Abstract
Genetic modifier loci influence the phenotypic expression of many Mendelian traits; insight into disease pathogenesis gained from their identification in animal disease models may impact the treatment of human multigenic disorders. We previously described an innate immune-driven model of spontaneous ulcerative colitis in T-bet−/−.Rag2−/− double-deficient mice that resembles human ulcerative colitis. On a BALB/c background, this disease is highly penetrant and results in the development of colorectal cancer. However, we observed that colitis in T-bet−/−.Rag2−/− mice on a C57BL/6 background was significantly less severe. Quantitative trait locus analysis using an N2 backcross strategy revealed a single major quantitative trait locus on chromosome 3 that mapped to the Cdcs1 (cytokine deficiency-induced colitis susceptibility-1) locus previously identified in the Il10−/− and Gnai2−/− colitis models. Congenic introduction of the susceptible Cdcs1 interval from C3H/He into the C57BL/6 background restored colitis severity. Bone marrow reconstitution experiments further mapped the effect of host genetics on disease severity to the hematopoietic compartment. There were distinct differences in the expression of several Cdcs1 genes in bone marrow-derived dendritic cells from Cdcs1 congenic mice. We conclude that the Cdcs1 locus controls colitis severity in T-bet−/−.Rag2−/− mice through innate immune cells.
Genetic predisposition plays an important role in the pathogenesis of inflammatory bowel disease (IBD) (1). Significant progress has been made toward identifying genomic variants that confer an increased risk for developing IBD. Recent meta-analyses of genome-wide association studies increased the number of confirmed disease-associated risk loci to 71 for Crohn Disease (CD) (2) and 47 for ulcerative colitis (UC) (3). Genome-wide association studies have highlighted the importance of several distinct biological pathways, most notably Nod2-mediated bacterial sensing and autophagy in CD, and IL-23/Stat3 signaling in both CD and UC (4, 5). However, the risk-conferring variant marked by a particular SNP is often not known. The gene reported to be associated with increased IBD risk represents in many cases a “best guess” and its function in the context of IBD may be uncertain (6).
Mouse models provide useful systems to genetically interrogate the pathways identified in human studies and to discover additional genes and pathways (7). Quantitative trait locus (QTL) analysis of differences in disease phenotype between two inbred mouse strains followed by positional cloning of the underlying gene is one such approach. Differential susceptibility between inbred mouse strains has been noted in several established murine IBD models. QTL mapping studies performed in two spontaneous IBD models (Il10−/− and Gnai2−/−) identified the same Cdcs1 (cytokine deficienty-induced colitis susceptibility-1) locus on mouse chromosome 3 as the major QTL (8–10). The identity of the susceptibility-controlling genetic variants within this region and the cell type through which they exert their effects are still unknown.
We recently described that T-bet−/−.Rag2−/− mice on a BALB/c background in our colony develop spontaneous UC-like disease (TRUC). This disease is driven by colitogenic intestinal microbiota, mediated by TNF, and results in spontaneous development of colorectal cancer (11–13). We report here that colitis in T-bet−/−.Rag2−/− mice on a C57BL/6 background (B6.TRUC) is significantly less severe compared with TRUC animals on a BALB/c background (BALB/c.TRUC). QTL analysis performed in an N2 backcross mapped Cdcs1 as the major susceptibility locus in the TRUC model. This finding was confirmed in congenic mice generated by replacing the Cdcs1 interval in B6.TRUC mice with the susceptible Cdcs1 locus from C3H/He. We show further that Cdcs1 controls disease severity in the TRUC model through hematopoietically derived innate immune cells.
Results and Discussion
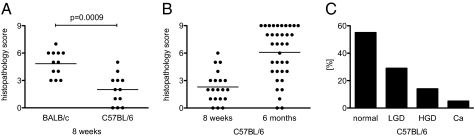
To make use of the growing library of genetic mutant mouse strains on a C57BL/6 background, we examined the colitis phenotype in our B6.TRUC colony. We found that B6.TRUC mice had significantly milder disease than BALB/c.TRUC animals. In contrast to BALB/c.TRUC mice with 100% penetrance of colitis at 8 wk of age, both penetrance and severity were significantly lower in B6.TRUC animals (Fig. 1A) (mean colitis score 4.8 vs. 2.0 in this cohort of animals, P = 0.0009). Disease progressed in severity over time and by 6 mo of age the majority of B6.TRUC animals had developed colitis. However, the distribution of colitis scores in 6-mo-old B6.TRUC mice was broad (range 0–9) (Fig. 1B), in contrast to what is observed in BALB/c.TRUC mice. The incidence of dysplasia and adenocarcinoma was also reduced. In a cohort of 36 B6.TRUC animals analyzed at 6 mo of age, 56% had no evidence of dysplasia, 29% had low-grade dysplasia, 15% high-grade dysplasia, and there were two cases (6%) of adenocarcinoma (Fig. 1C). This finding compares with <4% of animals without dysplasia, 89% with high-grade dysplasia, and 42% with adenocarcinoma in 6-mo-old BALB/c.TRUC mice (12).
