Abstract
Eukaryotic protein kinases evolved as a family of highly dynamic molecules with strictly organized internal architecture. A single hydrophobic F-helix serves as a central scaffold for assembly of the entire molecule. Two non-consecutive hydrophobic structures termed “Spines” anchor all the elements important for catalysis to the F-helix. They make firm, but flexible, connections within the molecule providing a high level of internal dynamics of the protein kinase. During the course of evolution, protein kinases developed a universal regulatory mechanism associated with a large Activation Segment that can be dynamically folded and unfolded in the course of cell functioning. Protein kinases thus represent a unique, highly dynamic and precisely regulated set of switches that control most biological events in eukaryotic cells.
Protein kinases: major signal transmitters in eukaryotic cells
Protein phosphorylation was discovered as a regulatory mechanism by Krebs and Fischer in the late 1950s through their classic studies of glycogen phosphorylase and their subsequent discovery of phosphorylase kinase [1]. The second protein kinase to be discovered was cAMP-dependent protein kinase (PKA) [2]. The tremendous diversity of the protein kinase family was later revealed when Src, the transforming protein from Rous Sarcoma Virus, was discovered to be a kinase that phosphorylates tyrosine rather than serine or threonine [3]. Comparing the sequences of Src and PKA established that they had evolved from a common precursor [4], and this defined the boundaries of what we now recognize as one of the largest gene families encoded by eukaryotic genomes.
Based on sequence similarities alone, in an era of “pre-BLAST'” science, Hanks and Hunter discovered a host of conserved sequence motifs embedded within the kinase core, which they classified into 12 subdomains [5]. Their analysis has stood the test of time. Sequencing of whole genomes allowed us to define complete kinomes and only then was the full magnitude and diversity of the family revealed. The human kinome, for example, encodes over 500 protein kinases, many more if one considers splice variants. This corresponds to nearly 2% of the entire genome and demonstrates the extraordinary importance of this family for regulating biological events [6].
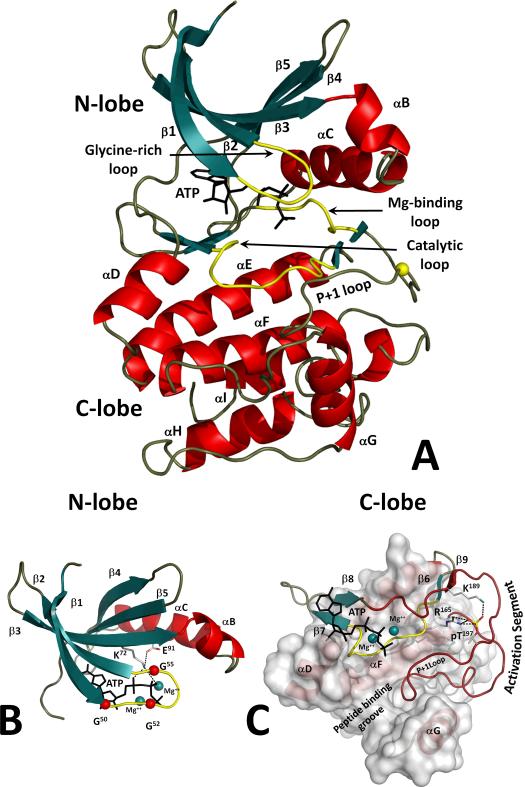
Although sequence was sufficient to define the conserved motifs, a structure was required to reveal what these conserved sequence motifs were doing and how they related to the overall structure and function of the protein. The PKA catalytic subunit was the first protein kinase to be crystallized, and that structure, co-crystallized with a high affinity inhibitor peptide derived from the small heat stable protein kinase inhibitor (PKI), revealed not only the overall architecture of the kinase core including where all the conserved residues were located [7], but also the way in which a kinase can recognize a peptide/protein substrate [8]. The subdomains defined by Hanks and Hunter could now be mapped onto a structure, and the sequence motifs immediately took on functional identities such as the Glycine-rich Loop, the Catalytic Loop and the P+1 Loop (Figure 1).
Figure 1. Structure of the conserved protein kinase core.
A. Protein kinases have a characteristic bilobal fold. The N-terminal lobe (N-lobe) contains five β strands (1 through 5; colored teal) and a universally conserved αC-helix. The C-lobe is mostly helical (colored red). An ATP molecule is bound to a deep cleft between the lobes. Major catalytically important loops are colored yellow. B. N-lobe structure. The Glycine-rich Loop coordinates the ATP phosphates. Three conserved glycines are shown as red spheres. Lys72 from β3 couples the phosphates and the C-helix. C. Catalytic and regulatory machinery is bound to the rigid helical core of the C-lobe. The extended Activation Segment (colored dark red) contains a phosphorylation site that is bound to β9 (K189) and the HRD-arginine (R165). The P+1 Loop accommodates the P+1 residue of the peptide substrate that is docked to the peptide binding groove.
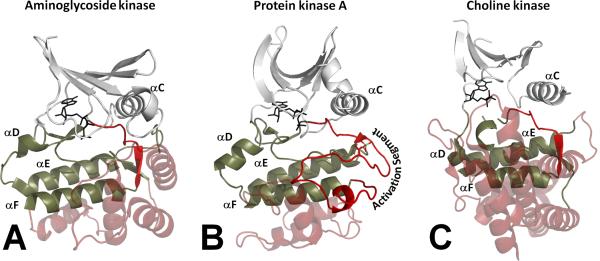
What are the origins of the eukaryotic protein kinases (EPKs) and what, if anything, makes them unique compared to other enzymes? The EPKs evolved in a divergent manner from much simpler eukaryotic-like kinases (ELKs) that are abundant, but poorly understood, in prokaryotes [9, 10]. Like the EPKs, they have an N-Lobe and a C-Lobe, and the adenine ring of ATP is buried at the base of the cleft between the two lobes. The complex catalytic machinery is also largely conserved in the ELKs. What then is different? Unlike the ELKs, the EPKs are highly regulated. They are molecular switches that are turned on and off by very precise biological cues, typically initiated by an extracellular signal such as a hormone, neurotransmitter, nutrient deprivation, or some other kind of stress. The structural features of the EPKs that distinguish them clearly from the ELKs correlate with two subdomains that are fused to the C-Lobe (Figure 2). One is the Activation Segment that is typically ordered by a critical phosphorylation event, and the other is a helical subdomain that provides docking sites for protein/peptide substrates. We describe here how the internal architecture of the EPK core is organized and then define how this architecture contributes in a unique way to the dynamic regulation of the EPKs and to catalysis. In conclusion, we will return to the evolution of the highly regulated and dynamic protein kinase molecule.
Figure 2. General structural differences between Eukaryote Like Kinases and EPKs.
EPKs are represented by Protein kinase A (panel B). Structural elements conserved in all kinases are shown as white (N-lobe) and tan (C-lobe) cartoons. Non-conserved helical C-terminal regions of the C-lobe are shown as red transparent helices. All EPKs have a helical motif comprising three short helices: G, H and I (GHI-domain). ELKs (panels A and C) contain multiple non-conserved helices that accommodate non-peptide substrates and are specific for each kinase. Another radical difference of EPKs from ELKs is the presence of an extended Activation Segment that lies between the DFG-motif and the F-helix. This segment is the latest evolutionary feature developed by EPKs that allows dynamic regulation of their activity.
