Abstract
Transcriptional activity of a gene is governed by transcriptional regulatory complexes that assemble/disassemble on the gene and control the chromatin architecture. How cytoplasmic components influence the assembly/disassembly of transcriptional regulatory complexes is poorly understood. Here we report that the budding yeast Saccharomyces cerevisiae has a chromatin architecture-modulating mechanism that is dependent on the endosomal lipid PI(3,5)P2. We identified Tup1 and Cti6 as new, highly specific PI(3,5)P2 interactors. Tup1—which associates with multiple transcriptional regulators, including the HDAC (histone deacetylase) and SAGA complexes—plays a crucial role in determining an activated or repressed chromatin state of numerous genes, including GAL1. We show that, in the context that the Gal4 activation pathway is compromised, PI(3,5)P2 plays an essential role in converting the Tup1-driven repressed chromatin structure into a SAGA-containing activated chromatin structure at the GAL1 promoter. Biochemical and cell biological experiments suggest that PI(3,5)P2 recruits Cti6 and the Cyc8–Tup1 corepressor complex to the late endosomal/vacuolar membrane and mediates the assembly of a Cti6–Cyc8–Tup1 coactivator complex that functions to recruit the SAGA complex to the GAL1 promoter. Our findings provide important insights toward understanding how the chromatin architecture and epigenetic status of a gene are regulated by cytoplasmic components.
Keywords: PI(3,5)P2 lipid; Cti6–Cyc8–Tup1; GAL1 transcriptional induction; lysosomal vacuolar endosome
Upon binding to its receptor, an extracellular ligand triggers signal transduction events at the cell surface (Schlessinger 2000; Cantley 2002). This leads to activation of cytoplasmic signaling cascades that often involve activation of transcriptional regulators. This cytoplasmic signaling ultimately controls the transcription of a distinctive set of genes in the nucleus, which primarily determines the output of the signaling pathway (Madhani and Fink 1998). Transcription of a gene is controlled by diverse mechanisms, including epigenetic regulation (Kornberg 1999; Roeder 2005; Fuda et al. 2009). Transcriptional regulatory complexes that assemble/disassemble at specific chromatin sites control the chromatin architecture and epigenetic status, and thus the transcriptional activity of a gene (Lemon and Tjian 2000; Cairns 2009). The assembly/disassembly of chromatin-associated transcriptional regulatory complexes may be influenced by cytoplasmic components through close communications between the nucleus and cytoplasm, but this remains poorly understood.
Phosphatidylinositol phosphates (PIPs, or phosphoinositide [PI] lipids) are lipids generated in small amounts at specific cellular membranes by distinct PIP kinases and function in cell signaling events. A specific PIP interacts with a specific set of effector proteins that contain the appropriate PIP-binding domain, thereby regulating the localization and activity of the signaling proteins. For example, Akt and PDK contain PH domains that bind PI(3,4,5)P3. Acute increases of PI(3,4,5)P3 at the plasma membrane by receptor tyrosine kinase-mediated activation of PI 3-kinase can recruit Akt and PDK to the plasma membrane, where the phosphorylation and activation of Akt by PDK takes place (Engelman et al. 2006). In another case, Tubby protein, a transcription factor for fat metabolism genes, binds PI(4,5)P2 at the plasma membrane. Upon stimulation by activated G-protein-coupled receptors, tubby is released from the membrane and shuttles into the nucleus to activate its target genes (Santagata et al. 2001).
Recently, it has been observed that endosomes play a key role in cytoplasmic signaling events (Tsukazaki et al. 1998; Coumailleau et al. 2009; Scita and Di Fiore 2010). Endocytosis generates vesicles, which fuse with early endosomes. The early endosomes mature into late endosomes and multivesicular bodies (MVBs) that fuse with the lysosome/vacuole (Scita and Di Fiore 2010). These endosomes constitute a dynamic endosomal system in the cell. It is at the endosomal membrane that the endocytosed activated TGF-β receptor kinase phosphorylates a transcriptional activator, Smad2—a key event in the TGF-β/Smad signaling pathway (Tsukazaki et al. 1998). PI(3)P, an endosomal PIP, plays a crucial role in the TGF-β/Smad signaling pathway (Tsukazaki et al. 1998). SARA (Smad anchor for receptor activation), a Smad-interacting protein, contains a FYVE domain that specifically binds PI(3)P. The endosomes containing both the endocytosed TGF-β receptor kinase and PI(3)P can recruit SARA, which in turn recruits Smad2. Smad2 is phosphorylated by TGF-β receptor kinase at the endosome. The phosphorylated Smad2 forms a transcriptional activator complex, the Smad2–Smad4 complex, which shuttles into the nucleus to activate its target genes.
The budding yeast Saccharomyces cerevisiae generates four PIPs: PI(3)P, PI(4)P, PI(3,5)P2, and PI(4,5)P2 (Strahl and Thorner 2007). PI(3)P and PI(3,5)P2 are generated at endosomal membranes. PI(3)P is synthesized from phosphatidylinositol by the PI 3-kinase Vps34 (Schu et al. 1993). PI(3,5)P2 is synthesized from PI(3)P by the PI(3)P 5-kinase Fab1 (Dove et al. 1997). Fab1 contains a FYVE domain at the N terminus and a phosphoinositide kinase domain at the C terminus (Gary et al. 1998). The FYVE domain binds PI(3)P. Fab1 localizes primarily at the endosomal/vacuolar membranes, but it also was detected in the cytosol (Gary et al. 1998). Fab1 has two positive regulators: Vac7 and Vac14. Cells without Vac7 cannot generate any detectable PI(3,5)P2, and cells without Vac14 generate 10%∼20% of the wild-type level of PI(3,5)P2 (Bonangelino et al. 2002; Rudge et al. 2004). Although PI(3,5)P2 is the rarest PIP in budding yeast, it functions in the regulation of several fundamental cellular processes, such as cell and vacuole size control, vacuolar acidification, membrane trafficking, and heat-shock response (Gary et al. 1998). At present, however, the molecular mechanisms by which PI(3,5)P2 regulates these processes are not well understood. Identification of the effector proteins that interact with PIPs has been critical to unravel the mechanisms by which they regulate cell signaling pathways. However, identification of PI(3,5)P2-binding proteins has been particularly elusive. To date, only one PI(3,5)P2-binding protein has been identified: Atg18 (Dove et al. 2004).
Here we report the identification of Tup1 and Cti6 as new PI(3,5)P2-binding proteins. Tup1 and Cti6 bind PI(3,5)P2 with high specificity. Tup1 is a highly conserved transcriptional regulator (human TLE [transducin-like enhancer of split] and Drosophila Groucho) (Smith and Johnson 2000). It forms a complex with Cyc8 (Ssn6) (Williams et al. 1991). The Cyc8–Tup1 complex functions as a transcriptional corepressor by interacting with gene-specific DNA-binding repressors such as Mig1, Rox1, and α2 (Smith and Johnson 2000). The Cyc8–Tup1 complex represses >150 genes, including FLO1, MATa-specific genes (STE2, STE6, MFA1, and MFA2), and GAL genes (GAL1, GAL3, and GAL4) (DeRisi et al. 1997; Smith and Johnson 2000; Rubio-Texeira 2005). The Cyc8–Tup1 complex mediates the formation of a compact repressive chromatin structure through its interaction with multiple transcriptional regulators—including HDAC (histone deacetylase) and histones—which prevents access of TBP and polymerase II (Pol II) to the promoter (Malave and Dent 2006).