Fig. 1.
Colitis severity in TRUC mice is influenced by genetic background. (A) Cohorts of B6.TRUC and BALB/c.TRUC mice were killed at 8 wk of age (n = 12, the horizontal bar represents the group mean). Standard H&E sections of the entire colon were scored in a blinded fashion. (B) Colitis scores in B6.TRUC mice at 8 wk (n = 20) and 6 mo of age (n = 36). (C) The colon sections of the same cohort of 6-mo-old animals were evaluated for evidence of dysplasia. LGD, low-grade dysplasia; HGD, high-grade dysplasia without carcinoma; Ca, carcinoma.
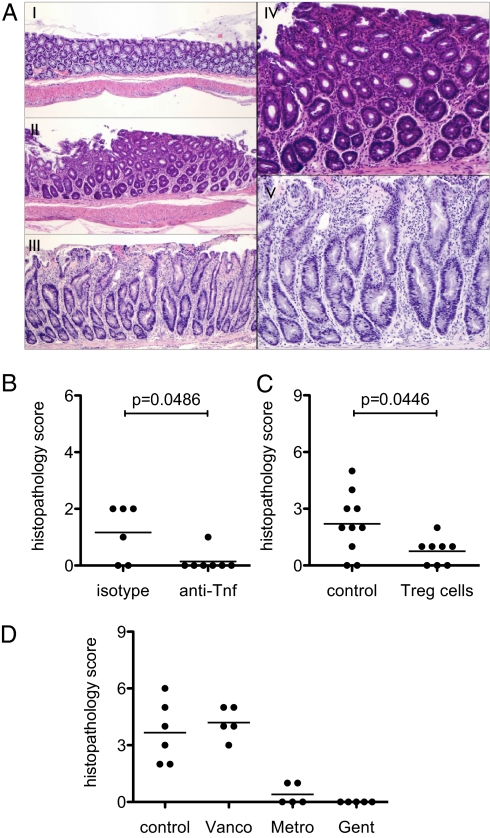
Despite the difference in severity, colitis in B6.TRUC mice appeared to be qualitatively the same as in BALB/c.TRUC animals. The mucosal inflammatory changes consisted of a mixed infiltrate of mononuclear and polymorphonuclear cells, crypt regeneration, variable architectural distortion, and injury ranging from crypt dropout to surface erosions. The changes were usually limited to the distal third of the colon, but could also focally spread to more proximal regions (Fig. 2A). Dysplasia, when present, was characterized by nuclear atypia, stratification, glandular crowding, and failed surface maturation. Systemic administration of anti-TNF antibodies or adoptive transfer of CD4+CD25+ regulatory T cells prevented the development of colitis (Fig. 2 B and C), and the disease was ameliorated by treating the animals with oral Metronidazole or Gentamicin, but not Vancomycin, as reported previously for BALB/c.TRUC mice (Fig. 2D) (11, 13).
Fig. 2.
Colitis in B6.TRUC mice is less severe but qualitatively similar to BALB/c.TRUC disease. (A) Representative colon histology of 8 wk-old B6.TRUC mice with score 0 (normal, panel I), score 3 (panels II and IV), and a rare animal with score 7 (panels III and V). H&E, panel I to III 40×, panels IV and V 100×. The higher magnification views show mild crypt regeneration, lamina propria mononuclear and neutrophilic inflammation (panel IV, score 3), and marked crypt regeneration with distortion, surface erosion, and mixed inflammatory infiltrate (panel V, score 7). (B) B6.TRUC animals were treated with four weekly intraperitoneal injections of a monoclonal hamster anti-TNF antibody or isotype control (n = 6–7) starting at weaning. (C) CD4+CD62L+CD25+ Treg cells were isolated from wild-type B6 donors and 75,000 cells were injected intraperitoneally at weaning (n = 8); control animals received no cells. (D) B6.TRUC mice (n = 5 per group) were treated with Vancomycin, Metronidazole, or Gentamicin in the drinking water beginning at weaning. Colon histopathology was analyzed at 8 wk of age for experiments (B–D).