Internal architecture of protein kinases
Because the kinases are so important not only for biology but also for disease phenotypes, many kinase structures have now been solved (Box 1). What can we learn from this structural kinome that we could not learn from in depth analysis of a single kinase? By comparing many protein kinase structures and searching, in particular, for spatially conserved residues, the internal architecture that allows for the assembly of an active protein kinase was revealed. It is an architecture that enables distal parts of the enzyme to be linked by conserved hydrophobic elements. Moreover, it is an architecture that is fully assembled in the active conformation of the kinase but broken in the most inactive kinases. This complex regulatory machinery not only distinguishes the EPKs from the ELKs, but also from most metabolic enzymes. Although metabolic enzymes can be highly regulated by allosteric mechanisms and by post-translational modifications, they do not function as dynamic switches and are not typically associated with the complex regulatory mechanisms that define the EPKs. The EPKs have evolved not to efficiently turn over lots of product, but rather to be transiently activated.
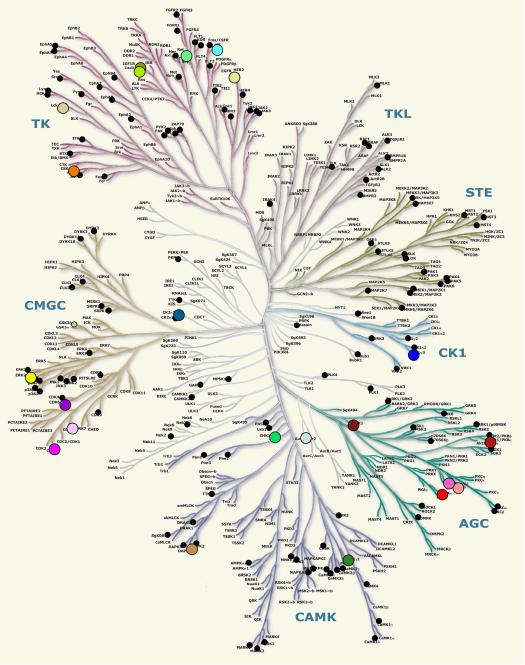
Box 1. Structural coverage of the human kinome.
The human kinome includes 518 protein kinases which were divided into seven major subfamilies [53]. The 155 publicly available human protein kinase structures were known by August 2010 (Figure I). Many of these proteins were crystallized in different conformations, bound to different ligands and proteins. A comparison of their structures demonstrates a high level of structural plasticity of protein kinases.
Fig I. The human kinome. Publicly available human protein kinase structures are shown as black circles. Colored circles represent the 23 EPKs aligned in Figures 3 and 4. Kinome illustration reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com).
Local Spatial Pattern (LSP) alignment was developed as a method to rapidly compare any two structures and identify spatially conserved residues (Box 2) [11, 12]. This led to the identification of spatially conserved hydrophobic motifs, termed Spines, that explain how a protein kinase is assembled into an active enzyme and revealed the internal architecture of protein kinase core.
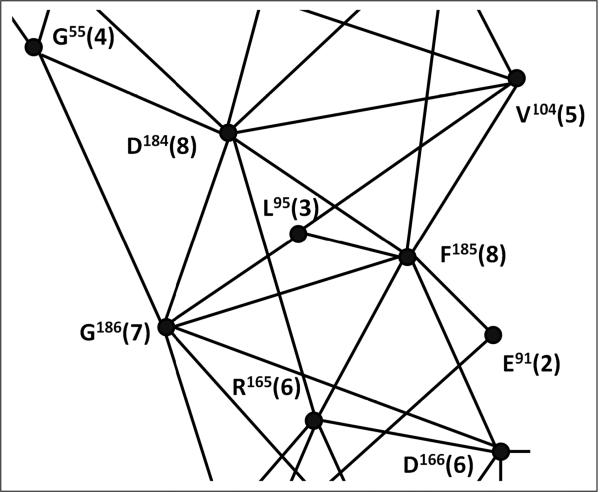
Box 2. Detecting functionally important residues by Local Spatial Pattern alignment.
Local Spatial Pattern (LSP) alignment is a new method to compare a pair of protein molecules and to detect residues that form similar spatial patterns in both proteins. This method disregards both the sequences and main chain geometry of the proteins. All residues are considered to be individual vectors that are arrayed in space. Proteins, thus, are represented by graphs with residues as vertices and edges describing their mutual orientation. The main advantage of the method is that each residue in the detected pattern is scored according to its involvement in the formation of the pattern (Figure I). The Involvement score corresponds to the number of connections on the similarity graph. This approach allows the detection of single residues that do not form any sequential motifs, but whose spatial positions are strictly conserved. It is especially effective in recognition of spatial motifs formed by hydrophobic residues because they can easily substitute for each other and, thus, are not rigorously conserved in the protein sequence. LSP alignment played a key role in the detection of the non-consecutive hydrophobic Spines.
Figure I. Involvement score is used to predict functional importance of residues. A fragment of similarity graph between PKA and CDK2 [11] showing conserved spatial relations between residues around the DFG-motif. A line (edge) between two residues (vertices) indicates that mutual spatial orientation of these residues in PKA and CDK2 are similar. Different residues can have different numbers of connections on the graph (shown in parentheses). The higher the number, the more conserved spatial relations this residue has in the molecule. This number, termed the “Involvement score”, can be used as a predictor of functional importance for the residues.
N-lobe
Each kinase consists of two structurally and functionally distinct lobes that contribute in unique ways to both catalysis and regulation, and the concerted way in which these two lobes interface is quite distinct compared to most metabolic kinases such as hexokinase or the ATPases. For simplicity in describing the domains and the motifs we shall use PKA numbering of individual residues, but the residues are conserved in every kinase. The smaller N-lobe is dominated by a five stranded β-sheet, which is coupled to a helical subdomain that typically consists of the C-helix (Figure 1). The αC-β4 Loop is the only part of the N-lobe that is functionally and constitutively anchored to the C-lobe [13, 14].
Two highly conserved sequence motifs are embedded within the first three β-strands. The first motif (GxGxxG) is the Glycine-rich Loop between β1 and β2. This loop folds over the nucleotide and positions the γ-phosphate of ATP for catalysis and is the most flexible part of the N-lobe. The tip of this loop is only fully closed when its backbone is anchored to the γ-phosphate of ATP in a ternary complex [15]. The Gly-rich Loop is quite distinct from the P-Loop that is often referred to as the Walker-A motif (GxxxxGKT/S) [16]. Even though both anchor nucleotide bound phosphates, their interaction with the adenine ring of ATP is different. While the P-loop does not have any contacts with the purine moiety of ATP, the Gly-rich Loop connects two β-strands that harbor the adenine ring (Figure 1b). Accordingly, instead of invariant Ser/Thr in the Walker-A motif, that binds to the γ-phosphate, the Gly-rich Loop is followed by a highly conserved Val57, that makes a hydrophobic contact to the base of ATP.
Another important motif, (AxK), is in the β3 strand. Lys72 from this motif couples the phosphates of ATP to the C-helix. The C-helix is a unique, very dynamic regulatory element in the protein kinase molecule. In terms of sequence it belongs to the N-lobe but occupies a strategically important position between the two lobes. The C-helix connects to many different parts of the molecule thus serving as a “Signal Integration Motif” [17]. Its C-terminus is anchored to the rigid body of the C-lobe by the αC-β4 Loop whereas its N-terminus interfaces with the Activation Loop. Positioning the N-terminus of the C-helix for efficient catalysis is one of the critical steps that must be achieved by activation of a kinase, and the distance between the N-terminus of the C-helix and the Activation Loop is a gauge that defines the open and closed conformations that are essential for catalysis [18]. In inactive kinases the N-terminus of the C-helix is sometimes disordered, but in other cases it is simply rotated into a position that is suboptimal for catalysis. The C-helix contains another conserved residue, Glu91, that bridges to Lys72 in β3-strand. The Lys72-Glu91 salt bridge is highly conserved through the entire protein kinase family and is often considered as a hallmark of the activated state.