In addition to its transcriptional corepressor function, the Cyc8–Tup1 complex also functions as a transcriptional coactivator during GAL1 induction (Papamichos-Chronakis et al. 2002). Distinct chromatin architecture and epigenetic statuses are established at the GAL1 promoter in the induced and repressed states (Lohr 1997; Lo et al. 2005; Pinskaya et al. 2009). In the repressed state, the GAL1 promoter is occupied by the Mig1–Cyc8–Tup1 repressor complex, HDAC Rpd3, and nucleosomes. In the induced state, the SAGA and SWI/SNF coactivator complexes and Snf1 are recruited to the promoter and the nucleosomes are disrupted. The two states also have different histone modification statuses (Lo et al. 2005; Pinskaya et al. 2009). The Cyc8–Tup1 complex is associated with the GAL1 promoter in both the repressed and activated states (Papamichos-Chronakis et al. 2002). During GAL1 induction, the Cyc8–Tup1 complex as well as the potent Gal4 activator participate in SAGA recruitment, one of the crucial steps for GAL1 transcriptional activation (Papamichos-Chronakis et al. 2002). Cti6, a Cyc8-interacting protein, plays a key role in the coactivator function of Cyc8–Tup1 by physically linking the Cyc8–Tup1 and SAGA complexes (Papamichos-Chronakis et al. 2002). Thus, Cti6 mediates the Cyc8–Tup1 complex-dependent SAGA recruitment to the GAL1 promoter (Papamichos-Chronakis et al. 2002). In addition, there are reports that the Cyc8–Tup1 complex can function as a transcriptional coactivator for genes like GRE2, AHP1, ARG1, FET3, and ARN1 (Proft and Struhl 2002; Kim et al. 2005; Crisp et al. 2006). The dual functionality seen for Tup1 also has been observed for other transcriptional regulators, such as Myc and REST/CoREST (Ballas and Mandel 2005; Eilers and Eisenman 2008; Abrajano et al. 2010). These transcriptional regulators can interact with multiple transcriptional regulatory proteins and assemble diverse complexes that function as either an activator or a repressor in a context-dependent manner.
We show here that S. cerevisiae has a PI(3,5)P2-dependent chromatin architecture-modulating mechanism that converts the Tup1-driven repressed state to a SAGA-containing activated state at the GAL1 promoter. In the context that the Gal4 activation pathway is compromised, this PI(3,5)P2-dependent mechanism becomes essential in establishing an activated chromatin state at the GAL1 promoter. Tup1 is recruited to the late endosomal/vacuolar membrane in a PI(3,5)P2-dependent manner. In galactose (Gal) medium, nuclear localization of Cti6 requires PI(3,5)P2, Cyc8, and Tup1. Furthermore, PI(3,5)P2 is required for the Cti6 interaction with the SAGA complex and Cti6–Cyc8–Tup1-mediated SAGA recruitment to the GAL1 promoter. We propose that, during GAL induction, the endosomal lipid PI(3,5)P2 converts the Cyc8–Tup1 corepressor complex to a coactivator complex by recruiting Cti6 and Cyc8–Tup1 and mediating the assembly of the Cti6–Cyc8–Tup1 complex, a transcriptional coactivator that functions to recruit the SAGA complex to the GAL1 promoter. Our results suggest that cytoplasmic PI(3,5)P2 lipid signaling is closely linked with nuclear regulatory events in remodulating the chromatin architecture and epigenetic status of a gene.
Results
A strategy for identifying candidate PI(3,5)P2 interactors
To better understand the molecular mechanisms of PI(3,5)P2-regulated cellular processes, we searched for potential PI(3,5)P2 interactors in S. cerevisiae based on the phenotypes of fab1 mutants and the amino acid sequence of Atg18, the first PI(3,5)P2 interactor to be identified. BLAST analysis of Atg18 identified proteins that share sequence similarity with Atg18, such as Atg21, Hsv2, Taf5, Tup1, Crn1, Lst8, Gid7, Sec12, Pwp2, and Erb1 (top 10 proteins). We focused our attention on Tup1 because the phenotypes of tup1 and fab1 mutants appear to indicate a genetic relationship between TUP1 and FAB1. First, we observed that deletion of TUP1 and FAB1 exhibited opposite cell flocculation phenotypes. Tup1 represses transcription of FLO genes, including the FLO1 gene, and deletion of TUP1 results in a strong cell flocculation phenotype. We observed that fab1 mutants exhibit a weaker flocculation phenotype than wild-type cells, suggesting that the genes that promote cell flocculation (like FLO1) may be poorly expressed in fab1Δ cells. Second, mutations in ste12+, the FAB1 homolog of Schizosaccharomyces pombe, result in a defect in transcriptional induction of certain mating genes (mat1-Pm, fus1, and sxa2) (Morishita et al. 2002). It is well known that MATa-specific mating genes are repressed by Tup1 in S. cerevisiae (Smith and Johnson 2000). These observations suggested that FAB1 antagonizes TUP1's repression function (Supplemental Fig. S1A).
Importantly, Atg18 was predicted to contain seven WD40-like domains and form a seven-blade β-propeller structure that is similar to the C-terminal β-propeller structure of Tup1 (Supplemental Fig. S1B) (Michell et al. 2006). Based on the genetic relationship (FAB1 and TUP1) and the structural similarity (Atg18 and Tup1 proteins), we hypothesized that (1) Tup1 may interact with PI(3,5)P2, and (2) PI(3,5)P2 may antagonize the repression function of Cyc8–Tup1 (Supplemental Fig. S1A).
Tup1 binds PI(3,5)P2 lipid
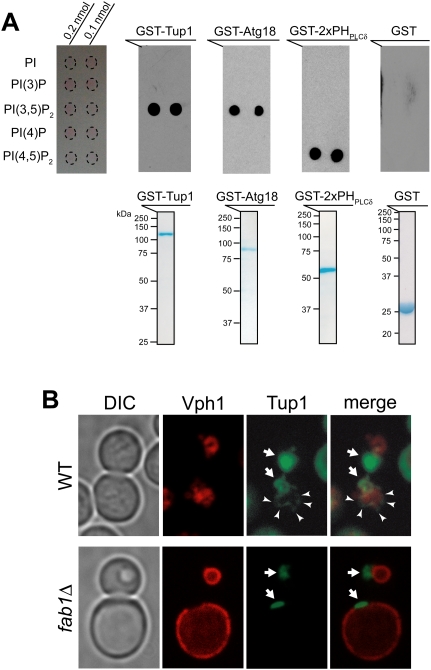
To test directly whether Tup1 binds PI(3,5)P2 lipid, we performed protein–lipid overlay assays. GST-Tup1 protein exclusively bound PI(3,5)P2 with high specificity, as did GST-Atg18, a known PI(3,5)P2 interactor, while a known PI(4,5)P2 interactor, GST-PHPLCδ domain, bound exclusively to PI(4,5)P2 (Fig. 1A). Thus, we identified Tup1 as a specific PI(3,5)P2 interactor. The C-terminal seven-blade β-propeller region of Tup1 (amino acids 301∼713) is sufficient for the PI(3,5)P2-specific interaction (Supplemental Fig. S2A).
Figure 1.
Tup1 binds PI(3,5)P2 lipid with a high specificity. (A, top panel) GST-Tup1 and GST-Atg18 bound PI(3,5)P2 with a high specificity, while GST-PHPLCδ specifically bound PI(4,5)P2. GST protein alone did not bind any PI lipid. (Bottom panel) Equivalent amount of purified, bacterially expressed GST fusion proteins (50 μg) were used for protein–lipid overlay assay. (B) Vph1-mCherry marks the limiting membrane of vacuoles (red). A pool of GFP-Tup1 localized at the vacuolar membrane (arrowhead), in addition to the nuclear Tup1 (arrow), was observed in some wild-type (WT) cells (see Supplemental Fig. S2B). In contrast, the vacuolar localization of Tup1 was not observed in fab1Δ cells.
To determine whether PI(3,5)P2 influences the cellular distribution of Tup1, we carefully examined the localization of GFP-Tup1 in wild-type and fab1Δ cells containing an mCherry-tagged Vph1, an integral vacuolar membrane protein. The fab1Δ cells have large vacuoles, and the cytoplasmic compartment is restricted to a small ring under the plasma membrane. The vast majority of Tup1 is found in the nucleus (Fig. 1B; Supplemental Fig. S2B). However, we also observed a small pool of cytoplasmic Tup1 that associated with the vacuolar endosomal membrane in some wild-type cells but not in fab1Δ cells (Figs. 1B, 3). Thus, PI(3,5)P2 appears to recruit a small pool of cytoplasmic Tup1 to the vacuolar endosomal membrane.
