Abstract
Vascular endothelial (VE)–cadherin, a divergent member of the type II classical cadherin family of cell adhesion proteins, mediates homophilic adhesion in the vascular endothelium. Previous investigations with a bacterially-produced protein suggested that VE-cadherin forms cell surface trimers which bind between apposed cells to form hexamers. Here we report studies of mammalian-produced VE-cadherin ectodomains which suggest that, like other classical cadherins, VE-cadherin forms adhesive trans-dimers between monomers located on opposing cell surfaces. Trimerization of the bacterially-produced protein appears to be an artifact that arises from a lack of glycosylation. We also present the 2.1Å resolution crystal structure of the VE-cadherin EC1-2 adhesive region which reveals homodimerization via the strand swap mechanism common to classical cadherins. In common with type II cadherins, strand swap binding involves two tryptophan anchor residues, but the adhesive interface resembles type I cadherins in that VE-cadherin does not form a large non-swapped hydrophobic surface. Thus, VE-cadherin is an outlier among classical cadherins, with characteristics of both type I and type II subfamilies.
Keywords: Cell-cell adhesion, N-glycosylation, cadherin adhesive binding, domain swapping
INTRODUCTION
Proteins of the cadherin superfamily are found in both invertebrate and vertebrate animals and are thought to function primarily in intercellular adhesion. More than 350 cadherins have been identified, and these are classified into several distinct subfamilies 1. The best characterized is the vertebrate classical cadherin subfamily, which includes the type I and type II classical cadherins which are defined by distinct sequence and structural characteristics 2. In mice and humans five type I cadherins and 13 type II cadherins have been described 3. These are expressed in specific and often overlapping patterns throughout solid vertebrate tissues where they function in calcium dependent cell-cell adhesion 4. Despite well-defined differences (see below), type I and type II cadherins share several common structural features. They both contain an ectodomain region composed of five tandem extracellular cadherin-like (EC) domains each of about 110 amino acids length, which is rigidified by the binding of calcium ions between successive EC domains 5; 6. Classical cadherins are anchored by a single transmembrane region, and have a short cytoplasmic domain with conserved binding sites for β- or γ,- and p120 catenins 7; 8; 9; 10; 11, which help to mediate attachment to the cytoskeleton and control cadherin trafficking 12; 13.
The mechanism by which cadherins mediate cell adhesion through interaction of their ectodomains has been intensively studied. It is now understood that both type I and type II cadherins bind via a 3D domain swapping mechanism involving their N-termini 2; 14; 15. Membrane distal cadherin EC1 domains from apposed cells bind across the intercellular space by a reciprocal exchange of N-terminal β-strands to form “strand swapped” dimers. The principle of strand exchange is shared by type I and type II cadherins, but it differs in detail at the molecular level 2. Structures of type I cadherin dimers reveal that residue Trp2 in the swapped strand is docked into a hydrophobic ‘acceptor’ pocket in the partner EC1 domain, anchoring the swapped dimer 5; 14; 16; 17; 18. In type II cadherins, two anchoring tryptophan residues are observed: Trp2 and Trp4, which are accommodated by an acceptor pocket that is larger in type II than in type I cadherins 2. Type II cadherins, unlike type I counterparts, also have a large non-swapped region in their adhesive interfaces, which is assembled by conserved hydrophobic residues thus extending the hydrophobicity of the acceptor pocket towards the base of the EC1 domain. This region contributes almost a third more buried accessible surface area in the dimer interface compared with type I cadherins, which lack this extended hydrophobic interface surface 2.
VE-cadherin is found exclusively in the cells of the vascular endothelium in vertebrate species where it is concentrated in adherens junctions 19; 20; 21; 22. Endothelial cells also express type I N-and P-cadherin, however only VE-cadherin strongly localizes to adherens junctions in the endothelium, while N-cadherin is dispersed over the cell surface 11; 23; 24; 25; 26. Gene deletion studies show that VE-cadherin is essential for embryonic angiogenesis, vascular maintenance and restoration of vascular integrity after injury 11; 27. VE-cadherin knock-out mice die during development at E9.5 due to disintegration of the vasculature 28. VE-cadherin gene null mutations in murine embryonic stem cells lead to a dispersed endothelium lacking organized vasculature in embryonic bodies 22. Similarly, wild type adult mice injected with antibodies directed against VE-cadherin died within 24 hours due to disassembly of the vasculature 29; 30. These results suggest a specific non-redundant role for VE-cadherin mediated adhesion in the vasculature. VE-cadherin also regulates cellular processes such as leukocyte trafficking 31 and control of vascular permeability 32, playing a central role in the formation of a controlled barrier between blood and underlying tissues.
Early structural studies on VE-cadherin using a bacterially expressed ectodomain fragment comprising domains EC1-4 identified a cylindrical hexameric structure in cryo electron microscopy (EM), and biophysical behavior consistent with hexamer formation 33; 34. Results from single particle EM reconstructions led to a unique model for VE-cadherin binding in which three cadherin molecules assemble on one cell surface to form a cis trimer via EC4 domain interactions. The cis trimer would then form an EC1-domain mediated trans dimer with a trimer from the opposing cell, resulting in a hexameric structure.
We set out to investigate the adhesive mechanism of VE-cadherin. Biophysical characterization of the complete VE-cadherin ectodomain produced in mammalian cells reveals dimers in solution, but not higher order oligomers. Furthermore, analysis of non-glycosylated bacterially expressed fragments reveals non-biological multimer formation involving the EC3-4 region, which is absent from similar mammalian-expressed fragments with natural glycosylation.
Crystallographic analysis of the adhesive EC1-2 domain fragment of chicken VE-cadherin reveals strand swap binding involving two tryptophan anchor residues, characteristic of type II cadherins. However, the adhesive interface also resembles type I cadherins in that it lacks a large non-swapped hydrophobic surface. Together, these findings show that VE-cadherin forms strand swapped dimers like other classical cadherins via an adhesive interface with characteristics of both type I and type II subfamilies.
RESULTS
Production of natively glycosylated whole VE-cadherin ectodomains in mammalian cells
Previous work on a bacterially-produced human VE-cadherin EC1-4 fragment revealed formation of hexameric structures in which two cis trimers appear to associate via strand swap binding involving each EC1 domain 33; 34; 35. Initially, we wished to examine this novel hexameric structure at high resolution by x-ray crystallography. Since the proteins used in these experiments lacked EC5, native glycosylation, and were purified from bacterial inclusion bodies, we endeavored to use a more native-like protein in our study. We therefore used a mammalian expression system to produce the complete soluble EC1-5 ectodomain of VE-cadherin, from human (Asp1-542) and chicken (Asp1-545), without the transmembrane and cytoplasmic regions. These proteins were secreted from human embryonic kidney (HEK) 293 F cells, resulting in native proteins which were glycosylated and could be purified from conditioned media.
Both human and chicken VE-cadherin ectodomains migrate on SDS polyacrylamide gels ~10kDa above their predicted masses suggesting the presence of significant glycosylation. We determined precise molecular masses for these proteins by MALDI-TOF mass spectrometry, and calculated the mass of glycan by the difference from molecular masses calculated from amino acid sequence. This procedure suggested the presence of 9,503Da and 13,144Da of glycan for the chicken and human proteins, respectively. To assess the contributions of N- and O-linked glycosylation we then produced VE-cadherin in HEK 293 GNTI− cells, which lack the enzyme N-acetyl-glucosaminyl-transferase I, limiting N-linked glycans to minimal sugar trees of Man5GlcNac2 which can then be removed by Endoglycosidase H treatment. Mass spectrometry analysis after treatment with Endoglycosidase H to remove the N-linked glycosylation, suggested that VE-cadherin ectodomain from chicken contained 1,836Da of O-linked sugar and 7,667Da of N-linked sugar. Similarly, human VE-cadherin ectodomain appeared to contain 2,731Da of O-linked sugar and 10,336Da of N-linked sugar. Five sites of N-linked glycosylation were mapped in the human VE-cadherin ectodomain by mass determination of tryptic peptides: Asn14 and Asn65 in EC1, Asn110 in EC2, Asn315 in EC3 and Asn395 in EC4 (Figure 1a). No N-linked glycosylation sites were found in domain EC5.
Figure 1.
N-linked glycosylation sites in the VE-cadherin ectodomain. (a) N-linked glycosylation sites were determined experimentally as described in the text and are depicted on a homology model of VE-cadherin encompassing domains EC1-5. Molecular surface is colored by residue type: negatively charged residues red, positively charged blue, hydrophobic in grey, semipolar in yellow, polar in light blue and aromatic residues in rose. Magenta spheres mark the positions of N-linked glycosylation. (b) Close up stereo image of the homology model of VE-cadherin showing N-linked glycosylation site Asn395 in the EC4 domain within a region with hydrophobic and aromatic surface residues. Residue side chains with solvent exposed surface area greater than 20% are shown as sticks. Surface is colored as for panel (a). (c) Protein sequence alignments of VE-, type I and type II cadherins showing conservation of the glycosylation site (N-X-S/T) at Asn395 in the EC4 domain.