In the active state, when the C-helix is bound to the β-sheet core, the N-lobe moves as a rigid body that opens and closes as part of the catalytic cycle. The Gly-rich loop, which is quite flexible, even when the kinase is activated, moves together with the N-lobe independently of whether the kinase is open or closed. The only exception in the N-lobe is the αC-β4 Loop, that is tightly anchored to the C-lobe [13, 14].
C-lobe
The large lobe contains mostly helices plus a β sheet (Figure 1). The helical subdomain, which is extremely stable, forms the core of the kinase and also serves as a tethering surface for protein/peptide substrates. Based on H/D exchange, the backbone amides of the core helices (D, E, F, and H) are well shielded from solvent [19, 20]. The exception is the G-helix, which is solvent exposed. The β subdomain, comprising four short β strands 6–9, contains much of the catalytic machinery associated with transfer of the phosphate from ATP to the protein substrate and is anchored through hydrophobic residues to the helical core. The Catalytic Loop that bridges β6 and β7 has most of the catalytic machinery whereas β strands 8 and 9 flank the DFG motif where Asp184 is critical for recognizing one of the ATP-bound Mg++ ions.
The Activation Segment extends from the DFG motif to Asp220 at the beginning of the F-helix (Figure 1c). Its length and sequence are the most variable part of the kinase core, and this segment is responsible for precisely turning the kinase on and off. The highly dynamic regulation of this segment is a unique feature of the EPKs, and, although the mechanism for achieving this regulation is often extremely complex and unique to each kinase, there are common themes.
The F-Helix is conserved in both ELKs and EPKs, but the extended helical element that follows the F-helix is unique to the EPKs (Figure 2). Because this segment includes the G-helix through the I-helix, we subsequently refer to it as the GHI-domain. Many substrate proteins and regulatory proteins are tethered to this domain, and it is thought to be important not only for stabilizing the active kinase core, but also for its allosteric sites [21]. Two conserved residues that lie far from the active site, Asp208 in the APE motif and Arg280 in the αH-αI Loop, anchor the GHI domain to the Activation Segment.
Regulatory Spine
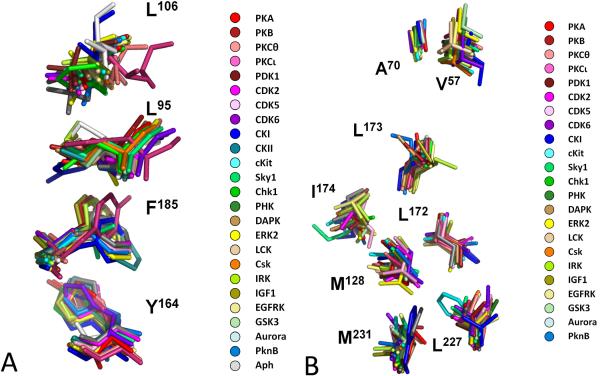
The initial application of LSP alignment to the protein kinases compared active and inactive kinases and considered residues that were at least partially exposed on the surface[11]. This analysis revealed a highly conserved spatial motif that was found in every active kinase but missing in inactive kinases (Figure 3a). This motif comprises four non-consecutive hydrophobic residues, two from the N-lobe: Leu106 from the β4 strand and Leu91 from the C-helix; and two from the C-lobe: Phe185 from the Activation Loop and Tyr164 from the Catalytic Loop. It can be thought of as a hydrophobic “spine” that links the two lobes. It cannot be identified from sequence alone, and the roles of these four residues had never been considered in previous analyses of protein kinase structure and function.
Figure 3. Multiple alignment of residues comprising the two Spines.
All structures of active EPKs and some ELKs have conserved structural motifs in their core. They form highly conserved spatial patterns. These patterns, termed “spines”, do not form sequential motifs coming from different parts of protein kinases sequence. A. Four residues from 25 human kinases and two ELKs (PknB and Aph) form the R-spine. B. Seven residues from 24 human kinases and one ELK (PknB) form the C-spine. PKA numbering is shown. Distribution of the human kinases on the kinome tree is shown in Box 1.
Because the middle part of this motif, the C-helix and the Activation Loop, can be very mobile [17, 22], the hydrophobic spine can be dynamically assembled or disassembled thereby regulating the protein kinase activity. This spine was thus designated as a Regulatory (R) Spine. One of the R-Spine residues in the C-lobe is in the HRD motif in the Catalytic Loop. In most kinases this is a histidine whereas in PKA and most of the AGC subfamily the His is replaced by Tyr [23]. The backbone of the His/Tyr is anchored to the F-helix through a conserved aspartate (Asp220), which serves as the base of the R-Spine.
Catalytic Spine
When all residues in the conserved kinase core were compared using the LSP alignment, another hydrophobic spine was identified [12]. Like the R-Spine, it comprises residues from both lobes; however, what distinguishes it from the R-Spine is that this Spine is completed by the adenine ring of ATP (Figure 3b). It was thus termed as the Catalytic (C) Spine. The two C-Spine residues in the N-Lobe, Val57 in β2 and Ala70 from the “AxK” motif in β3, are docked directly onto the adenine ring of ATP whereas in the C-Lobe it is Leu173 that docks directly onto the adenine ring. Leu173 lies in the middle of β7 and is always flanked by two hydrophobic residues, Leu172 and Leu174 in PKA [23]. These two residues rest on a hydrophobic residue from the D-helix, Met128, which is bound in turn to the F-helix (Leu227 and Met231). The role of the very short D-helix was not previously appreciated. Although it is present in all protein kinases, its sequence does not contain any conserved residues. Identification of the C-Spine shows that this helix contributes to the positioning of ATP with respect to the rigid hydrophobic core of the C-lobe.
Hydrophobic F-helix
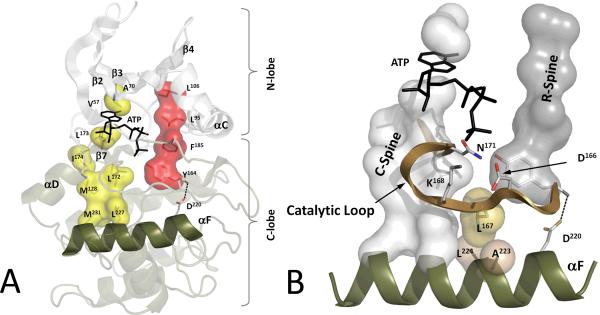
The other surprising feature that emerged from LSP alignment is the F-helix where every residue scores very high (Box 2). Although it was anticipated that residues in the Catalytic Loop would score highly, it was not immediately apparent why every residue in the F-helix should score so high. The F-helix is quite unusual because it is so hydrophobic; indeed, by many scoring algorithms it is predicted to be a transmembrane helix. Further analysis of the F-helix shows that it is the organizing element for the entire kinase core. Both spines are anchored to the F-helix (Figure 4a), the C-Spine directly to the hydrophobic C-terminus of the F-helix and the R-Spine to the N-terminus. Most other motifs in the C-lobe including the Catalytic Loop, the P+1 Loop, the Activation Segment, and the αH-αI Loop, are anchored firmly through hydrophobic contacts to the F Helix.