FAB1 regulates GAL induction
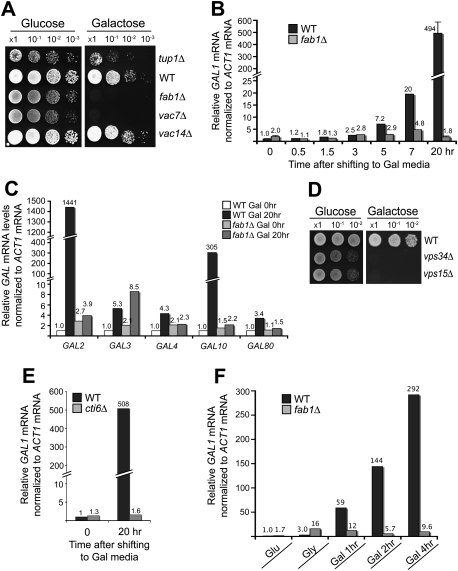
Next, we addressed the biological significance of the interaction between Tup1 and PI(3,5)P2. Among the many genes that are regulated by Tup1, the transcriptional regulation of the GAL genes is one of the best characterized. We analyzed GAL induction in fab1, vac7, and vac14 mutant cells. Remarkably, fab1Δ and vac7Δ cells exhibited a Gal− phenotype (Fig. 2A). The results are consistent with our hypothesis that PI(3,5)P2 antagonizes the repression function of Tup1 (Supplemental Fig. S1A). Interestingly, vac14Δ cells, which still contain ∼10% of wild-type levels of PI(3,5)P2, showed no discernible delay in Gal growth compared with wild-type cells.
Figure 2.
PI(3,5)P2 and Cti6 are required for transcriptional induction of GAL1. (A) SEY6210 fab1Δ and vac7Δ cells exhibited the Gal− phenotype. SEY6210 vac14Δ cells exhibited the Gal+ phenotype. SEY6210 tup1Δ cells showed slower Gal growth than wild-type (WT) cells. (B) RT-qPCR analysis showed gradual induction of GAL1 mRNA in SEY6210 wild-type cells. At 20 h after Gal shift, a 490-fold induction of GAL1 mRNA was observed in wild-type cells. No detectable increase in GAL1 mRNA was observed in fab1Δ cells at 20 h after Gal shift. The results that are shown with a standard deviation are from three independent analyses. (C) The mRNA levels of GAL2 and GAL10 were highly induced in SEY6210 wild-type cells, but remained constitutively repressed in SEY6210 fab1Δ cells at 20 h after Gal shift. The mRNA levels of GAL3, GAL4, and GAL10 were induced a few-fold in SEY6210 wild-type cells, while, in fab1Δ cells, the levels of GAL4 and GAL80 mRNA remained repressed (see a caveat in the text for the GAL3 mRNA results of fab1Δ cells). (D) SEY6210 vps34Δ and vps15Δ cells, which produce no PI(3)P and PI(3,5)P2, exhibited the Gal− phenotype. (E) SEY6210 cti6Δ cells exhibited a severe defect in GAL1 mRNA induction. (F) SEY6210 cells grown in Glu medium were shifted to Gly medium and grown for several hours to derepress GAL genes. When shifted to Gal medium, GAL1 mRNA was induced very rapidly in wild-type cells, whereas GAL1 remained constitutively repressed in fab1Δ cells.
We then examined whether the Gal− phenotype of fab1Δ cells is caused by inhibition of transcriptional induction of the GAL genes or by an indirect effect due to vacuolar dysfunction. We chose to analyze gene expression of GAL1, one of a few GAL genes whose transcription is regulated by Tup1. GAL1 transcription has served as a model for gene transcription studies and the underlying molecular mechanisms are well characterized (Johnston 1987; Winston and Carlson 1992; Ptashne and Gann 1997). The GAL1 promoter contains an upstream activating sequence (UAS) and an upstream repressor sequence (URS). The potent transcriptional activator Gal4 binds the UAS, and the transcriptional repressor Mig1 binds the URS. In glucose (Glu) medium, GAL1 is repressed: The GAL1 promoter is occupied by the Mig1–Cyc8–Tup1 repressor complex and Gal4 is inhibited by physical interaction with Gal80. Upon a shift into Gal medium (Gal shift), Gal3 antagonizes Gal80, freeing Gal4 to recruit the SAGA coactivator complex and Mediator complex (Bhaumik and Green 2001; Larschan and Winston 2001; Bryant and Ptashne 2003; Reeves and Hahn 2005). The SAGA and Mediator complexes, together with the SWI/SNF complex, transform the repressive chromatin architecture to an activated promoter structure that is accessible to TBP and Pol II.
Gal1 protein was strongly induced in SEY6210 wild-type cells at 20 h after Gal shift, while the Gal1 protein was not detectable in the fab1Δ cells (Supplemental Fig. S2C). RT-qPCR analysis showed gradual induction of GAL1 mRNA transcripts in SEY6210 wild-type cells after shifting to Gal medium (Fig. 2B). At 20 h after Gal shift, we observed ∼500-fold induction of GAL1 mRNA, but, remarkably, no detectable increase in GAL1 transcripts was observed in fab1Δ cells (Fig. 2B). The results suggest that PI(3,5)P2 plays a crucial role during GAL1 transcriptional induction in SEY6210.
RT-qPCR analysis of other GAL genes showed highly induced mRNA levels of GAL2 (∼1400-fold) and GAL10 (∼300-fold) in SEY6210 wild-type cells in Gal medium, whereas transcription of GAL2 and GAL10 remained constitutively repressed in fab1Δ cells (Fig. 2C). The mRNA levels of GAL3, GAL4, and GAL80 were induced a few-fold in SEY6210 wild-type cells, while, in SEY6210 fab1Δ cells, their mRNA levels were not significantly induced, except GAL3 (Fig. 2C). One caveat with the GAL3 results of fab1Δ cells is that, because ACT1 mRNA was used as an internal reference and the carbon and energy metabolism is inhibited in SEY6210 fab1Δ cells in Gal medium due to the GAL transcriptional defect, ACT1 mRNA levels could change and distort the GAL3 results of fab1Δ cells (giving a larger-fold increase in fab1Δ cells). The results suggest that transcriptional induction of the GAL regulon genes, particularly GAL genes (GAL1, GAL2, and GAL10) that are highly induced, depends on PI(3,5)P2 in SEY6210.
We examined whether PI(3)P, the substrate of Fab1 kinase, is also required for GAL induction. The SEY6210 vps34Δ and vps15Δ cells cannot generate PI(3)P [and PI(3,5)P2], and each strain exhibited a Gal− phenotype (Fig. 2D).
Next, we tested whether the strain background of SEY6210 might influence the predominant dependency of GAL1 transcription on PI(3,5)P2. The BY4742 strain induced high levels of GAL1 mRNA much faster than the SEY6210 strain (Fig. 2B; Supplemental Fig. S3A). Moreover, in a BY4742 background, fab1Δ cells induced GAL1 mRNA levels comparable with wild-type cells (Supplemental Fig. S3A), suggesting that the unique genetic makeup of SEY6210 makes FAB1 indispensable for GAL gene induction. SEY6210 contains the trp1Δ-901 mutation, a deletion that extends from TRP1 into the UAS region of GAL3, the neighboring gene. This mutation appears to make the strain highly sensitized to the function of FAB1. Reduced and delayed expression of Gal3, which is required for Gal4's activation function, will compromise the dynamics of the Gal4 activation pathway during GAL induction. Thus, the SEY6210 strain depends on PI(3,5)P2 for GAL induction.
The results suggest that S. cerevisiae has two activating forces [Gal4-dependent and PI(3,5)P2-dependent activation] that can function to override the repression by the Cyc8–Tup1 complex at the GAL1 promoter during GAL1 induction (Supplemental Fig. S3B). In the context that the Gal4 activation pathway is robust (BY4742), Gal4 drives to rapidly establish activated chromatin architecture. BY4742 fab1Δ cells appear to have only a modest GAL induction phenotype (Supplemental Fig. S3A), but, in the context that the Gal4 pathway is compromised (SEY6210), the PI(3,5)P2-dependent activation pathway becomes the major driving force to establish activated chromatin architecture by converting the Cyc8–Tup1 corepressor to the Cti6–Cyc8–Tup1 coactivator complex (Supplemental Fig. S3B). Although the PI(3,5)P2-dependent pathway is slower than the Gal4 pathway in establishing an activated state, it becomes essential to override the repressed chromatin structure when the Gal4 pathway is compromised. Thus, SEY6210 fab1Δ cells exhibit a drastic defect in GAL transcriptional induction (Fig. 2B).