Natively glycosylated whole VE-cadherin ectodomain forms dimers, but not higher order multimers
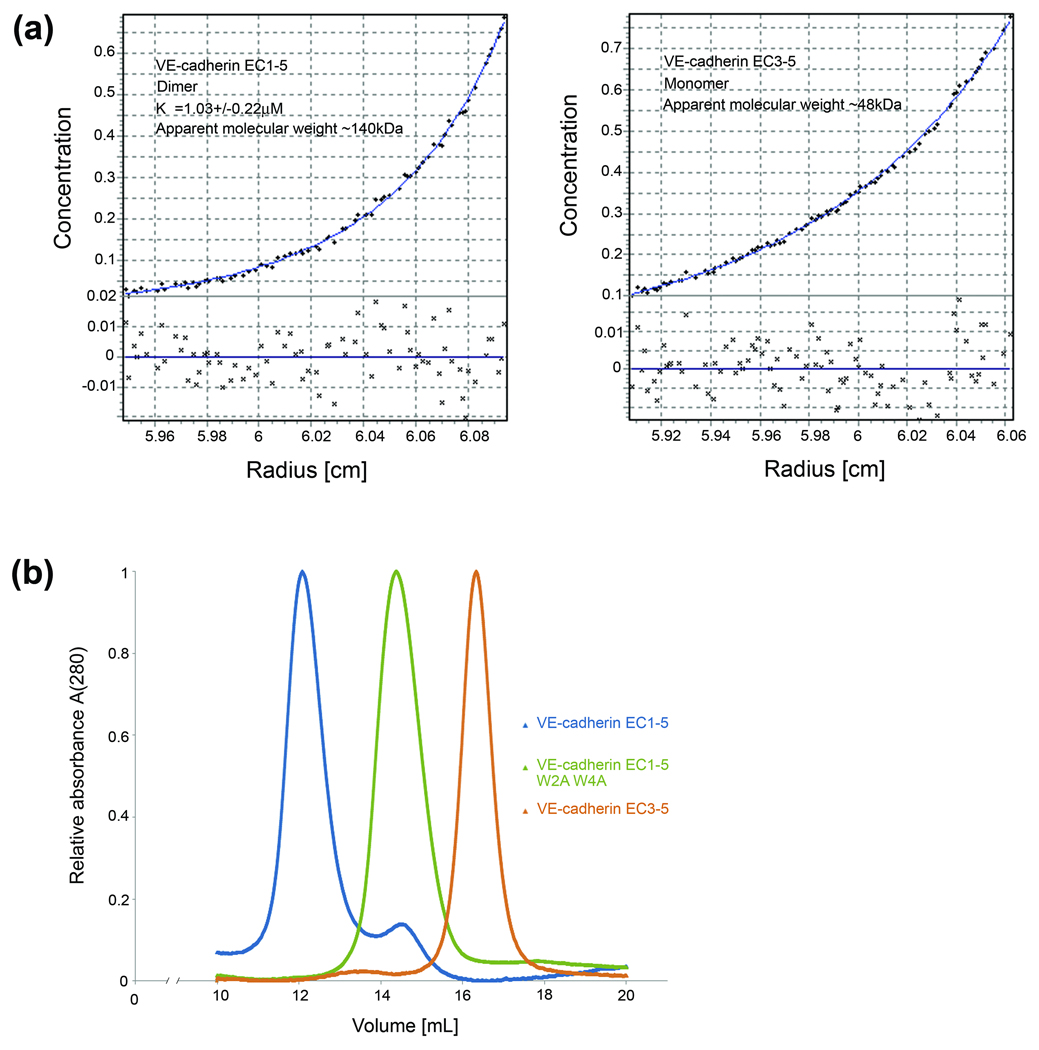
To determine the oligomerization behavior of human and chicken VE-cadherin ectodomains in solution, we used sedimentation equilibrium analytical ultracentrifugation (AUC). Sedimentation equilibrium experiments, which yield accurate masses independent of molecular shape, revealed a monomer/dimer equilibrium for VE-cadherin full ectodomains with dissociation constants (KD) values for dimerization of 1.14µM and 1.03µM for chicken and human, respectively (Table 1, Figure 2). These values are similar to those obtained for two-domain EC1-2 fragments, which encompass the adhesive binding region (Table 1). No evidence for higher order structures was found in any of the AUC experiments performed. Comparison of the KD value for dimerization of wild type human and chicken VE-cadherin EC1-5 fragments with KDs previously reported for type I cadherins (mouse, human and chicken E- and N-cadherin EC1-2 and frog C-cadherin EC1-5) shows that VE-cadherin dimerization is markedly stronger by between 20- and 150-fold (Table 1, 18; 36; 37).
Table 1.
Dissociation constants (KD) of cadherin ectodomain fragments determined by equilibrium analytical ultracentrifugation.
| Protein | Description | Dimer KD [µM] |
|---|---|---|
| VE-cadherin | ||
| ck VE-cadherin EC1-5 | Wild type, glycosylated | 1.14±0.04 |
| h VE-cadherin EC1-5 | Wild type, glycosylated | 1.03±0.22 |
| h VE-cadherin EC1-5 | Wild type, deglycosylated | Aggregation |
| h VE-cadherin EC3-5 | Wild type, glycosylated | Monomer |
| h VE-cadherin EC3-5 | Wild type, deglycosylated | Aggregation |
| h VE-cadherin EC3-4 | Wild type, E. coli | 1.93±0.26a |
| h VE-cadherin EC1-2 | Wild type, E. coli | 4.38 ±1.2 |
| ck VE-cadherin EC1-2 | Wild type, E. coli | 1.63±0.19 |
| m VE-cadherin EC1-2 | Wild type, E. coli | 2.22±0.11 |
| ck VE-cadherin EC1-2 W2A W4A | Strand swap mutant, E. coli | Monomer |
| m VE-cadherin EC1-2 W2A W4A | Strand swap mutant, E. coli | Monomer |
| Type I cadherins | ||
| m N-cadherin EC1-2 | Wild type, E. coli | 25.8±1.5b |
| m E-cadherin EC1-2 | Wild type, E. coli | 96.5±10.6b |
| Type II cadherins | ||
| ck cadherin-6b EC1-2 | Wild type, E. coli | 9.2±0.6 |
| m cadherin-6 EC1-2 | Wild type, E. coli | 3.13±0.09c |
| m cadherin-8 EC1-3 | Wild type, E. coli | 15±0.4 |
| m cadherin-9 EC1-2 | Wild type, E. coli | 17±1.1 |
| m cadherin-10 EC1-2 | Wild type, E. coli | 42.2±2.7 |
| m cadherin-11 EC1-2 | Wild type, E. coli | 33.8±0.2 |
Human VE-cadherin domains EC3-4 were also found to aggregate in some experiments.
Reported in Katsamba et al., 2009.
Reported in Harrison et al., 2010a.
Figure 2.
Biophysical characterization of the VE-cadherin ectodomain (a) Sedimentation equilibrium AUC profiles showing dimerization for human VE-cadherin EC1-5 and ideal monomeric behavior for the truncated EC3-5 fragment. Dissociation constants (KD) from the analyses are listed in Table 1; calculated molecular weights are given in the plots. (b) Comparison of elution profiles from analytical size-exclusion experiments with human VE-cadherin EC1-5 (blue), double tryptophan mutant W2A W4A (green) and EC3-5 (orange) at a concentration of 0.5mg per mL. VE-cadherin EC1-5 elutes as two peaks: a major peak of higher molecular size and a minor peak representing a smaller species whereas for the strand swap mutant W2A W4A only the lower molecular size peak is observed. The EC3-5 fragment elutes as one peak with the smallest molecular size of the three proteins. Void volume of the column is approximately 8mL.
To further analyze the biophysical behavior of human VE-cadherin ectodomains, we employed size exclusion chromatography and passed human VE-cadherin EC1-5 over an analytical Superose 6 column. The elution profile shows two distinct peaks which, given our AUC results, are likely to represent VE-cadherin dimer and monomer (Figure 2b). The same experiment was performed with the double tryptophan mutant W2AW4A, in which strand swapped dimerization is ablated by mutation of the docking tryptophans. The mutant elutes as a single peak with an elution volume corresponding to that of the suggested monomer peak of the wild type (Figure 2b).
The binding properties of the wild type and W2A W4A mutant proteins were also assessed by liposome aggregation experiments. Both proteins include a hexa histidine tag at the C-terminus which can bind by affinity to Nickel NTA-lipids incorporated into liposomes that act as artificial cells in the aggregation assay. The human VE-cadherin ectodomain EC1-5 aggregates liposomes efficiently, whereas the W2A W4A mutation ablates binding, consistent with analytical size exclusion chromatography results (Supplementary Figure 1).
Overall, our solution biophysics experiments suggest that VE-cadherin ectodomain fragments form only dimers in solution with no evidence of higher ordered structures, behaving identically to other classical cadherin adhesive dimers. Further, given that dimerization is ablated by the well characterized W2A W4A strand swapping mutation, VE-cadherin appears to adopt a strand swapped dimer in common with other classical cadherins.
Although extensive efforts to crystallize full length VE-cadherin ectodomain fragments did not yield diffracting crystals, we were able to image the overall arrangement of human VE-cadherin complexes by atomic force microscopy (AFM). Images of VE-cadherin ectodomain on poly-lysine coated mica surfaces revealed two distinct reoccurring forms (Figure 3). A crescent shaped form (Figure 3a) with a length of 28±2nm, is strikingly similar to the individual protomers observed in the published C-cadherin ectodomain crystal structure, and is therefore likely to represent the monomer (Figure 3a, 5). A larger structure with a length of approximately 48±9nm was also frequently observed, resembling two crescent shaped forms overlapping at their termini with a corresponding increase in measured sample thickness at the overlapping region (Figure 3b). The overall shape is reminiscent of the C-cadherin dimer observed by X-ray crystallography 5; 38; 39. No structures similar to the cylindrical hexameric structures reported for the bacterially expressed EC1-4 fragment of VE-cadherin 33 were observed. These data suggest that full length native VE-cadherin ectodomains form strand swapped dimers similar to other classical cadherins.