Figure 4. The F-helix and two hydrophobic Spines define the internal architecture of the protein kinase molecule.
A. The R-spine (red surface) and C-spine (yellow surface) are anchored to the F-helix (tan) in the middle of the rigid C-lobe. The C-spine is completed by the ATP molecule and, together with the R-spine, traverses the entire molecule, providing a firm, but flexible, connection between the two lobes. B. Major elements of the catalytic machinery are also anchored to the F-helix directly or via the Spines.
Protein kinase catalytic machinery is also anchored to the F-helix (Figure 4b). Because two residues in the Catalytic Loop dock directly onto the F-helix, Leu167 and Pro169, the backbone of the Catalytic Loop is fairly rigid even in the apo state or in the inactive state [24]. The Catalytic Loop is flanked by two important motifs. At the C-terminus is β7, which is a part of the C-Spine. At the N-terminus is the HRD motif, Tyr164, Arg165, and Asp166. Tyr164 is part of the R-Spine, Arg165 is anchored to the phosphate in the Activation Loop of active kinases, and Asp166 is positioned to serve as a weak catalytic base or a proton trap for the proton that is released from the peptide substrate [25, 26]. The backbone of the HRD motif is anchored to the F-Helix through Asp220, which lies at the beginning of the F-helix. Asp220 is highly conserved even in ELKs and is the only electrostatic link in the hydrophobic network [23]. The reason for the high conservation of the F-helix now becomes clear given that it coordinates the R-and C-Spines with the rest of the molecule.
Activation/inactivation of a protein kinase
One of the most important features that distinguishes protein kinases from ELKs, and from many other more classical metabolic enzymes, is that they are highly regulated in a manner that typically involves a dynamic reorganization of the molecule [22, 27]. The complicated machinery that is added to the simpler ELK scaffold to achieve these two functions is exceptional [23], not only for its complexity, but also for its dynamic properties. The function of EPKs is not simply to efficiently turnover product, but rather to initiate a cascade of events whether it be opening or closing of a channel or initiation of transcription. They toggle between active and inactive states, and the process by which an inactive kinase is converted into an active kinase is typically complex and highly regulated [28]. Kinase cascades, for example, such as the mitogen-activated protein kinases (MAPKs) where a series of activating kinases act in parallel, are common. In the case of the AGC kinases such as protein kinase C (PKC), p90 Ribosomal S6 kinase (Rsk) and S6K1, it often takes multiple kinases to activate one kinase. However, the identification of the R-Spine provides a unifying mechanistic explanation for the Activation Loop and what must be achieved by activation.
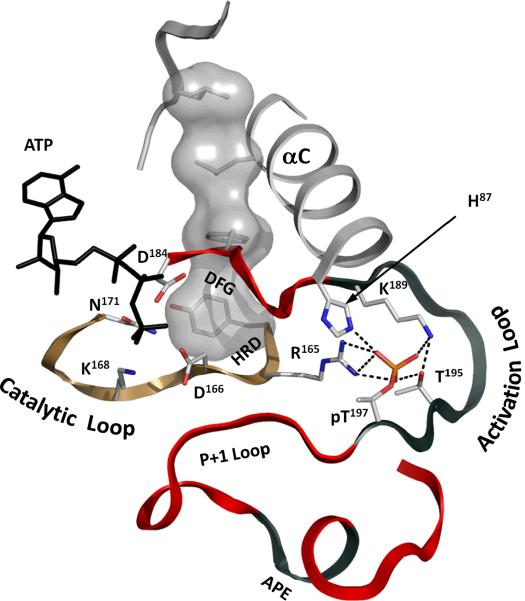
We begin here with the Activation Segment. Almost every residue in the Activation Segment plays a critical role beginning with the DFG motif (Figure 5). β Strand 9 in the active kinase (residues 189–190) typically contains a basic residue (K189) that is anchored to the Activation Loop phosphate whereas this strand is often disordered or ordered differently in inactive kinases. The Activation Loop (residues 191–197) contains the phosphorylation site (Thr197 in PKA). The P+1 Loop provides a docking site for the P+1 hydrophobic residue in the peptide whereas the other residues in this loop each contribute to different interactions with the peptide substrate. The APE motif and the following APE-αF linker anchor the Activation Segment to the F-helix and to the EPK-specific helical segment that follows the F-helix. Assembly of the Activation Segment into an active conformation is typically mediated by phosphorylation either by cis or trans autophosphorylation or by another activating kinase. In some cases this involves a disordered segment becoming ordered whereas in other cases it is a major reorganization of the conformation of the Activation Segment.
Figure 5. Phosphorylated residues in the Activation Segment control the catalytic elements of the kinase.
The Activation Segment consists of a short “Magnesium binding Loop” with the DFG-motif in its N-terminus. It is followed by the most diverse part of the segment, the “Activation Loop” that contains the primary phosphorylation site (pT197). The following “P+1 Loop” forms a pocket that accommodates residues in the peptide substrate positioned immediately after the serine, threonine or tyrosine to be phosphorylated. The highly conserved Ala-Pro-Glu (APE) motif stabilizes the Activation Segment by docking to the F-helix. The primary phosphate is anchored to the immobilized HRD-arginine (R165) from the Catalytic Loop (sand ribbon). It also forms multiple bonds inside the Activation Loop and to the C-helix thereby stabilizing the active configuration of the segment that is conserved through all EPKs. Correct positioning of the DFG-phenylalanine (F185) and the C-helix assembles the Regulatory Spine (shown as a white surface), thus providing efficient catalytic activity.
The Asp in the DFG motif has dominated our thinking so far because in the active state it is positioned to interact with the activating Mg++ ion, but what about the Phe? The driving force for shifting the equilibrium of the Activation Loop from its inactive or disordered conformation to its active conformation is most likely electrostatic, initiated by the phosphate bridging to Arg165. As a consequence of activation, Asp184 is now positioned for catalysis. However, the alignment of Phe185 with Tyr164 in the H/YRD motif and with Leu95 in the C-helix not only links the N- and C-Lobes but also provides a mechanism for coordinating the two lobes leaving them poised for catalysis. Alignment of Phe185 with the R-Spine residues thus stabilizes the active conformation and provides a contiguous pathway for entropy driven communication between the N- and C-lobes [29].
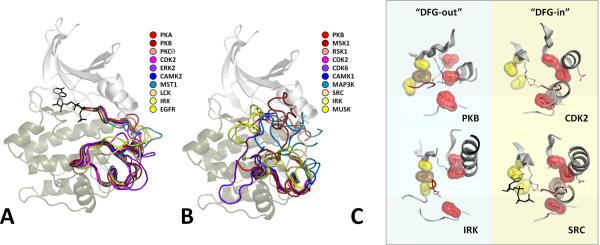
The geometry of the Activation Segment in different active protein kinases is remarkably well conserved with minor variations in the portion of the Activation Loop that is most solvent exposed (Figure 6a). By contrast, a comparison of various inactive kinases shows that the conformations of the Activation Segment are very different (Figure 6b). In some cases the Activation Segment also appears to be quite disordered. Thus we have a variety of activation mechanisms and a wide variety of inactive conformations of the Activation Segment; however, all kinases converge to a relatively conserved conformation following activation. To gauge whether a kinase is active requires examination of the R-Spine. An active kinase can be in an open or closed conformation, but a kinase will never be in an active conformation if its R-spine is broken.