Next we examined whether restoration of the robust Gal4 pathway in SEY6210 fab1Δ cells can shift the repression-oriented balance toward activation. When GAL3, with its intact endogenous promoter, was transformed into SEY6210 fab1Δ cells, it rescued the Gal growth defect in the fab1Δ cells (Supplemental Fig. S3C), consistent with the notion that the PI(3,5)P2-dependent pathway and the Gal4 pathway function in parallel during GAL induction (Supplemental Fig. S3B).
Cti6 is required for Cyc8–Tup1 complex-dependent SAGA recruitment to the GAL1 promoter, although it has not been shown how significantly the Cti6–Cyc8–Tup1 complex contributes to SAGA recruitment and GAL1 induction relative to the potent Gal4 activator (Papamichos-Chronakis et al. 2002). If the SEY6210 strain depends predominantly on the activation function of Cyc8–Tup1 for GAL induction as we postulated, it would depend primarily on Cti6 for GAL induction (Supplemental Fig. S3B). Indeed, SEY6210 cti6Δ cells exhibited a severe defect in transcriptional induction of GAL1, while BY4742 cti6Δ cells did not (Fig. 2E; Supplemental Fig. S3D). These results are consistent with the crucial activation function of the Cti6–Cyc8–Tup1 complex for GAL induction in the context that the Gal4 activation pathway is compromised.
GAL1 transcription was slowly induced in SEY6210 wild-type cells after shifting from Glu to Gal medium (Fig. 2B). We tested whether the slow GAL1 induction kinetics could be altered if SEY6210 cells are grown for several hours in nonrepressive glycerol (Gly) medium to derepress GAL genes before shifting into Gal medium. SEY6210 wild-type cells induced GAL1 transcription very rapidly, whereas, in SEY6210 fab1Δ cells, GAL1 still remains constitutively repressed (Fig. 2F). The rapid GAL1 induction could be driven by just the PI(3,5)P2-dependent activation pathway or together with the Gal4 activation pathway that may become functional after GAL gene derepression in Gly medium. Given that Gal4 and its regulators, Gal3 and Gal80, regulate transcription of their own genes, and thus form feedback regulatory loops, more careful analysis may be needed for a full understanding of these results.
Gal and hyperosmotic stress redistribute Tup1
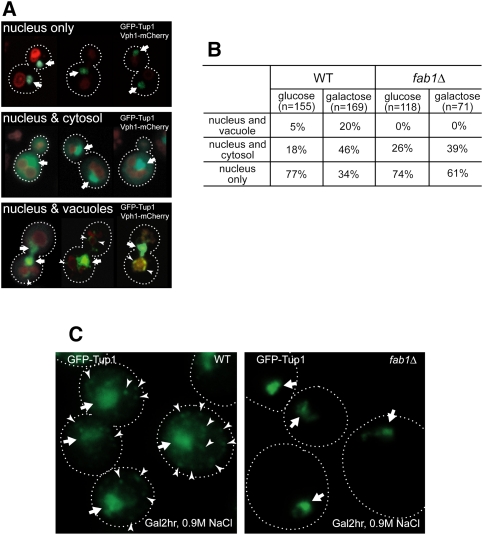
Our results suggest that PI(3,5)P2-mediated recruitment of Tup1 to the late endosomal/vacuolar membrane is required for the conversion of Cyc8–Tup1 from a transcriptional corepressor to a coactivator. We examined whether Tup1 localization changes in Gal medium. Nuclear GFP-Tup1 exhibits a bright signal, and every cell contains nuclear GFP-Tup1 (Fig. 3A). Cytosolic GFP-Tup1 exhibits a dispersed signal, and vacuolar membrane-localized GFP-Tup1 exhibits puncta at the vacuolar membranes (Fig. 3A). We scored the nuclear, vacuolar, and cytosolic localization of GFP-Tup1 in wild-type or fab1Δ cells either grown in Glu medium or shifted to Gal medium (Fig. 3B). In Gal medium, the cytoplasmic localization of Tup1 significantly increased in wild-type cells (Fig. 3B). Importantly, the recruitment of Tup1 to the vacuolar endosomal membrane also significantly increased in wild-type cells in Gal medium. However, the vacuolar recruitment of Tup1 could not be observed in fab1Δ cells in either Glu or Gal medium (Fig. 3B).
Figure 3.
Gal and hyperosmotic stress redistribute Tup1. (A) Montages of cells containing nuclear GFP-Tup1 only (top panel), nuclear and cytosolic GFP-Tup1 (middle panel), or nuclear and vacuolar GFP-Tup1 (bottom panel) and Vph1-mCherry. Nuclear GFP-Tup1 is indicated by an arrow and vacuolar membrane GFP-Tup1 is marked by an arrowhead. (B) Cells grown in Glu or Gal (2 h after shift) medium were imaged, analyzed, and scored for one of the three categories of GFP-Tup1 localization (nuclear only, nuclear and cytosolic, or nuclear and vacuolar membrane). Gal medium significantly enhanced cytosolic and vacuolar membrane localization of Tup1 in wild-type (WT) cells. In contrast, although Gal medium increased cytosolic localization of Tup1 in fab1Δ cells, vacuolar membrane localization of Tup1 was not observed. (C) When wild-type cells in Gal medium were treated with 0.9 M NaCl, Tup1 formed puncta (arrowhead) in the cytoplasm in most of the wild-type cells in addition to the nuclear Tup1 (arrow), whereas Tup1 did not form cytoplasmic puncta in fab1Δ cells.
The subset of wild-type cells containing Tup1 localized at the vacuolar membrane was a relatively minor portion of the total population. Even in the cells that exhibit vacuolar membrane-localized Tup1, only a small pool of Tup1 localized at the vacuolar membrane, presumably because Tup1 cycles rapidly from the vacuolar membrane back into the nucleus and at any given time only a small pool of nuclear Tup1 may undergo nucleocytoplasmic shuttling. We tested several conditions that could produce more robust localization of Tup1 at the late endosomal/vacuolar membrane. Upon hyperosmotic shock (0.9 M NaCl), PI(3,5)P2 levels are acutely elevated (20-fold) and the vacuoles fragment into many smaller compartments (Duex et al. 2006). Gal medium enhances the cytoplasmic localization of Tup1 (Fig. 3B). Therefore, we tested whether combining these two conditions altered Tup1 distribution. Remarkably, upon hyperosmotic treatment, cytoplasmic Tup1 formed puncta in most of the wild-type cells, but not in fab1Δ cells, suggesting a more robust response when PI(3,5)P2 levels are elevated (Fig. 3C). The Tup1 puncta are likely to correspond to endosomal membranes that contain the excess PI(3,5)P2.
Cti6 binds PI(3,5)P2 with high specificity, and the Cys residues in the PHD domain of Cti6 are important for nuclear localization of Cti6 in Gal medium
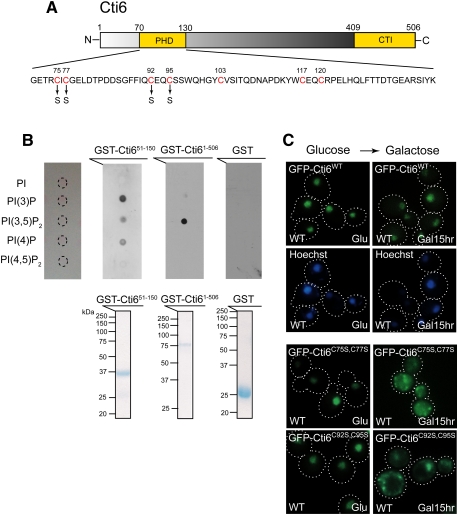
Next, we asked how the PI(3,5)P2–Tup1 interaction at the late endosomal/vacuolar membrane can contribute to the transcriptional activation function of Cyc8–Tup1. Interestingly, Cti6, which is required for Cyc8–Tup1's activation function, contains a PHD domain (Fig. 4A). A PHD domain is a general PIP-binding module (Gozani et al. 2003). Therefore, we tested whether Cti6 can bind any of the four PIPs that S. cerevisiae generates. The PHD domain of Cti6 has the capability of binding to PI(3)P, PI(3,5)P2, and PI(4)P (Fig. 4B). Remarkably, full-length Cti6 binds PI(3,5)P2 with high specificity (Fig. 4B). Cti6 is a new specific PI(3,5)P2-binding protein, structurally distinct from Atg18, a known PI(3,5)P2 interactor.