Figure 3.
AFM-imaging of full ectodomains of VE-cadherin deposited on poly-l-lysine mica reveals monomer and dimer forms. (a) Recurring crescent forms with a length of 28±2nm. These resemble C-cadherin ectodomain protomers in crystal structures (PDB: 1L3W), depicted in orange surface representation in bottom right panel. (b) Second recurring form in which two crescent shaped forms overlap at their tips with an overall length of 48±9nm. These resemble trans dimer structures observed in the crystal structure of C-cadherin (bottom right panel). Color scale indicates sample thickness above the mica surface. Scale bar for all images is 35nm.
The EC3-4 fragment forms multimers only when glycosylation is absent
The hexameric cryo-EM structures reported for the bacterially expressed VE-cadherin EC1-4 fragment suggested the presence of a cis trimer interface in or near the EC4 domain 33. To investigate the biological relevance of this finding, we produced human VE-cadherin ectodomain fragments encompassing the predicted trimer interface region using both bacterial and mammalian expression systems. A VE-cadherin EC3-4 fragment produced in bacteria and thus lacking glycosylation behaved as a dimer in size exclusion chromatography, and either as strong dimer with a KD of 1.9µM or an isodesmic aggregate in AUC (Table 1). Although this protein did not associate trimerically, it is known that proteins with non-specific hydrophobic associations can sometimes form complexes with unexpected numbers of protomers 40. We then used the mammalian HEK-293 F cell system to produce a similar but fully glycosylated protein, human VE-cadherin EC3-5. We included the EC5 domain in this construct since expression of the EC3-4 construct was poor in the HEK-293 system. Remarkably, the fully glycosylated EC3-5 protein behaves as a monomer in size exclusion chromatography, liposome aggregation assays, and equilibrium AUC (Figure 2 and Supplementary Figure 1). This result suggested that the multimeric association of the bacterial protein and the high affinity EC3-4 dimerization arose from its lack of glycosylation. To test this idea, we produced VE-cadherin EC3-5 in GNTI− cells which yield minimal sugars at N-linked glycosylation sites that can be removed by treatment with Endoglycosidase H. Consistent with our hypothesis, this minimally-glycosylated protein associated as dimers and isodesmic aggregates in AUC experiments and was able to efficiently aggregate liposomes (Table 1, Figure 2a and Supplementary Figure 1). When N-linked glycan was removed from this protein with Endoglycosidase H it behaved similarly. These results strongly suggest that the trimeric association previously observed in cryo-EM studies of the bacterially-produced EC1-4 fragment represents an artifact arising from the lack of glycosylation.
Mapping of the glycosylation sites we have identified onto the molecular surface of a VE-cadherin EC4 homology model reveals that the single N-linked glycan site in the domain is centered on a molecular surface dominated by hydrophobic residues (Figure 1b). Furthermore, sequence comparisons show that this glycosylation site is conserved among classical cadherins (Figure 1c). It is not unexpected that removal of the protruding hydrophilic glycan moiety to reveal a large hydrophobic surface could result in non-specific associations. Thus, structural data support the idea that trimerization of VE-cadherin in the EC4 domain as previously proposed is likely to be artifactual, which is supported by the high affinity for the EC3-4 protein association in comparison to the glycosylated counterpart monomers.
Biophysical characterization of VE-cadherin EC1-2 reveals strand swap-dependent homodimerization
Since classical cadherin adhesive interfaces have been localized to the EC1 domain, and adhesive binding is strongly affected by the Ca2+ binding sites between the EC1 and EC2 domains, we produced EC1-2 fragments of VE-cadherins from human, mouse, and chicken. We used equilibrium AUC analysis to assess the multimerization of these proteins. The results reveal a monomer-dimer equilibrium for each protein with dissociation constants (KD) for dimerization between 1.63 and 4.38 µM, values that are very similar to those reported for the full length EC1-5 constructs (Table 1).
To test if VE-cadherin dimerization is ablated by mutation of the Trp anchor residues, as for other cadherins, we substituted both W2 and W4 with alanine in the EC1-2 proteins. AUC analysis shows that these mutants fail to dimerize, consistent with the strand swap binding mode found for all classical cadherins tested to date (Table 1).
The binding affinities determined for VE-cadherin are at least one order of magnitude stronger than those of type I cadherins characterized previously 18; 36; 37. To address the question of whether higher affinity binding is a property of VE-cadherin or of type II cadherins in general, we performed sedimentation equilibrium analysis for wild type EC1-2 fragments of mouse cadherins-9, -10, and -11, and an EC1-3 fragment of cadherin-8 (Table 1). The KD values for dimerization of type II cadherins, including the previously reported value for cadherin-6 18, range from 3.13 to 42.2µM suggesting that, consistent with the larger size of their binding interfaces, type II cadherin dimerization is in general stronger than for type I cadherins. Nevertheless, significant variation is found within the type II cadherin family. VE-cadherin exhibits adhesive binding among the strongest of the type II cadherins.
Crystal Structure of chicken VE-cadherin EC1-2 reveals a strand-swapped dimer
To investigate the structural features of the adhesive interface of VE-cadherin we determined the crystal structure of chicken VE-cadherin domains EC1-2 (Asp1–Asp203 of the mature protein) at 2.1Å resolution. Data collection and refinement statistics are summarized in Table 2.
Table 2.
Crystallographic data and refinement statistics.
| Data collection | |
|---|---|
| Space group | P43212 |
| Cell dimensions a, b, c (Å); α, β, γ (°) | 99.97, 99.97, 105.99; 90, 90, 90 |
| Resolution (Å) | 80-2.1 |
| Rmerge | 13.7 (41.8) |
| I/σI | 1395.2/64.4 (339.1/48.2) |
| Completeness | 100 (100) |
| Redundancy | 14.1 (14.0) |
| Observed reflections | 450,866 |
| Unique reflections | 31,991 |
| Refinement | |
| Resolution (Å) | 72.7-2.1 |
| Number of reflections | 30,319 |
| Rwork | 18.3 |
| Rfree | 24.2 |
| Number of atoms | 3,841 |
| Protein | 3,247 |
| Ion | 6 |
| Water | 588 |
| R. m. s. deviations | |
| Bond length (Å) | 0.016 |
| Bond angles (°) | 1.59 |
| Mean B factors (Å2) | |
| Protein | 19.4 |
| Ion | 13.6 |
| water | 32.1 |
| Ramachandran plot | |
| Outliers (%) | 0 |
| Favored (%) | 97.1 |
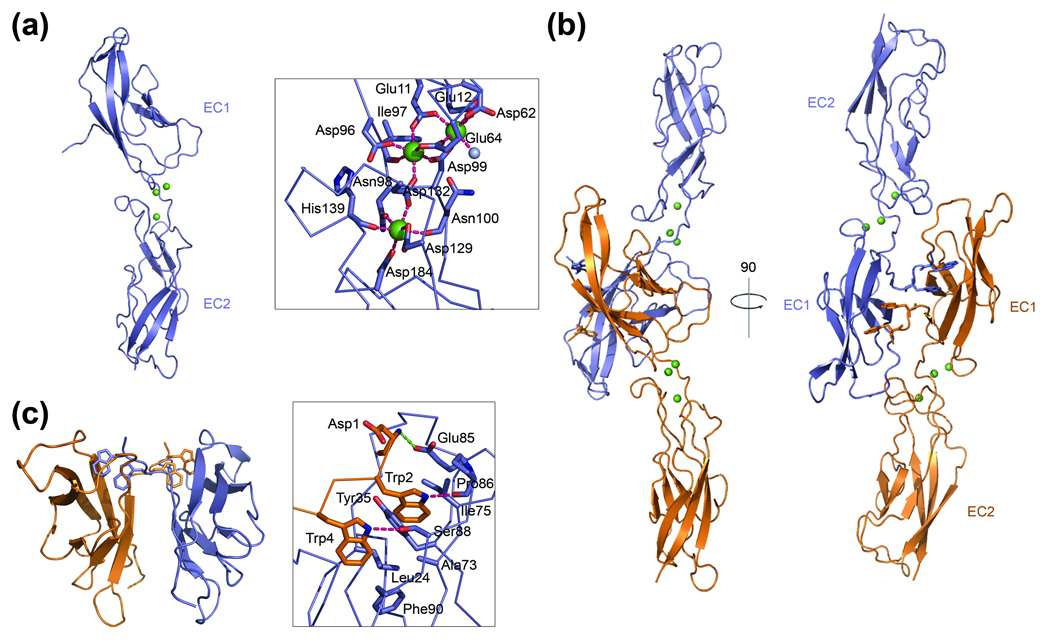
Two VE-cadherin molecules are present in the crystallographic asymmetric unit, each adopting an overall structure similar to that previously reported for structures of other type I and type II classical cadherins 2; 5; 17; 18. Two tandem EC domains, which each adopt a seven stranded β-barrel fold, are connected via a short linker region (Figure 4a). Three calcium ions are coordinated in this interdomain region by side chains of residues Glu11, Glu12, Asp62 and Glu64 from the EC1 domain, Asp96, Asn98 and Asp99 in the linker region, and Asp132 and Asp184 from the EC2 domain (Figure 4a). These residues are completely conserved within type II classical cadherins, and are also conserved in type I cadherins with the exception of Glu12 which in type I family members is replaced by an asparagine that does not directly coordinate calcium. In addition, the backbone carbonyl groups of residues Ile97, Asn100 and His139 as well as one water molecule contribute to the coordination of calcium in the VE-cadherin structure, precisely as observed in other type II cadherins (Figure 4a, 2).