Figure 6. Geometry of the Activation Segment in different EPK families (AGC, CMG, CAMK, STE and TK) changes significantly upon inactivation.
All active EPKs have a conserved internal architecture, but inactive structures can vary significantly. (A) Activation Segments of activated kinases typically have a highly conserved conformation. (B) Different kinases can be inactivated in different ways; therefore an Activation Segment in its inactivated state can accept diverse conformations. (C) The major target of the inactivation process is the disassembly of the Regulatory Spine (red surface) and stabilization of the inactive conformation. This can be achieved in two different ways. One of the most common mechanisms is to flip the DFG motif into a “DFG out” configuration [32, 33]. One variation of this is seen in the insulin receptor and PKB where the DFG-phenylalanine (brown surface) not only flips out, but also crosses over to fill the adenine pocket site in the C-Spine (yellow surface) thereby reinforcing the inactivation by blocking ATP binding as well. In this case, the C-Helix is also flipped into an “out” conformation. There are other examples where the DFG flips “out” but does not cross over to the C-Spine. Often, however, these conformations are associated with inhibitor-bound structures so that we do not know what the conformation is in an unliganded kinase, or even in the ATP-bound inactive conformation. In other cases, the DFG appears to be in a “DFG In” state although the configuration of the DFG backbone residues is not in an active conformation (e.g. CDK2 and Src). In these cases, the R-Spine is broken by the displacement of the C-Helix residue so that the C-Helix is in an “out” conformation. Thus there are many ways to break the R-Spine.
The R-spine can be disassembled in different ways (Figure 6C). An apparently common mechanism for breaking the R-spine is exemplified by the insulin receptor and PKB (also called Akt) where the DFG phenylalanine actually flips over and occupies the position in the C-spine that is filled by the adenine ring of ATP in the active conformation [30, 31]. In these cases the incorrect positioning of the DFG motif also sterically blocks binding of ATP. This has been designated as a “DFG-out” conformation, and this serves as a structural signature of the inactive form for many kinases [32, 33]. All kinases that fall into this category will be inactive. In other cases, however, movement of the C-helix might be sufficient to break the R-Spine. Inactive Src and cyclin-dependent kinase 2 (CDK2) [34, 35] (Figure 6C) are two such examples where the DFG motif is more-or-less in the correct “in” configuration, and there are no global changes in the Activation Segment. In Src and CDK2 the R-Spine is broken because the C-Helix is displaced. If Leu95 in the C-helix moves out the kinase is also inactive even though the Activation Segment has a “DFG-in” configuration. One thus needs to consider the integrity of the complete R-Spine to assess whether the conformation of the kinase is active or inactive. Open or closed is never a criteria for assessing whether a kinase is active or inactive. All kinases with a “DFG-out” configuration will be inactive, but each “DFG-in” configuration needs to be closely examined for the intactness of the rest of the R-spine.
Gatekeepers, Spines and kinase Inhibitors
An important role in protein kinase activation can be played by a so-called “gatekeeper” residue [36] positioned between the C- and the R-Spine (Figure 7a). A survey of the human kinases reveals that 77% of all human kinases have relatively large (Leu, Met, Phe) residues in this position whereas 21% of them, mostly tyrosine kinases, have smaller gatekeeper residues (Thr, Val) [37]. Mutagenesis of large gatekeepers to a smaller side chain allowed the engineering of mutant kinases that could accept bulky analogs of ATP [36]. Because the reduction of the gatekeeper side chain did not restrain protein kinase activity, it provided an opportunity to design selective inhibitors for a particular kinase thus identifying its substrates.
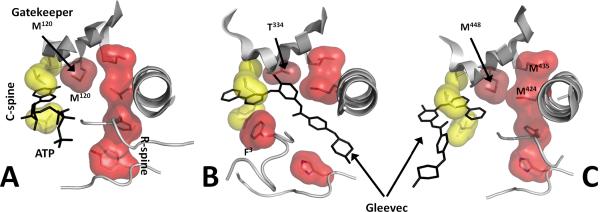
Figure 7. Gatekeeper residue can stabilize the R-spine and define the affinity or binding mode of inhibitors.
Gatekeeper residues are positioned at the end of the β5 strand, forming a part of the hydrophobic ATP-binding pocket and interacting with the two R-spine residues from the N-lobe. A. The gatekeeper residue (Met120) in the active form of PKA with two assembled spines (PDBID 1ATP). B. In Abl kinase the gatekeeper is a smaller threonine (Thr334) that is not an effective stabilizer of the R-spine. The latter is disassembled upon inactivation with the DFG phenylalanine flipped outside, forming an extended cavity for inhibitor (Gleevec) binding (PDBID 1OPJ). A substitution of the gatekeeper threonine to isoleucine or methionine can lock the R-spine in an active conformation [39]. C. Syk kinase not only has methionine as the gatekeeper (M448), but both residues from the R-spine that are bound to the gatekeeper are methionines (PDBID 1XBB). This, apparently, stabilizes the R-spine leading to an alternative mode of Gleevec binding.
Mutations that result in smaller gatekeepers changing to bulkier residues, however, appeared to have much more significant physiological consequences. For example, a set of such mutations were found to be responsible for drug resistance in patients with chronic myeloid leukemia and several solid tumors [38]. The drug, Gleevec (imatinib) usually binds to inactive “DFG-out” conformations (Figure 7b), but amino acid substitutions including T315I in BCR-Abl, T670I in Kit or T674I in PDGFRα lead to resistance to this drug. There could be several potential explanations of this resistance; however large gatekeeper residues stabilize the R-spine more efficiently than threonine, shifting the equilibrium to the active conformation [39]. Such stabilization, together with simple steric blocking of the binding site [40], prevents Gleevec binding thereby creating a constitutively active oncogenic kinase. Moreover, mutagenesis of any of the R-Spine residues generates an inactive kinase [39].
The region surrounding the R-spine is thus strategically of great importance and in the case of tyrosine kinases is especially malleable and important for activation which is typically achieved through the transient phosphorylation of the Activation Loop. Surprisingly, a spleen tyrosine kinase, Syk, which not only has a methionine in the gatekeeper position, but also two adjacent methionines in the R-spine, is capable of binding to Gleevec in an alternative configuration to the active (DFG-in) conformation of the kinase [41] (Figure 7c). Each kinase is thus unique, and it is dangerous to make general rules.
Completing the protein kinase core
As we discussed earlier, the F helix and two hydrophobic Spines serve as a scaffold for the assembly of the active protein kinase core. Is this sufficient for optimal catalysis? In some cases, it might be, but in most cases organizing the Activation Segment is an essential priming step, but is not sufficient for creating the most efficient catalyst. There is often also a requirement for optimal orientation of the N-Lobe. Each kinase will achieve this in a different way, but there are two requirements that must be met. First is that the Glycine-rich Loop be positioned to shield the ATP and position the γ-phosphate for transfer, and second is that the C-helix must be optimally positioned to interact with the Activation Segment thereby allowing the cleft to open and close. In many cases we have structures of only the kinase core, so it is not clear how linkers or other domains or other regulatory molecules influence the conformation and/or dynamic properties of the N-lobe.