Figure 4.
Cti6 binds PI(3,5)P2 lipid, and the Cys residues in Cti6's PHD domain are important for nuclear localization of Cti6 in Gal medium. (A) Cti6 contains a PHD domain and a CTI domain. The PHD domain contains several Cys residues (red) that are potentially involved in zinc binding and important for PIP binding. Two Cti6 mutants were generated, such as Cti6C75S,C77S (Cys75–Cys77 → Ser75–Ser77) and Cti6C92S,C95S (Cys92–Cys95 → Ser92–Ser95). (B, top panel) Full-length Cti6 bound PI(3,5)P2 with high specificity, while the region (amino acids 51∼150) encompassing the PHD domain of Cti6 bound PI(3)P, PI(3,5)P2, and PI(4)P. (Bottom panel) Bacterially expressed GST fusion proteins were purified, and the same amounts of proteins were used for the protein–lipid overlay assay. (C) Wild-type Cti6 localized primarily in the nucleus in wild-type (WT) cells in both Glu and Gal media. The Cti6 mutants Cti6C75S,C77S and Cti6C92S,C95S localized in the nucleus in wild-type cells in Glu medium, but a significant portion of the mutant Cti6 proteins accumulated in the cytoplasm upon Gal shift.
Zinc-coordinating Cys residues in a PHD domain are important for PIP-binding activity (Gozani et al. 2003). The PHD domain of Cti6 has several Cys residues that are potentially involved in coordinating zinc. We examined whether these Cys residues are important for the localization of Cti6. Cti6, which contains four putative nuclear localization sequences (NLSs), localized primarily in the nucleus in wild-type cells in both Glu and Gal media (Fig. 4C). Cti6C75S,C77S (Cys75–Cys77 → Ser75–Ser77) and Cti6C92S,C95S (Cys92–Cys95 → Ser92–Ser95) localized primarily in the nucleus in Glu medium; however, the mutant Cti6 proteins accumulated in the cytoplasm of wild-type cells upon Gal shift (Fig. 4C). The results suggest that there is dynamic nucleocytoplasmic shuttling of Cti6 in Gal medium, and PI(3,5)P2 binding by Cti6 may be important for its shuttling into the nucleus.
Nuclear localization of Cti6 depends on PI(3,5)P2, Cyc8, and Tup1 in Gal medium
Both Tup1 and Cti6 bind PI(3,5)P2 with high specificity (Figs. 1A, 4B). Cti6 interacts with Cyc8 through its C-terminal CTI domain (Papamichos-Chronakis et al. 2002). The Cys mutants of Cti6, which may lose PI(3,5)P2-binding activity, mislocalized in the cytoplasm in Gal medium (Fig. 4C). These observations raised the possibility that PI(3,5)P2 recruits Cyc8–Tup1 and Cti6 to the late endosomal/vacuolar membrane and mediates an event that is required for Cti6's shuttling into the nucleus. Therefore, we examined Cti6's localization in fab1Δ, cyc8Δ, and tup1Δ cells.
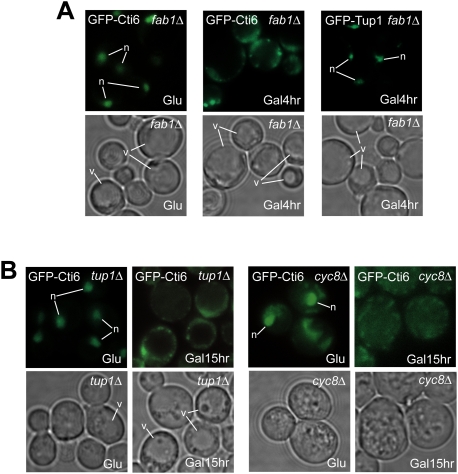
In fab1Δ cells grown in Glu medium, Cti6 localized primarily in the nucleus (Fig. 5A). However, Cti6 began to accumulate in the cytoplasm of fab1Δ cells upon Gal shift. At 4 h after Gal shift, most cells exhibited significant cytoplasmic accumulation of Cti6 (Fig. 5A). In contrast, Tup1 localized primarily in the nucleus at 4 h after Gal shift (Fig. 5A). Similarly, Cti6 started to accumulate in the cytoplasm of tup1Δ cells upon Gal shift (Fig. 5B). The kinetics of Cti6's cytoplasmic accumulation appeared to be slower in tup1Δ cells than in fab1Δ cells, perhaps because Tup1 may also affect Cti6's shuttling out of the nucleus. In cyc8Δ cells, the majority of Cti6 also localized in the cytoplasm in Gal medium (Fig. 5B). Even in Glu medium, a significant fraction of Cti6 often localized to the cytoplasm in cyc8Δ cells. These results show that PI(3,5)P2, Cyc8, and Tup1 are required for efficient nuclear localization of Cti6 in Gal medium.
Figure 5.
PI(3,5)P2 and the Cyc8–Tup1 complex are required for nuclear localization of Cti6 in Gal medium. (A) Cti6 localized primarily in the nucleus in fab1Δ cells in Glu medium, but Cti6 began to accumulate in the cytoplasm upon Gal shift, and a significant portion of Cti6 was observed in the cytoplasm at 4 h after Gal shift. Tup1 localized primarily in the nucleus at 4 h after Gal shift. (B) Cti6 localized in the nucleus in tup1Δ cells in Glu medium, but began to accumulate in the cytoplasm upon Gal shift. Similarly, in cyc8Δ cells, Cti6 localized in the nucleus in Glu medium, but a significant portion of Cti6 localized in the cytoplasm in Gal medium. Also, a significant portion of Cti6 often localized in the cytoplasm of cyc8Δ cells in Glu medium.
PI(3,5)P2 is required for Cti6-mediated recruitment of SAGA to the GAL1 promoter
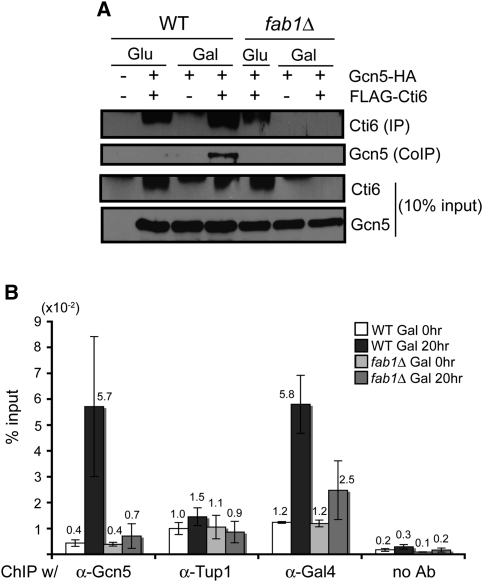
Cti6 links Cyc8–Tup1 with SAGA, thereby mediating Cyc8–Tup1-dependent SAGA recruitment (Papamichos-Chronakis et al. 2002). We examined whether SAGA recruitment depends on PI(3,5)P2. Using coimmunoprecipitation, we examined the interaction between Cti6 and Gcn5, a component of the SAGA complex, in wild-type and fab1Δ cells. The interaction between Cti6 and Gcn5 was significantly elevated in wild-type cells in Gal medium compared with Glu medium (Fig. 6A). The interaction of Cti6 with Gcn5 was not detectable in fab1Δ cells (Fig. 6A). In fact, in vivo, the levels of Cti6 in fab1Δ cells were significantly lower than in wild-type cells in Gal medium (Supplemental Fig. S4A). The amount of Cti6 further decreased during cell extraction from the cytosol of fab1Δ cells in Gal medium compared with the cytosol of wild-type cells (Fig. 6A). These results suggest that Cti6 is unstable in fab1Δ cells in Gal medium because Cti6 cannot form a proper complex.
Figure 6.