Figure 4.
Crystal structure of the EC1-2 domain of chicken VE-cadherin showing a strand swapped cadherin dimer. (a) Single protomer of VE-cadherin EC1-2 shown as ribbon diagram with calcium ions as green spheres. Boxed is a detailed view of calcium coordination in the linker region between EC1 and EC2. Three calcium ions (green spheres) are coordinated by side chains of Asp, Glu or Asn and by backbone carbonyl-groups, shown in stick representation in addition to one water molecule (grey sphere). Coordinating interactions are shown as magenta dotted lines. (b) Ribbon diagram in two rotations showing the symmetrical strand swapped dimer of VE-cadherin. One protomer is shown in orange and its binding partner in blue. Trp2 and Trp4 residues in the A-strand are exchanged between protomers in the dimer and are shown in stick representation. The dimer interaction involves only the EC1 domains. (c) Detailed view of the strand dimer interface. Docking of the A-strand of one protomer (shown in orange) into the pocket of the partnering molecule (blue) is mediated by: van der Waals interactions between the tryptophan side chains and Leu24, Tyr35, Ala73, Ile75, Phe90 in the acceptor pocket; two hydrogen bonds between the ε nitrogen of Trp2 and Trp4 and the backbone carbonyl of Pro86 and the hydroxyl-group of Ser88, respectively, and a salt bridge between the N-terminal amine group of Asp1 and the carboxylate side chain of Glu85. Residues mediating binding are shown in stick representation, hydrogen bonds are depicted as magenta colored dotted lines and salt bridge interactions as green dotted lines.
The two VE-cadherin molecules in the crystallographic asymmetric unit form strand-swapped dimers as observed for other classical cadherins (Figure 4b). The dimer interface is symmetrical and is formed between EC1 domains, which are oriented approximately perpendicular relative to each other and reciprocally swap a segment of the N-terminal β-strand, designated the A*-strand. The Trp2 and Trp4 residues in the A*-strand serve as anchors that are docked into a hydrophobic ‘acceptor’ pocket in the EC1 domain of the partner protomer. The tryptophan side chains partake in van der Waals interactions with hydrophobic residues lining the acceptor pocket: Leu24, Ala73, Phe90 at the base and Tyr35 and Ile75 towards the top of the pocket (Figure 4c).
Additionally, Trp2 and Trp4 engage in two intermolecular hydrogen bonds: one between the ε1 nitrogen of the Trp2 side chain and the backbone carbonyl of Pro86 and the other between the ε1 nitrogen of the Trp4 side chain and the side chain of Ser88. While the hydrogen bond involving Trp2 has been observed in other cadherin structures, the Trp4 hydrogen bond with Ser88 appears to be specific to VE-cadherin. The N-terminal amino group of the swapped A*-strand is held in place via a salt bridge with the side chain of Glu85, a conserved interaction among classical cadherins.
VE-cadherin has been classified as a type II family member since intron/exon boundaries are similar to the genes of other type II cadherins and it has two tryptophan anchor residues and a short pro-domain characteristic of the type II subfamily 41; 42. However, phylogenetic analysis indicates that it is an outlier within this group 42. Notably, mouse type II cadherins Cadherin-6, -8, -9, -10 and -11 share amino acid identities of 65–84% within their EC1–EC2 regions, whereas VE-cadherin in comparison is only 43–47% identical to these more representative type II family members (Supplementary Table 1). To determine how this sequence divergence impacts the structure of the VE-cadherin adhesive region, we superposed our VE-cadherin structure with the EC1-2 adhesive region structures available for type I and type II cadherins (Figure 5). The overall structures, including the angle between the EC1 and EC2 domains, appear to be quite similar. Root mean square deviations (r. m. s. d.) between superposed VE- and E-cadherin are of 1.52Å over 187 Cα-atom pairs and 1.86Å r. m. s. d. between VE-cadherin and cadherin-11 over 191 Cα-atoms. When comparisons are restricted to EC1 domains, VE-cadherin aligns more closely with type II cadherins than type I cadherins and notably lacks the quasi β-helix between strands C and D found in all type I cadherins, but absent from type II cadherins (Supplementary Table 2). However, the type II cadherins-8, -11 and MN-cadherin align more closely with each other than with VE-cadherin, indicating that VE-cadherin is, to some extent, a structural outlier within the group (Supplementary Table 2).
Figure 5.
Superposed α-carbon traces from crystal structures of VE-cadherin and type I and type II classical cadherins. Single protomer (left panel) and dimer (right panel) superpositions of chicken VE-cadherin EC1-2 (blue) with E-cadherin, a type I cadherin (red, upper panel, PDB: 2QVF) and cadherin-11, a type II cadherin (orange, lower left panel, PDB: 2A4E). The structures are represented as α-carbon traces. Note that overall structure and interdomain angles are closely similar between the three different classical cadherins, but the relative orientation of EC2 domains in the VE-cadherin dimer differs remarkably from that in dimers of the other cadherins. The angle between EC2 domains in VE-cadherin is 168°, compared to 129° for E-cadherin and 143° for cadherin-11.
Unique features of the VE-cadherin binding interface
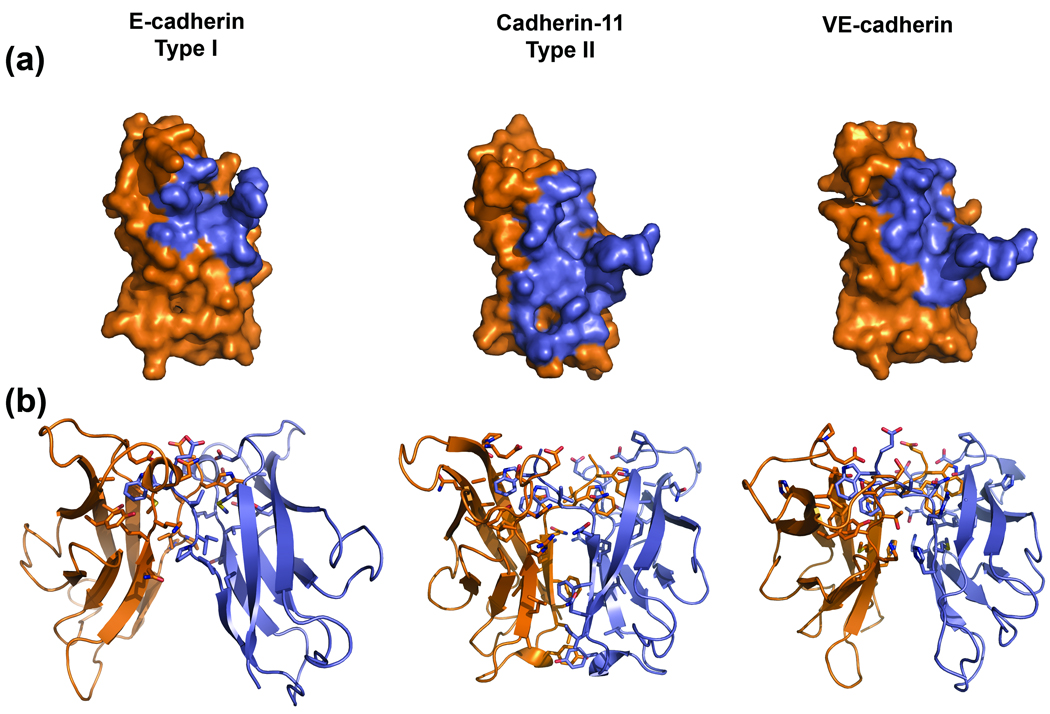
Despite a protomer structure similar overall to other classical cadherins, VE-cadherin shows remarkable differences in the arrangement of its adhesive interface (Figure 5, right panel). The VE-cadherin dimer appears to be almost linear with an angle of approximately 168° between the long axes of the opposing EC2 domains, whereas the N-cadherin and cadherin-11 dimers assemble with corresponding angles off 129° and 143°, respectively (Figure 5, right panel). Buried accessible surface (BSA) areas for type I cadherin adhesive interfaces average ~850 Å2 per protomer, whereas the area for type II cadherin protomers is significantly larger (~1250 Å2) (Supplementary Table 3). Interestingly, the VE-cadherin adhesive interface buries 1067 Å2 per protomer, which is intermediate between type I and type II cadherins. Figure 6a shows the footprint of the adhesive interface for VE-cadherin and representative type I and type II cadherins in a molecular surface representation. For the type I subfamily member E-cadherin the adhesive interface is restricted to the “upper” half of the EC1 domain around the swapped strand and acceptor pocket. However, the interface of type II cadherins, represented in Figure 6 by cadherin-11, extends over the whole face of the EC1 domain, owing to the participation of an extended region of non-swapped hydrophobic interaction surface at the base of the EC1 domain which is restricted to type II cadherins (Figure 6b and 7, 2). Remarkably, the surface involved in the VE-cadherin strand swapped dimer more closely resembles that of type I cadherins since the extended hydrophobic region is absent (Figure 6b and 7). The corresponding region in VE-cadherin does not contribute to the dimer and remains solvent exposed.
Figure 6.
