times with buffer D and eluted with 15 mM glutathione. For GST-CTD pull-down with recombinant U2AF65, 10 μg of His-U2AF65 was incubated with 5 μg of GST-CTD and incubated, washed, and eluted as above. For pull-downs performed with RNase pretreatment, extracts were preincubated in the presence of 20 μg/mL RNase A for 30 min at 37°C prior to incubation with GST-p-CTD.
Immunoprecipitations and depletions
For the co-IPs in Figure 2, 2 μg of each antibody (anti-Prp19 or CDC5L) was used to immunoprecipitate 100 μg of NF20–40. Beads were washed with buffer D before boiling and SDS-PAGE. For immunodepletions, 12 μg of each antibody (CDC5L or U2AF65) was used for each 50 μg of NF20–40 or Mono Q fraction. For each, 12 μg of anti-Flag M2 antibody served as a mock. Depletions were carried out for 2 h at 4°C in buffer D containing 500 mM NaCl. Following depletion, proteins were precipitated by addition of ammonium sulfate to 85%, redissolved in buffer D, and dialyzed against buffer D. Poly-U depletion of U2AF was performed as described (Page-McCaw et al. 1999). IP of spliceosomes was performed as described (Blencowe et al. 1994).
UV cross-linking
UV cross-linking U2AF65 IP was performed by setting up splicing reactions using 32P-UTP-labeled IgMA3 substrate as described (Millhouse and Manley 2005), but omitting PVA, and incubating for 10 min. The splicing reactions were placed on ice and then irradiated with UV light in a Stratalinker (Stratagene), followed by 30 min of RNase A treatment (20 μg/ mL). IP of cross-linked proteins was performed as described (Kashima and Manley 2003). For UV cross-linking with purified proteins, 10-μL reactions containing 32P-UTP-labeled IgMA3 substrate and 50 nM his-U2AF65 were incubated on ice for 10 min in the presence of buffer D or 200 nM SRSF1ΔRS/SRSF1ΔRS-CTD before exposure to UV light and RNase A treatment.
Acknowledgments
We thank B. Graveley, T. Maniatis, A. Roy, and R. Luhrmann for reagents. We thank N. Rao and M. Chen for NEs. J.-P. Hsin, P. Richard, M. Chen, Y. Shi, and other Manley laboratory members are acknowledged for helpful discussions. This work was supported by NIH grant R01-GM048259.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2038011.
Supplemental material is available for this article.
References
- Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will CL, Urlaub H, Stark H, et al. 2007. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J 26: 1737–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R 2008. Isolation of an active step I spliceosome and composition of its RNP core. Nature 452: 846–850 [DOI] [PubMed] [Google Scholar]
- Blencowe BJ, Nickerson JA, Issner R, Penman S, Sharp PA 1994. Association of nuclear matrix antigens with exon-containing splicing complexes. J Cell Biol 127: 593–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carty SM, Goldstrohm AC, Sune C, Garcia-Blanco MA, Greenleaf AL 2000. Protein-interaction modules that organize nuclear function: FF domains of CA150 bind the phosphoCTD of RNA polymerase II. Proc Natl Acad Sci 97: 9015–9020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC 2003. The Prp19p-associated complex in spliceosome activation. Science 302: 279–282 [DOI] [PubMed] [Google Scholar]
- Chung S, McLean MR, Rymond BC 1999. Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA 5: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Zhou Z, Huddleston KA, Harrison DA, Reed R, Coleman TA, Rymond BC 2002. Crooked neck is a component of the human spliceosome and implicated in the splicing process. Biochim Biophys Acta 1576: 287–297 [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Kornberg RD 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292: 1863–1876 [DOI] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R 2007. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell 26: 867–881 [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye MJ, Gromak N, Proudfoot NJ 2006. Exon tethering in transcription by RNA polymerase II. Mol Cell 21: 849–859 [DOI] [PubMed] [Google Scholar]
- Egloff S, Murphy S 2008. Cracking the RNA polymerase II CTD code. Trends Genet 24: 280–288 [DOI] [PubMed] [Google Scholar]
- Emili A, Shales M, McCracken S, Xie W, Tucker PW, Kobayashi R, Blencowe BJ, Ingles CJ 2002. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 8: 1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Bentley DL 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev 15: 1783–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A, Wiesner S, Martin-Malpartida P, Ramirez-Espain X, Ruiz L, Macias MJ 2006. The structure of Prp40 FF1 domain and its interaction with the crn-TPR1 motif of Clf1 gives a new insight into the binding mode of FF domains. J Biol Chem 281: 356–364 [DOI] [PubMed] [Google Scholar]
- Gornemann J, Kotovic KM, Hujer K, Neugebauer KM 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell 19: 53–63 [DOI] [PubMed] [Google Scholar]
- Graveley BR, Hertel KJ, Maniatis T 2001. The role of U2AF35 and U2AF65 in enhancer-dependent splicing. RNA 7: 806–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote M, Wolf E, Will CL, Lemm I, Agafonov DE, Schomburg A, Fischle W, Urlaub H, Luhrmann R 2010. Molecular architecture of the human Prp19/CDC5L complex. Mol Cell Biol 30: 2105–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Manley JL 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395: 93–96 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL 2000. RNA polymerase II and the integration of nuclear events. Genes Dev 14: 1415–1429 [PubMed] [Google Scholar]
- Hirose Y, Tacke R, Manley JL 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev 13: 1234–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Shuman S 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell 3: 405–411 [DOI] [PubMed] [Google Scholar]
- Hogg R, McGrail JC, O'Keefe RT 2010. The function of the NineTeen Complex (NTC) in regulating spliceosome conformations and fidelity during pre-mRNA splicing. Biochem Soc Trans 38: 1110–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell 12: 5–14 [DOI] [PubMed] [Google Scholar]
- Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8: 426–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan JL, Green MR 1999. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev 13: 462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima T, Manley JL 2003. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet 34: 460–463 [DOI] [PubMed] [Google Scholar]
- Kielkopf CL, Lucke S, Green MR 2004. U2AF homology motifs: protein recognition in the RRM world. Genes Dev 18: 1513–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadie SA, Tardiff DF, Kadener S, Rosbash M 2006. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev 20: 2055–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell 9: 1101–1111 [DOI] [PubMed] [Google Scholar]
- Listerman I, Sapra AK, Neugebauer KM 2006. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol 13: 815–822 [DOI] [PubMed] [Google Scholar]
- Lleres D, Denegri M, Biggiogera M, Ajuh P, Lamond AI 2010. Direct interaction between hnRNP-M and CDC5L/PLRG1 proteins affects alternative splice site choice. EMBO Rep 11: 445–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Legerski RJ 2007. The Prp19/Pso4 core complex undergoes ubiquitylation and structural alterations in response to DNA damage. Biochem Biophys Res Commun 354: 968–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL 1997a. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev 11: 3306–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL 1997b. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361 [DOI] [PubMed] [Google Scholar]
- Millhouse S, Manley JL 2005. The C-terminal domain of RNA polymerase II functions as a phosphorylation-dependent splicing activator in a heterologous protein. Mol Cell Biol 25: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DP, Greenleaf AL 2000. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 275: 39935–39943 [DOI] [PubMed] [Google Scholar]
- Munoz MJ, de la Mata M, Kornblihtt AR 2010. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem Sci 35: 497–504 [DOI] [PubMed] [Google Scholar]
- Page-McCaw PS, Amonlirdviman K, Sharp PA 1999. PUF60: a novel U2AF65-related splicing activity. RNA 5: 1548–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Wang D, Fu XD 2008. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol 20: 260–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley D 2009. ‘Cotranscriptionality’: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell 36: 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 20: 2922–2936 [DOI] [PubMed] [Google Scholar]
- Rosonina E, Ip JY, Calarco JA, Bakowski MA, Emili A, McCracken S, Tucker P, Ingles CJ, Blencowe BJ 2005. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol 25: 6734–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AL 2007. Signal-induced functions of the transcription factor TFII-I. Biochim Biophys Acta 1769: 613–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev 14: 2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL 2008. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat Struct Mol Biol 15: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Kan JL, Ghigna C, Biamonti G, Green MR 2004. A single polypyrimidine tract binding protein (PTB) binding site mediates splicing inhibition at mouse IgM exons M1 and M2. RNA 10: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ 2009. The SR protein family. Genome Biol 10: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Belotserkovskaya R, Reinberg D 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev 18: 2437–2468 [DOI] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM 2008. 'Nought may endure but mutability': spliceosome dynamics and the regulation of splicing. Mol Cell 30: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song EJ, Werner SL, Neubauer J, Stegmeier F, Aspden J, Rio D, Harper JW, Elledge SJ, Kirschner MW, Rape M 2010. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev 24: 1434–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Lee KR, Cheng SC 1993. The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol Cell Biol 13: 1883–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujvari A, Luse DS 2004. Newly Initiated RNA encounters a factor involved in splicing immediately upon emerging from within RNA polymerase II. J Biol Chem 279: 49773–49779 [DOI] [PubMed] [Google Scholar]
- Urano Y, Iiduka M, Sugiyama A, Akiyama H, Uzawa K, Matsumoto G, Kawasaki Y, Tashiro F 2006. Involvement of the mouse Prp19 gene in neuronal/astroglial cell fate decisions. J Biol Chem 281: 7498–7514 [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Jaladat Y 2010. The spliceosomal proteome: at the heart of the largest cellular ribonucleoprotein machine. Proteomics 10: 4128–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcarcel J, Gaur RK, Singh R, Green MR 1996. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science 273: 1706–1709 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402: 832–835 [DOI] [PubMed] [Google Scholar]
- Xiang K, Nagaike T, Xiang S, Kilic T, Behav MM, Manley JL, Tong L 2010. Crystal structure of the human symplekin–Ssu72–CTD phosphopeptide complex. Nature 467: 729–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin AJ 1997. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci 94: 12898–12903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Green MR 1989. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci 86: 9243–9247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Green MR 1991. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J 10: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Berget SM 2000. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol Cell Biol 20: 8290–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo P, Maniatis T 1996. The splicing factor U2AF35 mediates critical protein–protein interactions in constitutive and enhancer-dependent splicing. Genes Dev 10: 1356–1368 [DOI] [PubMed] [Google Scholar]