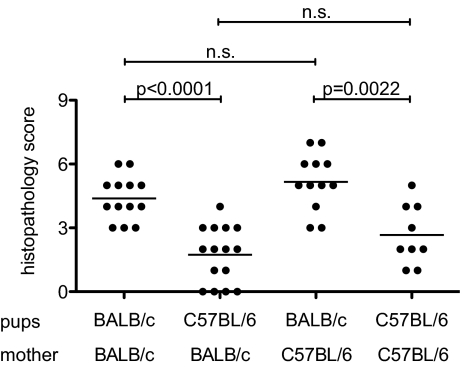
An important feature of TRUC disease is its dependence on a colitogenic intestinal flora derived from the mother and potentially other maternal factors. BALB/c.TRUC mice raised by a BALB/c wild-type mother do not display colitis at 8 wk of age (12). To evaluate whether maternal factors contributed to the observed difference in colitis severity between BALB/c and B6.TRUC animals, we performed a reciprocal cross-fostering experiment. BALB/c and B6.TRUC litters were removed from their mothers within 24 h after birth and raised by a foster mother of opposite genetic background. Cross-fostered animals, as well as BALB/c and B6.TRUC mice raised by their natural mothers, were killed at 8 wk of age and colon histology was analyzed (Fig. 3). Colitis scores were significantly higher in BALB/c.TRUC mice than in B6.TRUC animals, regardless of the genetic background of the mother (i.e., colitis severity correlated with the genomic background of the pups and not the genomic background of the mother). Effects of genetic background on intestinal microbiota have been previously described (14) and are likely to exist between BALB/c and B6. Nevertheless, the a priori colitogenicity of the microflora that a BALB/c or B6.TRUC mother transmits to the pups appears to be similar.
Fig. 3.
Reciprocal cross-fostering of BALB/c.TRUC and B6.TRUC mice demonstrates correlation between disease severity and genetic background of the offspring but not the mother. Newborn litters of BALB/c.TRUC and B6.TRUC mothers were exchanged within 24 h after birth. BALB/c.TRUC and B6.TRUC mice raised by their natural mothers served as controls. Colitis scores at 8 wk of age were pooled from three litters per experimental group.
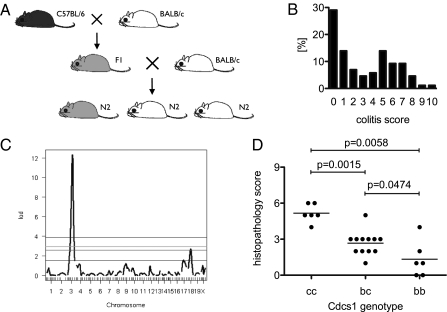
To understand which genetic factors control disease severity in the BALB/c vs. B6.TRUC animals, we performed a QTL analysis using the histopathology score as the phenotypic variable. F1 hybrids from BALB/c.TRUC × B6.TRUC matings were backcrossed to BALB/c.TRUC mice to generate a cohort of N2 backcross animals (Fig. 4A). A total of 86 N2 mice were analyzed at 8 wk of age. Colitis scores showed a bimodal distribution (range 0–10), with 51.2% of animals having a score <3 and 48.8% a score ≥3 (Fig. 4B). Of the N2 animals, 29.1% had no detectable colitis. All mice were genotyped for a panel of 99 markers and QTL analysis was performed using the colitis score either as a discrete numeric variable or as a binary variable (<3 colitis absent; ≥3 colitis present). The results from both approaches agreed well and revealed a strong QTL on chromosome 3 and a minor QTL on chromosome 18 (Fig. 4C). The QTL on chromosome 3 localized to 58 cM, with a LOD score of 12.3 for the normal model and 56 cM with a LOD score of 8.86 for the binary model. These positions convert to 126.4 Mbp and 129.0 Mbp, respectively, in National Center for Biotechnology Information build 37 (cgd.jax.org/mousemapconverter) (15). The minor QTL on chromosome 18 localized to 33.5 cM with a LOD score of 2.7 for the normal model and at 29.4 cM with a LOD score of 3.8 for the binary model. A third possible peak on chromosome 17 did not reach statistical significance with the relatively small number of animals examined.
Fig. 4.
The Cdcs1 locus on chromosome 3 is the major QTL in TRUC disease. (A) Breeding strategy. An N2 backcross generation was generated by mating (BALB/c × B6)F1 TRUC animals with BALB/c.TRUC males. (B) Frequency distribution of colitis scores in the N2 cohort (n = 86). (C) Each animal was genotyped for a panel of 99 genome-spanning markers and QTL analysis was performed. Results derived using a normal statistical model are depicted. The coordinates of the strong peak on chromosome 3 (LOD score 12.3) were 58 cM = 126.7 Mbp, mapping to the Cdcs1 locus. (D) BCR3 mice congenic for the colitogenic Cdcs1c interval from C3H/HeJBir on a B6 background were crossed into the B6.TRUC line. BALB/c and C3H/He are haploidentical across Cdcs1 (Cdcs1c), but different from B6 (Cdcs1b) (see also Fig. 1). TRUC.Cdcs1b/c × TRUC.Cdcs1b/c matings produced TRUC.Cdcs1b/b, TRUC.Cdcs1b/c, and TRUC.Cdcs1c/c offspring. Four litters with n = 24 mice total were scored for colitis severity at 8 wk of age.