Comparison of the strand swapped dimer interface of VE-cadherin with those of type I and II cadherins. (a) EC1 domains of single protomers of E-cadherin (left panel, PDB: 2QVF), cadherin-11 (middle panel, PDB: 2A4C) and VE-cadherin (right panel) are displayed as a molecular surface (orange). The “footprint” of the interaction partner in the strand swapped dimer, determined as regions with a per residue buried surface area in the dimer of >10Å2, is colored blue on the surface. The interface for type I cadherins (left panel) and VE-cadherin (right panel) is restricted to a smaller region than in type II cadherins (middle panel), in which the interface is not only localized around the A-strand but extends toward the base of the EC1 domain. (b) Ribbon diagrams of EC1 domains of the same dimers as in (a), with residues involved in the interface (BSA> 10Å2) shown as sticks. One protomer is colored orange, the other blue. Note the adhesive interface of VE-cadherin shares features of both subfamilies of classical cadherins.
Figure 7.
VE-cadherin uses a different set of residues for trans-dimerization than type II cadherins. (a) Sequence alignment of the EC1 domains of chicken VE- and MN-cadherin (PDB: 1ZVN) and mouse cadherin-8 (PDB: 1ZXK) and -11 (PDB: 2A4C), for which strand swapped dimer structures have been determined. Residues buried in the strand swapped interface (>5% buried) in all structures are boxed in purple. Residues buried in dimers of cadherin-8, -11 and MN cadherin but solvent exposed in VE-cadherin, which comprise the type II cadherin specific hydrophobic interface, are boxed in green. (b) Comparison of BSA values for EC1 domain residues in the strand swapped dimer interfaces of VE-cadherin and type II cadherins, as a percentage of the residue surface area. VE-cadherin shares the buried A-strand and acceptor pocket residues with type II cadherins (purple boxes in upper and lower panels), but residues in the type II cadherin hydrophobic interface, positions 10, 13, 19, 20, 21 and 22, are greater than 95% solvent exposed in the VE-cadherin dimer (green boxes in upper and lower panels).
The residues of VE-cadherin that map to the non-swapped hydrophobic interface of more typical type II cadherins are distinct in character (Figure 7a). In typical type II cadherins hydrophobic residues at positions 8, 10, 13 and 20 in the A and B strands pack together in the dimer interface (Figure 6b and 7a, 2). These are conserved in character in all type II cadherins except VE-cadherin, which like type I cadherins has hydrophilic residues at most of these positions (Figure 7a). These differences are likely to underlie the more type I-like dimer interface configuration found for VE-cadherin.
Overall, the structure of the VE-cadherin adhesive dimer shows that VE-cadherin is an outlier among the classical cadherins. While interactions between the swapped A*-strand and the acceptor pocket are almost identical between VE-cadherin and other type II cadherins, the VE-cadherin interface more closely resembles type I cadherins outside of the swapping region.
DISCUSSION
We employed biophysical characterization of natively glycosylated VE-cadherin EC1-5 to test a novel model for adhesive binding of VE-cadherin that differed substantially from the known structures of other classical cadherins. This model, based on solution biophysics and cryo-EM analysis of a bacterially-produced VE-cadherin EC1-4 ectodomain fragment, suggested that VE-cadherin forms cis cell surface trimers via an interface in EC4, which engage in EC1-mediated trans interactions between cells to form hexamers 33; 34; 35; 43; 44. By contrast, a wide array of evidence for other classical cadherins suggests that they form adhesive bonds through strand swap interactions between monomers. It is thought that adherent dimers then assemble to form adherens junctions via weak cis interactions which have recently been identified for type I cadherins 5; 45, but strong cis interactions like the trimer proposed for VE-cadherins have not been observed. While the known cadherin strand swap mechanism and the putative hexamer model both involve EC1-mediated trans dimers that are broadly compatible with the strand-swapped structure of chicken VE-cadherin EC1-2 reported here, they differ with respect to the presence of strong cis trimer interactions in EC3-4. Our biophysical studies showed that “naked”, bacterially produced EC3-4 exhibits high affinity for association and minimally glycosylated EC3-5 fragments of VE-cadherin aggregate liposomes through homophilic interactions. These findings provide an explanation for why a trimeric interface involving EC4, a phenomenon prevented by native glycosylation, could be observed in previous studies using bacterially expressed constructs of VE-cadherin33; 34; 35. In agreement with our data, Ahrens et al. (2003) found mammalian expressed VE-cadherin ectodomains to also associate only via their N-termini in rotary shadowing electron microscopy experiments. The biophysical experiments reported here clearly show that VE-cadherin, though a phylogenetic outlier, shares an adhesive mechanism similar to that of other classical cadherins.
We tested human VE-cadherin EC1-5 domain constructs, which were expressed in HEK 293F cells and purified from conditioned media, in equilibrium AUC and in size exclusion chromatography experiments. In all cases we observed exclusively a monomer-dimer equilibrium with no evidence of hexamer formation. By contrast, non-glycosylated bacterially-produced constructs of the EC1-4 region behave as multimers 33; 34; 35 and, in our experiments, bacterially expressed EC3-4 regions, encompassing the putative trimerization region, behave as dimers or aggregate. Furthermore, whereas isolated natively glycosylated VE-cadherin EC3-5 fragments were monomeric in solution, partially glycosylated or deglycosylated protein also showed multimer/aggregate behavior. The partially glycosylated EC3-5 fragments also showed binding activity in liposome aggregation assays whereas equivalent, natively glycosylated fragments did not. These results strongly suggest that the trimerization observed previously arises from the lack of glycosylation, inherent to bacterial protein production, which exposes a hydrophobic surface on EC4 which would be shielded in the natural protein by a conserved N-linked glycan. Furthermore, previous studies showed that the native glycan of human VE-cadherin carries negatively charged sialic acid in almost all branches that might contribute to repulsion mediated by the glycan46.
While our data exclude strong cis interactions such as those observed for bacterially expressed VE-cadherin EC1-4 fragments 33; 34; 35, other potential cis interactions cannot be excluded and could contribute to adhesive junction formation. Such cis interactions, which function in the context of membrane bound molecules, are often weak and can be undetectable by standard solution biophysics methods. Examples of weak cis interactions that have been characterized include type I cadherins 5; 45 and Eph/ephrin complexes 47; 48. In type I cadherins a cis oriented interaction common to the crystal structures of C-, E- and N-cadherins has recently been shown to facilitate assembly of adherens junctions in concert with the well characterized trans interaction (5, Harrison, adherens junction paper, submitted). VE-cadherin is also found in adherens junctions 49, but the cis interaction observed in type I cadherins is not present in the chicken VE-cadherin EC1-2 crystal lattice, possibly because it lacks the pseudo β-helix, a structural element contributing to the cis interface in type I cadherins. It remains to be determined whether VE-cadherin employs an alternative cis interface between neighboring proteins.
Here we have also presented the 2.1 Å resolution crystal structure of the adhesive dimer of chicken VE-cadherin EC1-2. The structure shows, that VE-cadherin adopts a strand swapped dimer stabilized by intermolecular docking of Trp2 and Trp4 residues, as observed for the other more typical type II cadherins 8, 11 and MN 2. However, VE-cadherin lacks the non-swapped hydrophobic interaction region of the adhesive interface, and thus the overall arrangement of the VE-cadherin adhesive dimer is more similar to type I cadherins. The absence of this contact region likely arises from the substitution of hydrophilic residues in VE-cadherin for the hydrophobic residues near the base of EC1 in the more typical type II cadherins. It has previously been suggested that the larger strand dimer interface in type II cadherins contributed by the non-swapped hydrophobic region may enhance binding affinities in comparison to those of type I cadherins 36. Prior to our study KD values from AUC analysis of type II cadherin dimerization were available only for EC1-2 fragments of mouse cadherin-6 18. We report here KD values for homodimerization of five further type II cadherins: mouse cadherin-8, -9, -10,-11 and chicken cadherin-6b, in addition to VE-cadherin from several species. The determined KD values span a range from 1.6–42.2µM, and in general indicate stronger adhesive binding than for type I cadherins (19.7–156µM for N-, E-, C-cadherin;36; 37; 45). It is surprising, however, that VE-cadherin is the strongest of these despite having a smaller buried surface area in the dimer interface. Other factors, such as strain in the closed monomer form must therefore play an important role in determination of homodimerization affinity 50; 51. The high affinity of VE-cadherin binding interactions, in comparison with other classical cadherins, may reflect its unique role in maintaining intercellular adhesion in the vascular endothelium, which is exposed to substantial hemodynamic stresses 52.
Classical cadherins often display homophilic dimerization but, at least within a subtype (e.g. type I cadherins) can bind heterophilically as well 36. Domain shuffling experiments have shown that the determinants of cellular specificity reside in the EC1 domains 2; 36; 53; 54; 55. Cell aggregation studies have shown that cells expressing individual type II cadherins, while displaying homophilic selectivity in general, bind to each other promiscuously to different degrees 53. Cells expressing E- and N-cadherins have been shown to display qualitatively similar behavior 36, however consistent with the differing structures of their adhesive regions cross interactions between type I and type II cadherins have not been observed either at the molecular 36 or cellular levels 2; 53. Accordingly, it has been demonstrated, that transfected cells expressing VE-cadherin do not heterophilically interact with transfected cells expressing type I N-cadherin 56. Furthermore, VE-cadherin excludes N-cadherin from adherens junctions in vascular endothelial cells, in which the two cadherins are co-expressed in vivo 11; 24; 25; 26. The differences we observe between the dimer interfaces of VE-cadherin and other classical cadherins, including the previously studied type II family members suggest that VE-cadherin is unlikely to bind heterophilically to other classical cadherin molecules due to incompatibility of the binding interfaces they present. The structural divergence of VE-cadherin, which has a critical, non -redundant role in the assembly of the vascular endothelium, is therefore likely to result in a high degree of homophilic specificity since cadherins with similar structural features have not been identified.