PI(3,5)P2 is required for Cti6's interaction with SAGA and for the recruitment of the SAGA complex to the GAL1 promoter in Gal medium. (A) Flag-tagged Cti6 was immunoprecipitated from the extracts prepared from the same number of cells grown in Glu and Gal media. Coimmunoprecipitated Gcn5 significantly increased from wild-type (WT) cell extract in Gal medium compared with in Glu medium. In contrast, coimmunoprecipitated Gcn5 was not detectable from fab1Δ cell extracts in both Glu and Gal media. Much less Cti6 was immunoprecipitated from fab1Δ cells in Gal medium, probably due to its instability in the cell extract (see the text). (B) ChIP-qPCR experiments were performed with wild-type and fab1Δ cells containing HA-tagged Gcn5 protein (n = 3). The amount of Gcn5 associated with the GAL1 promoter increased significantly (∼15-fold) in wild-type cells in Gal medium. In contrast, no significant increase of Gcn5 was detected in fab1Δ cells in Gal medium. The amount of Tup1 associated with the GAL1 promoter was not significantly changed in both wild-type and fab1Δ cells in Gal medium compared with Glu medium. A slightly increased amount of Tup1 associated with the GAL1 promoter was detected in wild-type cells in Gal medium. The amount of Gal4 bound to the GAL1 promoter increased a few-fold in wild-type cells (about fivefold) and fab1Δ cells (about twofold) in Gal medium.
Nuclear localization of Cti6, as well as its interaction with the SAGA complex in Gal medium, depends on PI(3,5)P2. We further examined whether recruitment of the SAGA complex to the GAL1 promoter, a crucial step during GAL1 induction, also depends on PI(3,5)P2. Chromatin immunoprecipitation (ChIP)-qPCR experiments were performed to analyze the recruitment of Gcn5 proteins, a component of the SAGA complex, and the association of Gal4 and Tup1 at the GAL1 promoter. The amount of Gcn5 associated with the GAL1 promoter increased significantly in wild-type cells in Gal medium (∼15-fold increase), while no significant increase was detected in fab1Δ cells (Fig. 6B). Gal4 binding increased a few-fold in wild-type cells (about fivefold) and fab1Δ cells (about twofold) in Gal medium (Fig. 6B). As reported before (Papamichos-Chronakis et al. 2002), the amount of Tup1 associated with the GAL1 promoter remained similar in Glu and Gal media (Fig. 6B). The results show that SAGA recruitment to the GAL1 promoter is dependent predominantly on PI(3,5)P2 when the Gal4 activation pathway is compromised.
Discussion
Transcriptional regulatory complexes that assemble/disassemble at specific chromatin sites regulate the chromatin architecture and epigenetic status, and thus the transcriptional activity of a gene. Extensive studies have been performed on the regulation of assembly/disassembly of transcriptional regulatory complexes at chromatin inside the nucleus, but regulation outside of the nucleus is not well characterized. Here we identified that S. cerevisiae has a chromatin architecture-modulating mechanism that is dependent on PI(3,5)P2 on the late endosomes/vacuoles. This finding provides an important and unique opportunity to understand how chromatin architecture and epigenetic status are influenced by cytoplasmic components.
We showed that (1) Tup1 binds PI(3,5)P2 lipid with high specificity; (2) a pool of Tup1 is recruited to the late endosomal/vacuolar membrane in a PI(3,5)P2-dependent manner; (3) when the Gal4 activation pathway is compromised (strain SEY6210, a GAL3 hypomorph), PI(3,5)P2 is indispensable for the transcriptional induction of GAL1, a new and unexpected role; (4) Cti6, a Cyc8–Tup1 interactor, is also essential for GAL induction in SEY6210; (5) Cti6 binds PI(3,5)P2 lipid with high specificity; (6) in Gal medium, nuclear localization of Cti6 depends on PI(3,5)P2, Cyc8, and Tup1; and (7) PI(3,5)P2 is required for the recruitment of the SAGA complex to the GAL1 promoter in SEY6210. Here we identified Tup1 and Cti6, key transcriptional regulators of GAL1, as new specific PI(3,5)P2-binding proteins. PI(3,5)P2 is required for nuclear localization of Cti6 and for Cti6–Cyc8–Tup1-mediated SAGA recruitment to the GAL1 promoter. Our results reveal that the endosomal lipid PI(3,5)P2 plays a key role in the transcriptional regulation of GAL1.
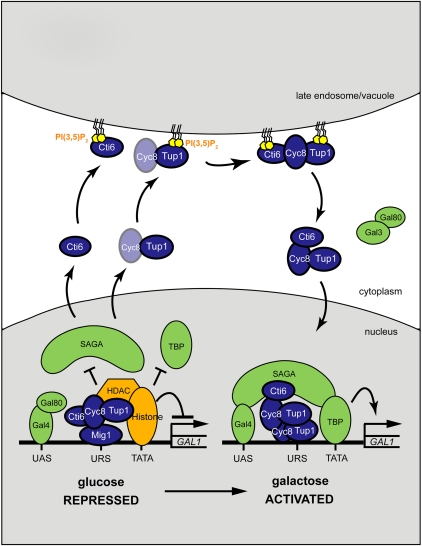
Based on these results, we propose that (1) upon Gal shift, there is dynamic nucleocytoplasmic shuttling of Cti6, Cyc8, and Tup1; (2) PI(3,5)P2 recruits Cyc8–Tup1 and Cti6 to the late endosomal/vacuolar membrane and mediates the assembly of the Cti6–Cyc8–Tup1 complex, an event that is required for Cti6's shuttling into the nucleus; and (3) once in the nucleus, the Cti6–Cyc8–Tup1 complex functions as a transcriptional coactivator by recruiting the SAGA complex to the GAL1 promoter (Fig. 7).
Figure 7.
A model for the PI(3,5)P2 function in GAL induction. We propose that (1) upon Gal shift, there is dynamic nucleocytoplasmic shuttling of Cti6, Cyc8, and Tup1; (2) PI(3,5)P2 recruits Cyc8–Tup1 and Cti6 to the late endosomal/vacuolar membrane and mediates the assembly of the Cti6–Cyc8–Tup1 complex that is required for Cti6's shuttling into the nucleus; and (3) once in the nucleus, the Cti6–Cyc8–Tup1 complex functions as a transcriptional coactivator by recruiting the SAGA complex to the GAL1 promoter.
To induce GAL genes upon Gal shift, the chromatin architecture at the GAL promoters has to transition from the repressed chromatin state to an activated chromatin state (Lohr 1997; Lo et al. 2005; Pinskaya et al. 2009). Because Mig1 and Gal80 rapidly translocate out of the nucleus upon Gal shift, significant changes at GAL promoters appear to take place at early times (De Vit et al. 1997; Peng and Hopper 2002). Our results suggest that a significant portion of Cti6 shuttles out of the nucleus upon Gal shift, and that PI(3,5)P2-containing endosomes/vacuoles mediate the assembly of a transcriptional coactivator, the Cti6–Cyc8–Tup1 complex. Similarly, in the TGF-β/Smad signaling in metazoan cells, PI(3)P-containing endosomes regulate the formation of a transcriptional activator, the Smad2–Smad4 complex, by recruiting the PI(3)P-binding protein SARA that anchors Smad2 at the endosome (Tsukazaki et al. 1998).
There are several advantages of functional regulation of transcriptional factors at the endosome instead of inside the nucleus. One obvious advantage is that the functional regulation of transcriptional factors can be tightly coupled to cytoplasmic signaling events, thus making nuclear regulation of gene expression more tightly intertwined with extracellular signals. Many of the genes regulated by Tup1 are required for stress responses. The recruitment of Tup1 to endosomes could allow it to detect cytoplasmic changes. In addition, this spatial separation of the functional regulation of these transcriptional regulators could permit more specific and tight transcriptional control of target genes.
We identified two new PI(3,5)P2-binding proteins: Tup1 and Cti6. Tup1 and Cti6 are known to form a complex, the Cti6–Cyc8–Tup1 complex (Papamichos-Chronakis et al. 2002). Our observations here suggest that PI(3,5)P2 recruits Cyc8–Tup1 and Cti6 to the late endosomal/vacuolar membrane and mediates the assembly of a transcriptional coactivator, the Cti6–Cyc8–Tup1 complex (Supplemental Fig. S4B). PI(3,4,5)P3 signaling at the plasma membrane recruits Akt and PDK, proteins containing PH domains that bind PI(3,4,5)P3. PDK then can phosphorylate and activate Akt (Supplemental Fig. S4B) (Engelman et al. 2006). PI(3,5)P2 and PI(3,4,5)P3 are among the rarest PIPs. Recruitment of two PIP-binding proteins by these rare PIPs to a specific locus at a specific time could provide a way to achieve a highly specific signaling event involving the two proteins in the complex intracellular environment.