Analysis of the surface geometry of protein kinase cores reveals that there are many spatially conserved pockets that can be filled in unique ways (Box 3) [42]. To position the C-helix, for example, there are two pockets, and docking to these pockets orients the C-Helix for optimal catalysis. In the case of CDKs, cyclin fills one of these pockets and positions the C-Helix into the correct orientation [43]. In other cases, such as Fes, an SH2 domain binds to the C-helix and not only orients the C-helix for catalysis but also provides a priming site for docking of the substrate protein [44]. The AGC kinases, such as PKA and PKC, are a bit unusual in that they are often assembled as active and fully phosphorylated kinases that are inhibited by regulatory domains or subunits and then activated by second messengers. The AGC kinases also share a conserved C-tail that extends about 50–60 residues beyond the core, and this C- tail primes the N-Lobe for catalysis. This tail wraps around both lobes and contains many elements that are essential for catalysis. The ATP binding pocket, for example, is completed by the C-Tail. The Glycine-rich Loop is positioned for catalysis. Docking of a hydrophobic motif near the C-terminus is also required for correct orientation of the C-helix. Critical phosphorylation sites are also embedded within the C-tail. In some cases, autophosphorylation and one activating kinase might be sufficient for this final processing, whereas in other cases, such as RSK, five or more kinases are required [45]. The tail for the AGC kinases is thus considered to be a cis-regulatory element [46]. Although each kinase is regulated in a unique way, it is important to recognize that regions that lie outside the core are often essential for optimizing the catalytic efficiency of the enzyme. This is especially important to appreciate given that many of the available kinase structures include only the core whereas the attached linkers, tails and/or domains are often a critical part of the functional kinase.
Box 3. Stabilization of the kinase core is achieved in a kinase specific manner.
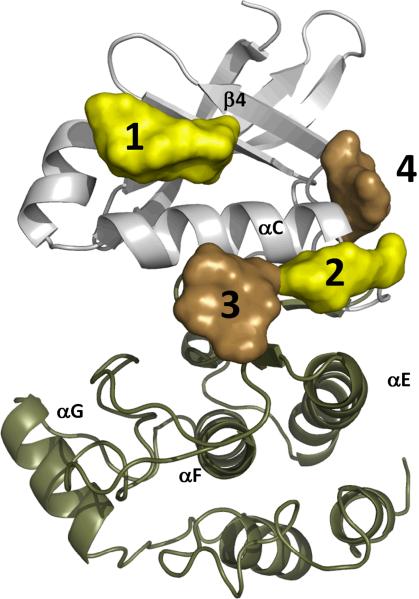
Following the realization that the geometry of the kinase core is strictly defined by the F-helix and the two Spines, an important question arose: to what extent is the exterior form of the core conserved? 10 protein kinases from different families were aligned by their F-helices, and cavities on their surfaces were identified and compared [42]. Among several conserved pockets detected on the protein kinases, four surrounded the C-helix (Figure I). Pocket #1 is well known as the Hydrophobic or PIF-pocket in AGC kinases; it must be filled with a conserved hydrophobic “FxxF” motif in the C-terminus of these kinases. It also is known to be an important binding site in EGFR kinases and a docking site for cyclins in CDKs. Pocket #2 is occupied by two hydrophobic residues in the PKA N-terminus and the ERK C-terminus. Pocket #3 is another docking site for cyclin in CDKs, whereas #4 is often used in tyrosine kinases such as Src or LCK. Apparently the conserved kinase core is not stable enough and requires reinforcement by additional elements that can come from N/C-terminal tails or from other proteins.
Figure I A set of conserved pockets surrounding the C-helix. The C-helix is a key structural element that, along with the Activation Segment, forms the R-spine (Figures 5, 7). Stability of this helix is vital for the kinase activity. Although the geometry and positions of these pockets can be conserved in different kinases, the way in which they are filled is a kinase-specific feature. However, the role of these interactions is the same - to stabilize the C-helix in a particular conformation.
Dynamic Integration of the kinase for catalysis
Once the kinase is activated by assembly of the R-Spine and the N-Lobe is correctly positioned, the enzyme is poised for catalysis. Identification of the C-Spine also provides a new framework for considering how the catalytic machinery is coordinated. Unlike many other enzymes such as the proteases where there is a catalytic triad that has evolved in both convergent and divergent ways that allow for the same positioning of the three catalytic residues, protein kinases that phosphorylate Ser, Thr, and Tyr have evolved in a strictly divergent manner from the ELKs. There are many conserved catalytic motifs that are scattered throughout the kinase core, and these are all linked through a conserved hydrophobic architecture that is also an integral conserved part of the active enzyme. An important feature of this architecture is that it is formed by hydrophobic residues. This provides strong, but flexible, connections between the lobes. Such flexibility is very important for protein kinases as dynamics is an intrinsic property of these enzymes [17, 22]. It has to be emphasized that the major signature of an active kinase is the assembly of the two hydrophobic spines in its core, not “open” or “closed” states or formation of the conserved K72-E91 salt bridge. Opening and closing of a kinase molecule is a part of catalytic cycle and the K72-E91 bond can be formed or broken during this cycle, but the spines in active kinase remain intact [47]. The misconception of equating open conformations with inactive kinases and closed conformation as the signature of an active kinase, apparently, comes from the earlier studies of enzymes that do not require flexibility for their activity and can be regulated by a simple open/closed transition. Once the R-spine is assembled and the C-helix is correctly oriented, the kinase is primed for catalysis. Binding of ATP, which completes the C-spine, then commits the kinase to proceed with catalysis.
Evolution of the Eukaryotic Protein Kinases
If we return now to the ELKs and compare them structurally and functionally to the EPKs, we can better appreciate what is so unique about the EPKs and how they have evolved to be such dynamic molecular switches. As a result of evolution, two additional segments have been added to the conserved core of the EPKs to achieve fast and efficient regulation (Figure 8). First is the insertion of the Activation Segment between β9 and the F-Helix. The second EPK-specific segment is the GHI-subdomain that follows the F-Helix. In the AGC kinases the GHI-subdomain is fused to the C-tail that lies outside the conserved core, and this C-tail also contributes to stabilizing the active conformation of the kinase [46, 48].
Figure 8. Activation Segment and the GHI-subdomain distinguish EPKs from ELKs.
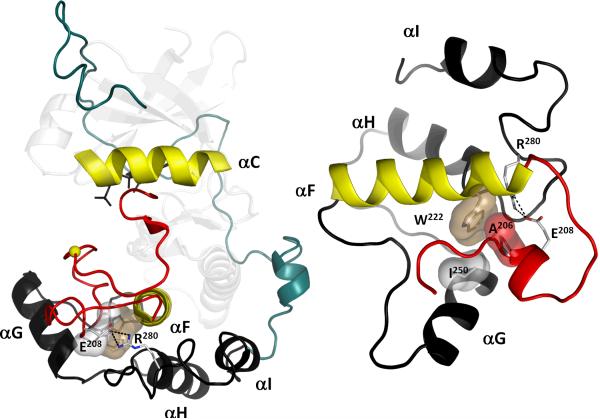
An extended Activation Segment (red) is a conserved feature in EPKs. It contains several loops that perform different functions (Figure 5). This segment contains the primary phosphorylation site (yellow sphere) and is missing in ELKs (Figure 2). The GHI-subdomain (black cartoon) contains three helices G, H and I. It is also missing in ELKs, but is highly conserved in EPKs. These two structural elements surround the large hydrophobic F-helix in the middle of the C-lobe that serves as a scaffold for the protein kinase core (Figure 4). The Activation Segment and the GHI-subdomain also interact with each other via a conserved APE motif (E208-R280) which is anchored to the F-helix by hydrophobic interaction (A206, P207-W222). The AGC-specific C-terminal tail (teal cartoon) reinforces the protein kinase core stability (Box 3).