MATERIALS AND METHODS
Bacterial protein production
Coding sequences of mouse, chicken and human VE-cadherin EC1-2, mouse cadherin -8 EC1-3 and cadherin-9, -10, -11 EC1-2 were amplified by PCR from cDNA libraries (Clontech; chicken VE-cadherin: Delaware Biotechnology Institute, clone pgp1n.pk007.i4) and cloned in frame with an N-terminal hexa histidine-tagged SUMO protein into the BamHI/NotI sites of the pSMT3 vector. Cleavage of SUMO-fusion-proteins with Ulp1 (Ubiquitin-like protease 1) after a Gly-Gly motif yields cadherin proteins with native N-termini. Extra amino acids occurring due to cloning after the cleavage site were removed by using the QuikChange site directed mutagenesis kit (Stratagene) to ensure native N-termini of all proteins used in our studies, unless specifically altered. We introduced the W2AW4A mutation using the same method.
For protein expression, E. coli Rosetta 2DE3 pLysS cells (Novagen) were transformed with prepared pSMT3 vector and grown at 37°C until OD600 reached 0.6 shaking at 200rpm. To induce protein expression, we added 100µM IPTG and lowered the temperature to 16°C. After 18h cells were harvested by centrifugation at 6,000g for 15min. Pelleted bacteria were resuspended in lysis buffer (500mM NaCl, 20mM Tris-Cl pH8.0, 20mM Imidazole pH8.0, 3mM CaCl2) and lysed for 6 minutes by sonication in 15 second intervals with 45 seconds rest in between pulsing. Cell debris was spun down at 4°C and 20,000g for 1 hour and His-tagged proteins were extracted from the clear lysate by flowing over nickel charged IMAC Sepharose 6 Fast Flow resin (GE Healthcare). Beads were subsequently washed with 40 column volumes of lysis buffer to remove contaminants and His-SUMO-fusion proteins were eluted with 250mM imidazole containing lysis buffer. The 6-His-SUMO tag of fusion proteins was cut enzymatically by adding Ulp-1 to a final concentration of 2µg/mL to the elution. Proteins were dialyzed into a low ionic strength buffer (100mM NaCl, 20mM Tris-Cl pH8.0 and 3mM CaCl2). For cadherin-8, -9, -10 and -11 we removed cleaved 6His-SUMO tags and uncut fusion protein on nickel charged IMAC resin, equilibrated in dialysis buffer. The cadherins were further purified by anion exchange chromatography (Mono Q 10/10 HR, GE Healthcare) and size exclusion chromatography (HiLoad 26/60 Superdex™ S75 prepgrade, GE Healthcare) leaving them in a final buffer of 150mM NaCl, 20mM Tris-Cl pH8.0 and 3mM CaCl2. Proteins were concentrated to a final concentration of approximately 5–10mg/mL using Amicon Spin concentrators (Milli Pore) and flash frozen.
Mammalian protein production
Coding sequences for human or chicken VE-cadherin domains EC1-5 (Asp1–Asp542, Asp1-Glu545 of the native protein, respectively) or the coding sequence for human VE-cadherin EC3-5 (Ile204 – Asp542) were amplified by PCR from cDNA libraries and, all preceeded by a Kozak sequence and the signal sequence for human CD33 (MPLLLLWAGALA), with the transmembrane domain and intracellular domain replaced by a hexa histidine tag on the C-terminus were cloned into the episomal expression vector pCEP4 (Invitrogen) using the restriction sites KpnI/NotI. For protein expression human embryonal kidney (HEK) 293 F cells or HEK 293 GNTI− cells were transfected in 6 well stage using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s manual. During continuous selection with 200µM Hygromycin B for stable expression, proteins were secreted and purified from 4 liters collected DMEM/F12 medium, supplemented with 10% newborn calf serum, 1:100 Penicillin/Streptomycin (Gibco), 1:50 L-Glutamine (Gibco) and 200µM Hygromycin B (Cellgro). After adding 500mM sodium chloride, 20mM Tris-Cl pH8.0, 3mM calcium chloride and 10mM imidazole pH8.0 to the supernatant His-tagged proteins were extracted from the medium by batch affinity purification by adding 20mL of nickel charged IMAC resin (GE), pre-equilibrated in 500mM sodium chloride, 20mM Tris-Cl pH8.0, 3mM calcium chloride and 10mM imidazole pH8.0. After 3 hours of incubation the resin was collected from supernatant by passing through Kontes columns and washed with 20 column volumes of binding buffer and 20 column volumes of the same buffer with increased imidazole concentration (12.5mM). Proteins were eluted with five column volumes 75mM imidazole in same buffer. Eluted protein is dialyzed over night at 4°C into a low ionic strength buffer of 100mM sodium chloride, 20mM Tris-Cl pH8.0 and 3mM CaCl2. Further purification by flowing through Mono S HR 10/10 (GE) and Mono Q HR 10/10 (GE) ion exchange columns, and size exclusion chromatography with HiLoad 26/60 Superdex™ S200 prepgrade (GE) leaves proteins in buffer of 150mM NaCl, 10mM Tris-Cl pH8.0, 3mM CaCl2. Proteins were concentrated and flash frozen at a concentration of approximately 7mg/mL. Proteins were N-terminally sequenced, to ensure the proper cleavage of the artificial signal sequence, ensuring native N-termini.
Analytical ultracentrifugation
Equilibrium analytical ultracentrifugation experiments were performed using a Beckman XLA/I ultracentrifuge, with a Ti50An or Ti60An rotor. Prior to each experiment, all proteins were diluted with buffer (150mM NaCl, 10mM Tris-Cl pH8.0, 3mM CaCl2) and dialyzed for 16 hours at 4°C in the same buffer. 120µL of proteins at three different concentrations 0.7mg/mL, 0.46mg/mL and 0.24mg/mL were loaded into six-channel equilibrium cells with parallel sides and sapphire windows. We performed all experiments at 25°C and collected data at wavelengths of 280 nm (UV) and 660 nm (interference). Five-domain proteins were spun for 20 hours at 8,800g and four scans (1 per hour) were collected, speed was increased to 12,300g for 10 hours and four scans (1per hour) were collected and speed was increased to highest speed of 16,400g for another 10 hours and four scans (1 per hour) were taken, which yields 72 scans per sample. Three-domain proteins were analyzed using the same protocol, except using 14,200g, 23,500g and 35,200g, respectively. For two-domain proteins, 23,500g, 35,200g and 49,100g were used. RCF:s are given at the measuring cell center at a radius of 65mm. We calculated the buffer density and protein v-bars using the program SednTerp (Alliance Protein Laboratories), and analyzed the retrieved data using HeteroAnalysis 1.1.0.28 (http://www.biotech.uconn.edu/auf). We fitted data from all concentrations and speeds globally by nonlinear regression to either a monomer-dimer equilibrium model or an ideal monomer model. All experiments were performed at least in duplicate.
Analytical size exclusion chromatography
Analytical size exclusion chromatography was performed at 4°C using a superose 12 10/20 column (GE Healthcare). After equilibrating the column in buffer (150mM NaCl, 10mM Tris-Cl pH8.0, 3mm CaCl2) at a flow rate of 0.3mL per min, 150µL of purified protein at a concentration of 6.7µM was injected and eluted with the same flow rate while monitoring the UV280nm signal. We collected 0.5mL fractions for SDS PAGE analysis with subsequent immunological detection (QIAGEN, Tetra-His antibody, anti mouse antibody with HRP, SantaCruz) against the C-terminal hexa histidine tag common to all proteins used in this experiment. Void volume was determined with Blue Dextran (GE healthcare) to be 8mL.
Liposome aggregation assay
We prepared Liposomes in a 9:1 molar ration of 1,2-dioleoyl-sn-glycero-3-phospohocholine (DOPC) and Nickel (II) 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid) succinyl]) (DGS-NTA) using the hydration and extrusion method. Lipids were prepared according to the manufacturer’s instructions (Avanti lipids), suspended in liposome aggregation buffer (25mM HEPES pH7.4, 0.1M KCL, 10% (v/v) glycerol, 3mM CaCl2) and passed through a 100nm filter membrane to yield liposomes with equal size.
Conduction of aggregation assays were based on the method described in 45; 57. Liposomes at a final concentration of ~5mg/mL and hexa histidine tagged protein at concentrations of 0.5mg/mL were mixed at the starting point zero and aggregation was monitored every 20 seconds by changes in light scattering at OD650 for a total time of 40 minutes. As negative control, a sample containing only liposomes and buffer was used. Experiments were performed as triplicates for VE-cadherin EC1-5 and duplicates for EC3-5 proteins.
Atomic force microscopy imaging
AFM sample pucks were prepared as follows; 10mm diameter disks of Muscovite mica (Agar Scientific) were stamped out from the as-received sheets. Each was fixed to a 13mm steel puck (agar Scientific) with cyanoacrylate super-glue and allowed to dry fo16 h. Poly-L-lysine (Sigma Aldrich) was diluted 1/100 into BPC-grade water (Sigma Aldrich) and 45µL pipetted onto freshly-cleaved mica, given 30min incubation at room temperature, washed 10× with 1mL BCP-grade water and dried under a gentle and steady stream of nitrogen.