We do not rule out the possibility that the PI(3,5)P2-dependent regulation of Tup1 and Cti6 might occur in the nucleus. For example, a water-soluble inositol 1,3,5-triphosphate [Ins(1,3,5)P3] that is cleaved from PI(3,5)P2 could diffuse into the nucleus and interact with Tup1 and Cti6, or a small pool of the Fab1 kinase might function in the nucleus. In fact, there are reports showing that a small pool of a PI 4-kinase and a PI(4)P 5-kinase exist in the nucleus (Audhya and Emr 2003; Strahl et al. 2005). In addition, inositol tetrakisphosphate and inositol pentakisphosphate modulate the activity of chromatin remodeling complexes, including the SWI/SNF complex (Shen et al. 2003; Steger et al. 2003). However, we do not favor these possibilities because (1) a water-soluble Ins(1,3,5)P3 has not been detected in yeast (Stevenson-Paulik et al. 2006); (2) nuclear Fab1 kinase has not been observed; and (3) Vac7, an integral vacuolar membrane protein, is essential for the function of Fab1 kinase (Gary et al. 2002).
Is the transcriptional induction of other Tup1-repressed genes regulated by a similar PI(3,5)P2-dependent mechanism? GRE2 is another Tup1-repressed gene whose transcription is activated by Cyc8–Tup1 and SAGA recruitment (Proft and Struhl 2002). RT-qPCR analyses of GRE2 mRNA showed that the transcriptional induction of GRE2 by hyperosmotic stress occurs normally in SEY6210 fab1Δ cells (Supplemental Fig. S4C). Importantly, the results suggest that PI(3,5)P2 is not generally required for the recruitment of general transcriptional regulators such as the SAGA complex or the Pol II complex. Although the transcriptional induction of GRE2 does not require PI(3,5)P2, we believe that there are additional Tup1-repressed genes whose induction depends on PI(3,5)P2. Interestingly, we found that SEY6210 fab1Δ cells have a sporulation defect similar to S. pombe fab1Δ cells (Morishita et al. 2002). Because several sporulation genes repressed by Cyc8–Tup1 (with a1–α2 repressors) have to be induced for sporulation to take place, it will be interesting to investigate whether the sporulation defect is due to a defect in transcriptional induction of some of these genes.
Tup1 is a highly conserved protein. Humans and Drosophila have TUP1 homologs—TLE and Groucho, respectively—that also function as transcriptional repressors (Buscarlet and Stifani 2007). It has been shown that Groucho plays a crucial role in Drosophila neurogenesis (Paroush et al. 1994). Recently, it was reported that mutations in the homologs of VAC14 and FIG4 cause neurodegeneration in humans and mice (Chow et al. 2007; Zhang et al. 2007). Given that Vac14 and Fig4 are required for normal synthesis and stability of PI(3,5)P2, and that TLE/Groucho proteins play an important role in neuronal development, the PI(3,5)P2-dependent regulatory pathway of GAL1 transcription that we identified here may provide important insights toward our understanding of these defects. The function of TLE/Groucho proteins may be regulated by PI(3,5)P2-dependent signaling analogous to our observations in yeast. Interestingly, cytoplasmic localization, as well as nuclear localization of TLE/Groucho, was reported in cultured human cells (Nuthall et al. 2002). An important test will be to examine whether PI(3,5)P2 is required for overriding or converting the repression function of a TLE/Groucho protein.
Materials and methods
Yeast strains and plasmids
Yeast strains and plasmids were generated by standard genetic and molecular methods. Supplemental Tables S1 and S2 summarize the yeast strains and plasmids used in this study.
Gal shift experiments
For all of the experiments performed in Glu medium, yeast cells were taken at the early exponential growth phase. For Gal shift experiments, yeast cells were grown in YPD or synthetic medium containing 2% Glu, collected at the exponential phase, washed twice with Glu-free medium, and resuspended in YP or synthetic medium containing 2% Gal (YPGal medium). For Gal shift experiments in Figure 2F, cells were grown in YPD, collected at the exponential phase, washed twice with Glu-free medium, and resuspended in YP containing 5% Gly and 0.01% Glu (YPGly medium). Cells grown in YPGly medium overnight were collected, washed twice with Glu-free medium, and resuspended in YPGal medium.
Protein–lipid overlay assay
Using commercial PIPs (Cell Signaling), protein–lipid overlay assays were performed based on Gozani et al. (2003). Fifty micrograms of bacterially expressed and purified proteins was used for each assay.
Bacterial expression and purification of protein
The proteins were expressed in C41 Escherichia coli (Lucigen) and purified by a single-step elution method according to the manufacturer's instructions.
Cell extraction, immunoprecipitation, and Western analysis
Yeast cells were lysed by standard bead-beating methods in 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.2% Tween 20, and protease inhibitor cocktail (Roche) (Han et al. 2005). For immunoprecipitation, soluble extract fractions were used after preparation by centrifuging the lysates at 13,000g for 10 min (twice). Proteins were analyzed by standard Western analysis methods (Han et al. 2003).
Dye staining and fluorescence microscopy
All imaging of yeast cells was performed using a Delta Vision RT system (Applied Precision). Hoechst 33342 (Invitrogen) was used to stain the nucleus in live yeast cells according to the manufacturer's instructions.
RT-qPCR analysis
Two-step RT-qPCR was performed using SYBR green (Petesch and Lis 2008). Total RNA was purified using RNeasy minikit (Qiagen), and first strand cDNA was synthesized using SuperScriptIII reverse transcriptase kit (Invitrogen). ACT1 mRNA was used as an internal reference calibrator. ΔΔCt analysis method was used to quantify mRNA. Supplemental Table S3 summarizes the primers used in RT-qPCR.
ChIP-qPCR analysis
Yeast cell cross-linking and lysis were performed based on Goldfarb and Alani's protocol (Goldfarb and Alani 2004). Five percent of input lysate was saved. The remaining lysate was divided into two aliquots, one of which was immunoprecipitated with α-HA antibody (12CA5, Roche) and the other with no antibody. qPCR was performed with the purified DNA from the immunoprecipitate based on Petesch and Lis's methods (Petesch and Lis 2008) using a pair of primers (TGATTTTTGATCTATTAACAGATA and CATTTGAATAAGAAGTAATACAAA) that amplify a region in the GAL1 promoter (Papamichos-Chronakis et al. 2002; Petesch and Lis 2008).
[H3]inositol labeling and HPLC analysis of PIPs
Quantification of in vivo PIP levels was performed as described previously (Gary et al. 1998).
Acknowledgments
We thank John Lis, Fred Winston, Michael Polymenis, and Damien Garbett for helpful comments on the manuscript; Robert Trumbly and Dimitris Tzamarias for sharing reagents; Chris Stephen, Jason MacGurn, Mike Henne, Yading Ling, Andy Manford, and Emr laboratory members for helpful discussions; John Lis, Leighton Core, Nick Fuda, and Steven Petesch for help and sharing instruments and reagents for RT-qPCR and ChIP-qPCR analysis; and Yuxin Mao for help in preparing Supplemental Figure S1B.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1998611.
Supplemental material is available for this article.