Although the dynamic properties of the Activation Segment and its regulation by phosphorylation were described earlier, little attention has been paid to the GHI-subdomain. The helical domain of the C-Lobe is essentially made up two parts, the DEF-subdomain and the GHI-subdomain. The DEF-subdomain is essential for catalysis and for coordination of the R- and C-Spines and is conserved in both ELKs and EPKs. The GHI segment, however, is unique to the EPKs. Although it can contribute to tethering of protein substrates [49], to allostery [21], and to protein–protein interactions [50, 51], it is not as dynamic as the Activation Segment.
Although the Activation Segment and the GHI-subdomain do not have extensive direct interactions with one another, they are both linked to the hydrophobic core of the kinase (Figure 8). Three conserved residues create a critical node for coupling these three elements: Glu208, Arg280, and Trp222. Glu208 is part of the APE motif and Ala206 and Pro207 both provide the hydrophobic anchor to the N-terminus of the F helix. Arg280 in the αH-αI linker forms an ion pair with Glu208. Trp222 anchors the two residues to the F helix. The Activation Segment and the GHI-subdomain are, thus, two coevolved structural features of EPKs with a twofold function: docking of substrate proteins or other regulatory proteins and effective regulation of kinase activity through an allosteric site. The importance of this module is reflected in somatic single nucleotide polymorphisms (SNPs) that are associated with disease [52]. Of these disease-associated residues, Glu208 was actually at the top of the list and Arg280 and Trp222 were among the top seven, together with such catalytically essential residues as Glu170 and Arg165. Here we demonstrate why perturbation of any one of these residues could severely interfere with the finely tuned function of the kinase.
Pseudokinases
About 10% of the human kinome contain most of the major sequence motifs that define the eukaryotic protein kinase structure, specifically the Activation Loop and the helical GHI subdomain, as discussed above, but harbor mutations in positions that were thought to be crucially important for catalysis [53]. In the beginning these were collectively referred as “pseudokinases” and were assumed to be inactive. As was demonstrated recently, however, pseudokinases come in many different flavors. Some of these pseudokinases do indeed appear to be scaffolds whereas others, can actually function as catalysts using non-standard mechanisms. One example is a kinase family that lacks the universally conserved lysine K72 in the β3 strand (called WNK kinases: With No K) [54]. Instead of lysine in the β3 strand, the WNKs recruit another lysine (in β2) to fill the same space and fulfill the same function. Another example is calcium/calmodulin-dependent serine protein kinase (CASK) which harbors four critical amino acid substitutions in the active site. It was thought to be catalytically incompetent because it lacked key residues that bind to the active site Mg++ ions. It was later shown, however, that CASK is a Mg++-independent kinase that selectively phosphorylates neurexin 1. In fact, CASK is actually inhibited by magnesium ions [55]. There are also examples of pseudokinases that are completely immobilized in a single conformation. Vaccinia related kinase 3 (VRK3), for example, has a series of hydrophobic mutant residues that fill the adenine binding pocket and complete the C-spine without ATP [56]. Thus, this kinase is locked in an active conformation without the capability to undergo catalysis. Such “mimicking” of the activated kinase conformation seems to be a common feature for different pseudokinases [57–59].
Concluding remarks and future perspectives
Recognizing the protein kinase internal architecture and the importance of a hydrophobic scaffold creates a new framework for analysis of the function and regulation of this biologically important enzyme family. The unique feature of these enzymes is that they are highly regulated, and their activation creates a conserved conformation that integrates the hydrophobic scaffold. The scaffold can be broken in a variety of ways and the activation, typically achieved by phosphorylation of the Activation Loop, is highly regulated in ways that are unique for each kinase. Activation is transient and requires integration and stabilization of the whole internal core. It is quite striking that this can be achieved by the strategic addition of a single phosphate, but it is consistent with the role of these enzymes as dynamic switches, a requirement that is not imposed on metabolic enzymes. Many questions, however, remain unanswered. Can the “spine paradigm” provide an explanation for the correlated evolution that relates surface sites that are far apart [60]. Can the “spine paradigm” provide an explanation for allosteric sites, which cannot be explained by rigid hydrophilic networks? Already we are seeing that the recognition of the spine motifs is helping to develop new strategies for designing highly specific protein kinase inhibitors. The spine paradigm provides a cautionary message about how we interpret our various crystal structures where typically we have an incomplete molecule. How do dynamic linkers, tails, and binding proteins influence the spines and hence the global architecture of the kinase? These are challenges that will require the concerted integration of crystallography and NMR with enzymologists and biophysicists. Most importantly, it will require the close integration of structural and computational biology.
References
- 1.Krebs EG. An accidental biochemist. Annu Rev Biochem. 1998;67:xii–xxxii. doi: 10.1146/annurev.biochem.67.1.0. [DOI] [PubMed] [Google Scholar]
- 2.Walsh DA, Perkins JP, Krebs EG. An adenosine 3',5'-monophosphate-dependant protein kinase from rabbit skeletal muscle. J Biol Chem. 1968;243(13):3763–5. [PubMed] [Google Scholar]
- 3.Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol. 2009;21(2):140–6. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker WC, Dayhoff MO. Viral src gene products are related to the catalytic chain of mammalian cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1982;79(9):2836–2839. doi: 10.1073/pnas.79.9.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. Faseb J. 1995;9(8):576–96. [PubMed] [Google Scholar]
- 6.Manning G, et al. Evolution of protein kinase signaling from yeast to man. Trends in Biochemical Sciences. 2002;27(10):514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- 7.Knighton DR, et al. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253(5018):407–14. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 8.Knighton DR, et al. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253(5018):414–20. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- 9.Kannan N, Neuwald AF. Did protein kinase regulatory mechanisms evolve through elaboration of a simple structural component? J Mol Biol. 2005;351(5):956–72. doi: 10.1016/j.jmb.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 10.Kannan N, et al. Structural and functional diversity of the microbial kinome. PLoS Biol. 2007;5(3):e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kornev AP, et al. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(47):17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kornev AP, Taylor SS, Ten Eyck LF. A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci U S A. 2008;105(38):14377–82. doi: 10.1073/pnas.0807988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shudler M, Niv MY. BlockMaster: partitioning protein kinase structures using normal-mode analysis. J Phys Chem A. 2009;113(26):7528–34. doi: 10.1021/jp900885w. [DOI] [PubMed] [Google Scholar]
- 14.Tsigelny I, et al. 600 ps molecular dynamics reveals stable substructures and flexible hinge points in cAMP dependent protein kinase. Biopolymers. 1999;50(5):513–24. doi: 10.1002/(SICI)1097-0282(19991015)50:5<513::AID-BIP5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 15.Madhusudan, et al. Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nat Struct Biol. 2002;9(4):273–7. doi: 10.1038/nsb780. [DOI] [PubMed] [Google Scholar]
- 16.Ramakrishnan C, Dani VS, Ramasarma T. A conformational analysis of Walker motif A [GXXXXGKT (S)] in nucleotide-binding and other proteins. Protein Eng. 2002;15(10):783–98. doi: 10.1093/protein/15.10.783. [DOI] [PubMed] [Google Scholar]