Samples were diluted in 150mM NaCl, 20mM Tris-Cl pH8.0, 3mM CaCl2 to a final concentration of 2nM and stored at 4°C which yielded workable and consistent surface concentrations of protein. 45µL of sample was pipetted onto poly-L-lysine coated mica and incubated at room temperature for 10min, then washed 10× with 1mL of BPC-grade water and dried under a gentle and steady stream of dry nitrogen.
Imaging was performed with a Multimode atomic force microscope with attached Nanoscope IIIa controller (both Veeco) under ambient conditions in tapping mode, using OMCL0AC160TS single-crystal silicon probes (Olympus, Japan) with a resonant frequency of ~300kHz, a nominal spring constant of ~42Nm and a radius of curvature <10nm. Images were collected at 3Hz with an integral gain of 0.2, a proportional gain of 0.35, a look-ahead gain of 0 and a set point of ~0.85 (to minimize vertical probe-induced sample deformation). Length determination of molecules was performed using SPIP (Scanning Probe Image Processor) version 3.3 from 3-dimensional AFM data using the full-width at half-maximal approach.
Crystallization and structure determination
We expressed and purified chicken VE-cadherin EC1-2 as described above and used it for crystallization studies at 8.6 mg/mL. Using the vapor diffusion method, protein crystals grew at 20°C after combining 1µL protein with 1µL well solution composed of 18% (w/v) PEG 8000, 200mM calcium acetate and 100mM sodium cacodylate pH6.5. The crystals grew within 20h and were flash frozen in liquid nitrogen after being immersed briefly in cryo protectant (30% (w/v) glycerol, 18% (w/v) PEG 8000, 200mM calcium acetate, 100mM sodium cacodylate pH6.5).
Data was collected on a single frozen crystal at the X4C beam line of the National Synchrotron Light Source, Brookhaven National Laboratory at a wavelength of 0.979Å. We processed the data using the HKL suite 58 and solved the structure by molecular replacement with mouse cadherin-11 (PDB code: 2A4E) as search model. Refinement was carried out by manual building in Coot 59 followed by automated refinement in Refmac 60. Ramachandran plot statistics for the final model are 96.1% favored, 100% allowed and 0% disallowed; data collection and refinement statistics are summarized in Table 2.
Supplementary Material
Acknowledgements
This work was supported by US National Institutes of Health grants R01 GM062270 (L.S.) and US National Science Foundation grant MCB-0918535 (B.H.). X-ray data was acquired at the X4C beamline of the National Synchrotron Light Source, Brookhaven National Laboratory; the X4 beamlines are operated by the New York Structural Biology Center.
Abbreviations
- VE-cadherin
vascular endothelial cadherin
- C-cadherin
Xenopus cleavage stage cadherin
- E-cadherin
epithelial cadherin
- N-cadherin
neural cadherin
- P-cadherin
placental cadherin
- AFM
atomic force microscopy
- HEK 293 cells
human embroyonal kidney 293 cells
- GNTI
N-acetyl-glucosaminyl transferase I
- AUC
analytical ultra centrifugation
- EC domain
extracellular cadherin domain
- BSA
buried surface area
- EM
electron microscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession number
Coordinates and structure factors for chicken VE-cadherin domains EC1-2 have been deposited in the Protein Data Bank with accession number 3PPE.
REFERENCES
- 1.Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol. 2009;41:349–369. doi: 10.1016/j.biocel.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 2.Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, Shapiro L. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 3.Posy S, Shapiro L, Honig B. Sequence and structural determinants of strand swapping in cadherin domains: do all cadherins bind through the same adhesive interface? J Mol Biol. 2008;378:954–968. doi: 10.1016/j.jmb.2008.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeichi M, Hatta K, Nose A, Nagafuchi A. Identification of a gene family of cadherin cell adhesion molecules. Cell Differ Dev. 1988;25 Suppl:91–94. doi: 10.1016/0922-3371(88)90104-9. [DOI] [PubMed] [Google Scholar]
- 5.Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 6.Nagar B, Overduin M, Ikura M, Rini JM. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380:360–364. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- 7.Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lampugnani MG, Corada M, Caveda L, Breviario F, Ayalon O, Geiger B, Dejana E. The molecular organization of endothelial cell to cell junctions: differential association of plakoglobin, beta-catenin, and alpha-catenin with vascular endothelial cadherin (VE-cadherin) J Cell Biol. 1995;129:203–217. doi: 10.1083/jcb.129.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- 10.Ishiyama N, Lee SH, Liu S, Li GY, Smith MJ, Reichardt LF, Ikura M. Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell. 2010;141:117–128. doi: 10.1016/j.cell.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Gentil-Dit-Maurin A, Oun S, Almagro S, Bouillot S, Courcon M, Linnepe R, Vestweber D, Huber P, Tillet E. Unraveling the distinct distributions of VE- and N-cadherins in endothelial cells: A key role for p120-catenin. Exp Cell Res. 2010 doi: 10.1016/j.yexcr.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Komiya S, Shimizu M, Fukunaga Y, Nagafuchi A. Involvement of p120 carboxy-terminal domain in cadherin trafficking. Cell Struct Funct. 2007;32:127–137. doi: 10.1247/csf.07023. [DOI] [PubMed] [Google Scholar]
- 13.Reynolds AB, Carnahan RH. Regulation of cadherin stability and turnover by p120ctn: implications in disease and cancer. Semin Cell Dev Biol. 2004;15:657–663. doi: 10.1016/j.semcdb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Shapiro L, Fannon AM, Kwong PD, Thompson A, Lehmann MS, Grubel G, Legrand JF, Als-Nielsen J, Colman DR, Hendrickson WA. Structural basis of cell-cell adhesion by cadherins. Nature. 1995;374:327–337. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- 15.Chen CP, Posy S, Ben-Shaul A, Shapiro L, Honig BH. Specificity of cell-cell adhesion by classical cadherins: Critical role for low-affinity dimerization through beta-strand swapping. Proc Natl Acad Sci U S A. 2005;102:8531–8536. doi: 10.1073/pnas.0503319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parisini E, Higgins JM, Liu JH, Brenner MB, Wang JH. The crystal structure of human E-cadherin domains 1 and 2, and comparison with other cadherins in the context of adhesion mechanism. J Mol Biol. 2007;373:401–411. doi: 10.1016/j.jmb.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haussinger D, Ahrens T, Aberle T, Engel J, Stetefeld J, Grzesiek S. Proteolytic E-cadherin activation followed by solution NMR and X-ray crystallography. EMBO J. 2004;23:1699–1708. doi: 10.1038/sj.emboj.7600192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison OJ, Bahna F, Katsamba PS, Jin X, Brasch J, Vendome J, Ahlsen G, Carroll KJ, Price SR, Honig B, Shapiro L. Two-step adhesive binding by classical cadherins. Nat Struct Mol Biol. 2010;17:348–357. doi: 10.1038/nsmb.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breier G, Breviario F, Caveda L, Berthier R, Schnurch H, Gotsch U, Vestweber D, Risau W, Dejana E. Molecular cloning and expression of murine vascular endothelialcadherin in early stage development of cardiovascular system. Blood. 1996;87:630–641. [PubMed] [Google Scholar]
- 20.Dejana E, Zanetti A, Del Maschio A. Adhesive proteins at endothelial cell-to-cell junctions and leukocyte extravasation. Haemostasis. 1996;26 Suppl 4:210–219. doi: 10.1159/000217301. [DOI] [PubMed] [Google Scholar]
- 21.Lampugnani MG, Resnati M, Raiteri M, Pigott R, Pisacane A, Houen G, Ruco LP, Dejana E. A novel endothelial-specific membrane protein is a marker of cell-cell contacts. J Cell Biol. 1992;118:1511–1522. doi: 10.1083/jcb.118.6.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vittet D, Buchou T, Schweitzer A, Dejana E, Huber P. Targeted null-mutation in the vascular endothelial-cadherin gene impairs the organization of vascular-like structures in embryoid bodies. Proc Natl Acad Sci U S A. 1997;94:6273–6278. doi: 10.1073/pnas.94.12.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liaw CW, Cannon C, Power MD, Kiboneka PK, Rubin LL. Identification and cloning of two species of cadherins in bovine endothelial cells. EMBO J. 1990;9:2701–2708. doi: 10.1002/j.1460-2075.1990.tb07456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salomon D, Ayalon O, Patel-King R, Hynes RO, Geiger B. Extrajunctional distribution of N-cadherin in cultured human endothelial cells. J Cell Sci. 1992;102(Pt 1):7–17. doi: 10.1242/jcs.102.1.7. [DOI] [PubMed] [Google Scholar]