References
- Abrajano JJ, Qureshi IA, Gokhan S, Molero AE, Zheng D, Bergman A, Mehler MF 2010. Corepressor for element-1-silencing transcription factor preferentially mediates gene networks underlying neural stem cell fate decisions. Proc Natl Acad Sci 107: 16685–16690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Emr SD 2003. Regulation of PI4,5P2 synthesis by nuclear-cytoplasmic shuttling of the Mss4 lipid kinase. EMBO J 22: 4223–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Mandel G 2005. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol 15: 500–506 [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev 15: 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino CJ, Nau JJ, Duex JE, Brinkman M, Wurmser AE, Gary JD, Emr SD, Weisman LS 2002. Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J Cell Biol 156: 1015–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant GO, Ptashne M 2003. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol Cell 11: 1301–1309 [DOI] [PubMed] [Google Scholar]
- Buscarlet M, Stifani S 2007. The ‘Marx’ of Groucho on development and disease. Trends Cell Biol 17: 353–361 [DOI] [PubMed] [Google Scholar]
- Cairns BR 2009. The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198 [DOI] [PubMed] [Google Scholar]
- Cantley LC 2002. The phosphoinositide 3-kinase pathway. Science 296: 1655–1657 [DOI] [PubMed] [Google Scholar]
- Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, Szigeti K, Shy ME, Li J, Zhang X, et al. 2007. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature 448: 68–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coumailleau F, Furthauer M, Knoblich JA, Gonzalez-Gaitan M 2009. Directional Delta and Notch trafficking in Sara endosomes during asymmetric cell division. Nature 458: 1051–1055 [DOI] [PubMed] [Google Scholar]
- Crisp RJ, Adkins EM, Kimmel E, Kaplan J 2006. Recruitment of Tup1p and Cti6p regulates heme-deficient expression of Aft1p target genes. EMBO J 25: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686 [DOI] [PubMed] [Google Scholar]
- De Vit MJ, Waddle JA, Johnston M 1997. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell 8: 1603–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH 1997. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature 390: 187–192 [DOI] [PubMed] [Google Scholar]
- Dove SK, Piper RC, McEwen RK, Yu JW, King MC, Hughes DC, Thuring J, Holmes AB, Cooke FT, Michell RH, et al. 2004. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J 23: 1922–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex JE, Nau JJ, Kauffman EJ, Weisman LS 2006. Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot Cell 5: 723–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN 2008. Myc's broad reach. Genes Dev 22: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 7: 606–619 [DOI] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT 2009. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461: 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD 1998. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol 143: 65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Sato TK, Stefan CJ, Bonangelino CJ, Weisman LS, Emr SD 2002. Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase pathway by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. Mol Biol Cell 13: 1238–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T, Alani E 2004. Chromatin immunoprecipitation to investigate protein–DNA interactions during genetic recombination. Methods Mol Biol 262: 223–237 [DOI] [PubMed] [Google Scholar]
- Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, et al. 2003. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 114: 99–111 [DOI] [PubMed] [Google Scholar]
- Han BK, Aramayo R, Polymenis M 2003. The G1 cyclin Cln3p controls vacuolar biogenesis in Saccharomyces cerevisiae. Genetics 165: 467–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BK, Bogomolnaya LM, Totten JM, Blank HM, Dangott LJ, Polymenis M 2005. Bem1p, a scaffold signaling protein, mediates cyclin-dependent control of vacuolar homeostasis in Saccharomyces cerevisiae. Genes Dev 19: 2606–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M 1987. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev 51: 458–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Swanson MJ, Qiu H, Govind CK, Hinnebusch AG 2005. Activator Gcn4p and Cyc8p/Tup1p are interdependent for promoter occupancy at ARG1 in vivo. Mol Cell Biol 25: 11171–11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD 1999. Eukaryotic transcriptional control. Trends Cell Biol 9: M46–M49 doi: 10.1016/S0962-8924(99)01679-7 [PubMed] [Google Scholar]
- Larschan E, Winston F 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev 15: 1946–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon B, Tjian R 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev 14: 2551–2569 [DOI] [PubMed] [Google Scholar]
- Lo WS, Gamache ER, Henry KW, Yang D, Pillus L, Berger SL 2005. Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J 24: 997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D 1997. Nucleosome transactions on the promoters of the yeast GAL and PHO genes. J Biol Chem 272: 26795–26798 [DOI] [PubMed] [Google Scholar]
- Madhani HD, Fink GR 1998. The riddle of MAP kinase signaling specificity. Trends Genet 14: 151–155 [DOI] [PubMed] [Google Scholar]
- Malave TM, Dent SY 2006. Transcriptional repression by Tup1-Ssn6. Biochem Cell Biol 84: 437–443 [DOI] [PubMed] [Google Scholar]
- Michell RH, Heath VL, Lemmon MA, Dove SK 2006. Phosphatidylinositol 3,5-bisphosphate: metabolism and cellular functions. Trends Biochem Sci 31: 52–63 [DOI] [PubMed] [Google Scholar]
- Morishita M, Morimoto F, Kitamura K, Koga T, Fukui Y, Maekawa H, Yamashita I, Shimoda C 2002. Phosphatidylinositol 3-phosphate 5-kinase is required for the cellular response to nutritional starvation and mating pheromone signals in Schizosaccharomyces pombe. Genes Cells 7: 199–215 [DOI] [PubMed] [Google Scholar]
- Nuthall HN, Joachim K, Palaparti A, Stifani S 2002. A role for cell cycle-regulated phosphorylation in Groucho-mediated transcriptional repression. J Biol Chem 277: 51049–51057 [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Petrakis T, Ktistaki E, Topalidou I, Tzamarias D 2002. Cti6, a PHD domain protein, bridges the Cyc8–Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol Cell 9: 1297–1305 [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RL Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D 1994. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79: 805–815 [DOI] [PubMed] [Google Scholar]
- Peng G, Hopper JE 2002. Gene activation by interaction of an inhibitor with a cytoplasmic signaling protein. Proc Natl Acad Sci 99: 8548–8553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134: 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinskaya M, Gourvennec S, Morillon A 2009. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J 28: 1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Struhl K 2002. Hog1 kinase converts the Sko1–Cyc8–Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell 9: 1307–1317 [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A 1997. Transcriptional activation by recruitment. Nature 386: 569–577 [DOI] [PubMed] [Google Scholar]
- Reeves WM, Hahn S 2005. Targets of the Gal4 transcription activator in functional transcription complexes. Mol Cell Biol 25: 9092–9102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG 2005. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett 579: 909–915 [DOI] [PubMed] [Google Scholar]
- Rubio-Texeira M 2005. A comparative analysis of the GAL genetic switch between not-so-distant cousins: Saccharomyces cerevisiae versus Kluyveromyces lactis. FEM Yeast Res 5: 1115–1128 [DOI] [PubMed] [Google Scholar]
- Rudge SA, Anderson DM, Emr SD 2004. Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14–Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol Biol Cell 15: 24–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L 2001. G-protein signaling through tubby proteins. Science 292: 2041–2050 [DOI] [PubMed] [Google Scholar]
- Schlessinger J 2000. Cell signaling by receptor tyrosine kinases. Cell 103: 211–225 [DOI] [PubMed] [Google Scholar]
- Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD 1993. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260: 88–91 [DOI] [PubMed] [Google Scholar]
- Scita G, Di Fiore PP 2010. The endocytic matrix. Nature 463: 464–473 [DOI] [PubMed] [Google Scholar]
- Shen X, Xiao H, Ranallo R, Wu WH, Wu C 2003. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science 299: 112–114 [DOI] [PubMed] [Google Scholar]
- Smith RL, Johnson AD 2000. Turning genes off by Ssn6–Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem Sci 25: 325–330 [DOI] [PubMed] [Google Scholar]
- Steger DJ, Haswell ES, Miller AL, Wente SR, O'Shea EK 2003. Regulation of chromatin remodeling by inositol polyphosphates. Science 299: 114–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson-Paulik J, Chiou ST, Frederick JP, dela Cruz J, Seeds AM, Otto JC, York JD 2006. Inositol phosphate metabolomics: merging genetic perturbation with modernized radiolabeling methods. Methods 39: 112–121 [DOI] [PubMed] [Google Scholar]
- Strahl T, Thorner J 2007. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim Biophys Acta 1771353–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl T, Hama H, DeWald DB, Thorner J 2005. Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J Cell Biol 171: 967–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL 1998. SARA, a FYVE domain protein that recruits Smad2 to the TGFβ receptor. Cell 95: 779–791 [DOI] [PubMed] [Google Scholar]
- Williams FE, Varanasi U, Trumbly RJ 1991. The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiae are associated in a protein complex. Mol Cell Biol 11: 3307–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Carlson M 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet 8: 387–391 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zolov SN, Chow CY, Slutsky SG, Richardson SC, Piper RC, Yang B, Nau JJ, Westrick RJ, Morrison SJ, et al. 2007. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc Natl Acad Sci 104: 17518–17523 [DOI] [PMC free article] [PubMed] [Google Scholar]