- 17.Johnson DA, et al. Dynamics of cAMP-dependent protein kinase. Chem Rev. 2001;101(8):2243–70. doi: 10.1021/cr000226k. [DOI] [PubMed] [Google Scholar]
- 18.Narayana N, et al. Crystal structure of a polyhistidine-tagged recombinant catalytic subunit of cAMP-dependent protein kinase complexed with the peptide inhibitor PKI(5–24) and adenosine. Biochemistry. 1997;36(15):4438–48. doi: 10.1021/bi961947+. [DOI] [PubMed] [Google Scholar]
- 19.Steichen JM, et al. Global consequences of activation loop phosphorylation on protein kinase A. J Biol Chem. 2010;285(6):3825–32. doi: 10.1074/jbc.M109.061820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, et al. Allosteric network of cAMP-dependent protein kinase revealed by mutation of Tyr204 in the P+1 loop. J Mol Biol. 2005;346(1):191–201. doi: 10.1016/j.jmb.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 21.Deminoff SJ, Ramachandran V, Herman PK. Distal recognition sites in substrates are required for efficient phosphorylation by the cAMP-dependent protein kinase. Genetics. 2009;182(2):529–39. doi: 10.1534/genetics.109.102178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109(3):275–82. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 23.Scheeff ED, Bourne PE. Structural evolution of the protein kinase-like superfamily. PLoS Comput Biol. 2005;1(5):e49. doi: 10.1371/journal.pcbi.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akamine P, et al. Dynamic features of cAMP-dependent protein kinase revealed by apoenzyme crystal structure. J Mol Biol. 2003;327(1):159–71. doi: 10.1016/s0022-2836(02)01446-8. [DOI] [PubMed] [Google Scholar]
- 25.Adams JA. Kinetic and catalytic mechanisms of protein kinases. Chem Rev. 2001;101(8):2271–90. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- 26.Valiev M, et al. Phosphorylation reaction in cAPK protein kinase-free energy quantum mechanical/molecular mechanics simulations. Journal of Physical Chemistry B. 2007;111(47):13455–13464. doi: 10.1021/jp074853q. [DOI] [PubMed] [Google Scholar]
- 27.Adams JA. Activation loop phosphorylation and catalysis in protein kinases: Is there functional evidence for the autoinhibitor model? Biochemistry. 2003;42(3):601–607. doi: 10.1021/bi020617o. [DOI] [PubMed] [Google Scholar]
- 28.Johnson LN, Lewis RJ. Structural basis for control by phosphorylation. Chemical Reviews. 2001;101(8):2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- 29.Masterson LR, et al. Role of internal dynamics in substrate recognition and catalysis by protein kinase A. Nat Chem Biol. 2010 doi: 10.1038/nchembio.452. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubbard SR, et al. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature. 1994;372(6508):746–54. doi: 10.1038/372746a0. [DOI] [PubMed] [Google Scholar]
- 31.Yang J, et al. Molecular mechanism for the regulation of protein kinase B/Akt by hydrophobic motif phosphorylation. Mol Cell. 2002;9(6):1227–40. doi: 10.1016/s1097-2765(02)00550-6. [DOI] [PubMed] [Google Scholar]
- 32.Levinson NM, et al. Structural basis for the recognition of c-Src by its inactivator Csk. Cell. 2008;134(1):124–34. doi: 10.1016/j.cell.2008.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shan Y, et al. A conserved protonation-dependent switch controls drug binding in the Abl kinase. Proc Natl Acad Sci U S A. 2009;106(1):139–44. doi: 10.1073/pnas.0811223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown NR, et al. Effects of phosphorylation of threonine 160 on cyclin-dependent kinase 2 structure and activity. J Biol Chem. 1999;274(13):8746–56. doi: 10.1074/jbc.274.13.8746. [DOI] [PubMed] [Google Scholar]
- 35.Xu W, et al. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3(5):629–38. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, et al. A molecular gate which controls unnatural ATP analogue recognition by the tyrosine kinase v-Src. Bioorg Med Chem. 1998;6(8):1219–26. doi: 10.1016/s0968-0896(98)00099-6. [DOI] [PubMed] [Google Scholar]
- 37.Zuccotto F, et al. Through the “gatekeeper door”: exploiting the active kinase conformation. J Med Chem. 2010;53(7):2681–94. doi: 10.1021/jm901443h. [DOI] [PubMed] [Google Scholar]
- 38.Tamborini E, et al. A new mutation in the KIT ATP pocket causes acquired resistance to imatinib in a gastrointestinal stromal tumor patient. Gastroenterology. 2004;127(1):294–9. doi: 10.1053/j.gastro.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 39.Azam M, et al. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat Struct Mol Biol. 2008;15(10):1109–18. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daub H, Specht K, Ullrich A. Strategies to overcome resistance to targeted protein kinase inhibitors. Nat Rev Drug Discov. 2004;3(12):1001–10. doi: 10.1038/nrd1579. [DOI] [PubMed] [Google Scholar]
- 41.Atwell S, et al. A novel mode of Gleevec binding is revealed by the structure of spleen tyrosine kinase. J Biol Chem. 2004;279(53):55827–32. doi: 10.1074/jbc.M409792200. [DOI] [PubMed] [Google Scholar]
- 42.Thompson EE, et al. Comparative surface geometry of the protein kinase family. Protein Sci. 2009;18(10):2016–26. doi: 10.1002/pro.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeffrey PD, et al. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376(6538):313–20. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 44.Filippakopoulos P, et al. Structural coupling of SH2-kinase domains links Fes and Abl substrate recognition and kinase activation. Cell. 2008;134(5):793–803. doi: 10.1016/j.cell.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008;9(10):747–58. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 46.Kannan N, et al. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc Natl Acad Sci U S A. 2007;104(4):1272–7. doi: 10.1073/pnas.0610251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dixit A, Verkhivker GM. Hierarchical modeling of activation mechanisms in the ABL and EGFR kinase domains: thermodynamic and mechanistic catalysts of kinase activation by cancer mutations. PLoS Comput Biol. 2009;5(8):e1000487. doi: 10.1371/journal.pcbi.1000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romano RA, et al. A chimeric mechanism for polyvalent trans-phosphorylation of PKA by PDK1. Protein Sci. 2009;18(7):1486–97. doi: 10.1002/pro.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122(6):887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 50.Kim C, et al. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell. 2007;130(6):1032–43. doi: 10.1016/j.cell.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 51.Wu J, et al. PKA type IIalpha holoenzyme reveals a combinatorial strategy for isoform diversity. Science. 2007;318(5848):274–9. doi: 10.1126/science.1146447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torkamani A, et al. Congenital disease SNPs target lineage specific structural elements in protein kinases. Proc Natl Acad Sci U S A. 2008;105(26):9011–6. doi: 10.1073/pnas.0802403105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manning G, et al. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 54.Xu B, et al. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem. 2000;275(22):16795–801. doi: 10.1074/jbc.275.22.16795. [DOI] [PubMed] [Google Scholar]
- 55.Mukherjee K, et al. CASK Functions as a Mg2+-independent neurexin kinase. Cell. 2008;133(2):328–39. doi: 10.1016/j.cell.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheeff ED, et al. Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure. 2009;17(1):128–38. doi: 10.1016/j.str.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fukuda K, et al. The pseudoactive site of ILK is essential for its binding to alpha-Parvin and localization to focal adhesions. Mol Cell. 2009;36(5):819–30. doi: 10.1016/j.molcel.2009.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Labesse G, et al. ROP2 from Toxoplasma gondii: A virulence factor with a protein-kinase fold and no enzymatic activity. Structure. 2008 doi: 10.1016/j.str.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 59.Zeqiraj E, et al. Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science. 2009;326(5960):1707–11. doi: 10.1126/science.1178377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suel GM, et al. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10(1):59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]