- 25.Navarro P, Ruco L, Dejana E. Differential localization of VE- and N-cadherins in human endothelial cells: VE-cadherin competes with N-cadherin for junctional localization. J Cell Biol. 1998;140:1475–1484. doi: 10.1083/jcb.140.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaggi M, Wheelock MJ, Johnson KR. Differential displacement of classical cadherins by VE-cadherin. Cell Commun Adhes. 2002;9:103–115. doi: 10.1080/15419060214150. [DOI] [PubMed] [Google Scholar]
- 27.Gulino D, Delachanal E, Concord E, Genoux Y, Morand B, Valiron MO, Sulpice E, Scaife R, Alemany M, Vernet T. Alteration of endothelial cell monolayer integrity triggers resynthesis of vascular endothelium cadherin. J Biol Chem. 1998;273:29786–29793. doi: 10.1074/jbc.273.45.29786. [DOI] [PubMed] [Google Scholar]
- 28.Carmeliet P, Lampugnani MG, Moons L, Breviario F, Compernolle V, Bono F, Balconi G, Spagnuolo R, Oosthuyse B, Dewerchin M, Zanetti A, Angellilo A, Mattot V, Nuyens D, Lutgens E, Clotman F, de Ruiter MC, Gittenberger-de Groot A, Poelmann R, Lupu F, Herbert JM, Collen D, Dejana E. Targeted deficiency or cytosolic truncation of the VE-cadherin gene in mice impairs VEGF-mediated endothelial survival and angiogenesis. Cell. 1999;98:147–157. doi: 10.1016/s0092-8674(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 29.Corada M, Mariotti M, Thurston G, Smith K, Kunkel R, Brockhaus M, Lampugnani MG, Martin-Padura I, Stoppacciaro A, Ruco L, McDonald DM, Ward PA, Dejana E. Vascular endothelial-cadherin is an important determinant of microvascular integrity in vivo. Proc Natl Acad Sci U S A. 1999;96:9815–9820. doi: 10.1073/pnas.96.17.9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.May C, Doody JF, Abdullah R, Balderes P, Xu X, Chen CP, Zhu Z, Shapiro L, Kussie P, Hicklin DJ, Liao F, Bohlen P. Identification of a transiently exposed VE-cadherin epitope that allows for specific targeting of an antibody to the tumor neovasculature. Blood. 2005;105:4337–4344. doi: 10.1182/blood-2005-01-0010. [DOI] [PubMed] [Google Scholar]
- 31.Navarro P, Caveda L, Breviario F, Mandoteanu I, Lampugnani MG, Dejana E. Catenin-dependent and -independent functions of vascular endothelial cadherin. J Biol Chem. 1995;270:30965–30972. doi: 10.1074/jbc.270.52.30965. [DOI] [PubMed] [Google Scholar]
- 32.Corada M, Liao F, Lindgren M, Lampugnani MG, Breviario F, Frank R, Muller WA, Hicklin DJ, Bohlen P, Dejana E. Monoclonal antibodies directed to different regions of vascular endothelial cadherin extracellular domain affect adhesion and clustering of the protein and modulate endothelial permeability. Blood. 2001;97:1679–1684. doi: 10.1182/blood.v97.6.1679. [DOI] [PubMed] [Google Scholar]
- 33.Hewat EA, Durmort C, Jacquamet L, Concord E, Gulino-Debrac D. Architecture of the VE-cadherin hexamer. J Mol Biol. 2007;365:744–751. doi: 10.1016/j.jmb.2006.10.052. [DOI] [PubMed] [Google Scholar]
- 34.Legrand P, Bibert S, Jaquinod M, Ebel C, Hewat E, Vincent F, Vanbelle C, Concord E, Vernet T, Gulino D. Self-assembly of the vascular endothelial cadherin ectodomain in a Ca2+dependent hexameric structure. J Biol Chem. 2001;276:3581–3588. doi: 10.1074/jbc.M002667200. [DOI] [PubMed] [Google Scholar]
- 35.Bibert S, Jaquinod M, Concord E, Ebel C, Hewat E, Vanbelle C, Legrand P, Weidenhaupt M, Vernet T, Gulino-Debrac D. Synergy between extracellular modules of vascular endothelial cadherin promotes homotypic hexameric interactions. J Biol Chem. 2002;277:12790–12801. doi: 10.1074/jbc.M111597200. [DOI] [PubMed] [Google Scholar]
- 36.Katsamba P, Carroll K, Ahlsen G, Bahna F, Vendome J, Posy S, Rajebhosale M, Price S, Jessell TM, Ben-Shaul A, Shapiro L, Honig BH. Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proc Natl Acad Sci U S A. 2009;106:11594–11599. doi: 10.1073/pnas.0905349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chappuis-Flament S, Wong E, Hicks LD, Kay CM, Gumbiner BM. Multiple cadherin extracellular repeats mediate homophilic binding and adhesion. J Cell Biol. 2001;154:231–243. doi: 10.1083/jcb.200103143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahrens T, Lambert M, Pertz O, Sasaki T, Schulthess T, Mege RM, Timpl R, Engel J. Homoassociation of VE-cadherin follows a mechanism common to "classical" cadherins. J Mol Biol. 2003;325:733–742. doi: 10.1016/s0022-2836(02)01286-x. [DOI] [PubMed] [Google Scholar]
- 39.Tomschy A, Fauser C, Landwehr R, Engel J. Homophilic adhesion of E-cadherin occurs by a co-operative two-step interaction of N-terminal domains. EMBO J. 1996;15:3507–3514. [PMC free article] [PubMed] [Google Scholar]
- 40.Weis WI, Kahn R, Fourme R, Drickamer K, Hendrickson WA. Structure of the calcium-dependent lectin domain from a rat mannose-binding protein determined by MAD phasing. Science. 1991;254:1608–1615. doi: 10.1126/science.1721241. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki S, Sano K, Tanihara H. Diversity of the cadherin family: evidence for eight new cadherins in nervous tissue. Cell Regul. 1991;2:261–270. doi: 10.1091/mbc.2.4.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- 43.Al-Kurdi R, Gulino-Debrac D, Martel L, Legrand JF, Renault A, Hewat E, Venien-Bryan C. A soluble VE-cadherin fragment forms 2D arrays of dimers upon binding to a lipid monolayer. J Mol Biol. 2004;337:881–892. doi: 10.1016/j.jmb.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 44.Lambert O, Taveau JC, Him JL, Al Kurdi R, Gulino-Debrac D, Brisson A. The basic framework of VE-cadherin junctions revealed by cryo-EM. J Mol Biol. 2005;346:1193–1196. doi: 10.1016/j.jmb.2004.12.053. [DOI] [PubMed] [Google Scholar]
- 45.Harrison OJ, Jin X, Hong S, Bahna F, Ahlsen G, Brasch J, Wu Y, Vendome J, Felsovalyi K, Hampton CM, Troyanovsky R, Ben-Shaul A, Frank J, Troyanovsky S, Shapiro L, Honig B. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. 2010 doi: 10.1016/j.str.2010.11.016. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geyer H, Geyer R, Odenthal-Schnittler M, Schnittler HJ. Characterization of human vascular endothelial cadherin glycans. Glycobiology. 1999;9:915–925. doi: 10.1093/glycob/9.9.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seiradake E, Harlos K, Sutton G, Aricescu AR, Jones EY. An extracellular steric seeding mechanism for Eph-ephrin signaling platform assembly. Nat Struct Mol Biol. 2010;17:398–402. doi: 10.1038/nsmb.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Himanen JP, Yermekbayeva L, Janes PW, Walker JR, Xu K, Atapattu L, Rajashankar KR, Mensinga A, Lackmann M, Nikolov DB, Dhe-Paganon S. Architecture of Eph receptor clusters. Proc Natl Acad Sci U S A. 2010;107:10860–10865. doi: 10.1073/pnas.1004148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uehara K. Distribution of adherens junction mediated by VE-cadherin complex in rat spleen sinus endothelial cells. Cell Tissue Res. 2006;323:417–424. doi: 10.1007/s00441-005-0064-5. [DOI] [PubMed] [Google Scholar]
- 50.Vendome J, Posy S, Jin X, Bahna F, Ahlsen G, Shapiro L, Honig B. Molecular design principles underlying affinity and specificity in classical cadherins. 2010 doi: 10.1038/nsmb.2051. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baumgartner W, Drenckhahn D. Plasmalemmal concentration and affinity of mouse vascular endothelial cadherin, VE-cadherin. Eur Biophys J. 2002;31:532–538. doi: 10.1007/s00249-002-0248-9. [DOI] [PubMed] [Google Scholar]
- 53.Shimoyama Y, Takeda H, Yoshihara S, Kitajima M, Hirohashi S. Biochemical characterization and functional analysis of two type II classic cadherins, cadherin-6 and -14, and comparison with E-cadherin. J Biol Chem. 1999;274:11987–11994. doi: 10.1074/jbc.274.17.11987. [DOI] [PubMed] [Google Scholar]
- 54.Nose A, Tsuji K, Takeichi M. Localization of specificity determining sites in cadherin cell adhesion molecules. Cell. 1990;61:147–155. doi: 10.1016/0092-8674(90)90222-z. [DOI] [PubMed] [Google Scholar]
- 55.Shan WS, Tanaka H, Phillips GR, Arndt K, Yoshida M, Colman DR, Shapiro L. Functional cis-heterodimers of N- and R-cadherins. J Cell Biol. 2000;148:579–590. doi: 10.1083/jcb.148.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Breviario F, Caveda L, Corada M, Martin-Padura I, Navarro P, Golay J, Introna M, Gulino D, Lampugnani MG, Dejana E. Functional properties of human vascular endothelial cadherin (7B4/cadherin-5), an endothelium-specific cadherin. Arterioscler Thromb Vasc Biol. 1995;15:1229–1239. doi: 10.1161/01.atv.15.8.1229. [DOI] [PubMed] [Google Scholar]
- 57.He Y, Jensen GJ, Bjorkman PJ. Cryo-electron tomography of homophilic adhesion mediated by the neural cell adhesion molecule L1. Structure. 2009;17:460–471. doi: 10.1016/j.str.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 59.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 60.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallographica Section D-Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.