The coordinates of the strong QTL on chromosome 3 mapped to the Cdcs1 locus. Cdcs1 was originally identified as the major QTL in an F2 intercross of colitis-susceptible C3H/HeJBir.Il10−/− and -resistant B6.Il10−/− mice (8), which was replicated in a second study in C3H/HeJBir.Il10−/− and B6.Il10−/− mice using an N2 backcross strategy (10). Cdcs1 was independently identified in a QTL analysis in the Gnai2−/− colitis model (9) also comparing susceptible C3H/HeN and resistant B6 backgrounds. More recently, analysis of a panel of reciprocal congenic Il10−/− lines (derived from C3H/HeJBir and B6 animals) defined three Cdcs1 subloci that independently affect susceptibility to intestinal inflammation (16). The most proximal sublocus, Cdcs1.1 (located between 88.1 and 108.8 Mbp), controls cecal inflammation and the other two subloci, Cdcs1.2 (120–123 Mbp) and Cdcs1.3 (125.6–128 Mbp), affect colitis severity. Only the latter two subloci are relevant for the TRUC model, which lacks cecal inflammation (13). We compared the genomic sequence of B6, C3H/He, and BALB/c mice for the 120 to 128 Mbp region on chromosome 3 using publicly available data (phenome.jax.org). The “SNP landscape” (Fig. S1) demonstrates that the colitis susceptible BALB/c and C3H/He strains are haploidentical across this region, while the sequence of the colitis-resistant B6 reference strain differs significantly. To confirm the QTL association, we crossed the BCR3 congenic line containing the susceptible Cdcs1 interval from C3H/HeJBir on a B6 background (16) with B6.TRUC females to generate TRUC.Cdcs1 congenic animals. TRUC.Cdcsb/c mice (heterozygous for the Cdcs1b locus from B6 and the Cdcs1c locus from C3H/HeJBir) were then mated to produce litters with the three possible genotypes Cdcs1c/c, Cdcs1b/c, and Cdcs1b/b. The histopathological analysis of colon tissue at 8 wk of age showed that TRUC.Cdcs1c/c homozygous animals had significantly more severe disease (mean colitis score 5.2) than heterozygous TRUC.Cdcs1b/c mice (mean colitis score 2.7, P = 0.0015) or littermates homozygous for the B6-derived Cdcs1b locus (mean colitis score 1.3, P = 0.0058), proving that Cdcs1 controls colitis severity in the TRUC model. Disease in the heterozygous TRUC.Cdcs1b/c animals was also slightly more severe compared with TRUC.Cdcs1b/b littermates (P = 0.0474).
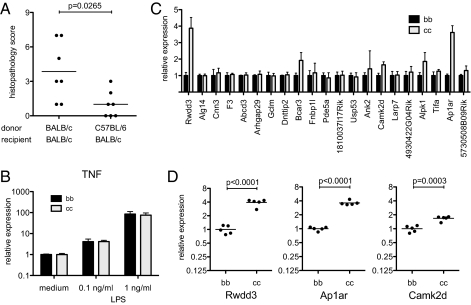
To determine how the Cdcs1 locus modified intestinal inflammation, we generated bone marrow chimeras transferring BALB/c and B6.TRUC bone marrow into lethally irradiated adult BALB/c.TRUC animals. Colitis severity was analyzed at 20 wk of age to allow for recovery of the colitis phenotype following bone marrow engraftment. Disease severity in recipients of BALB/c.TRUC bone marrow was significantly higher than in recipients of B6.TRUC bone marrow (mean colitis score 3.9 versus 1.0, P = 0.0265) (Fig. 5A). These data map the effect of host genetics on colitis severity to the hematopoietic compartment, although we have yet to establish whether there may also be a contribution from radioresistant intestinal cells. Given the central role of TNF produced by dendritic cells in the TRUC model (13), we compared the production of TNF by bone marrow-derived dendritic cells from Cdcs1 congenic animals. There was no difference in the induction of TNF mRNA in response to LPS stimulation (Fig. 5B). We also analyzed the expression of the genes located in the Cdcs1.2 and Cdcs1.3 subloci in bone marrow-derived dendritic cells by real-time quantitative PCR. Combined, these two regions contain 26 annotated genes and 4 expressed Riken sequences. Nine of the 30 genes were not expressed in dendritic cells. Messenger RNA levels for most of the remaining 21 genes were the same in Cdcs1c/c and Cdcs1b/b dendritic cells (Fig. 5C). However, there were a few notable exceptions: Rwdd3 in Cdcs1.2, and Ap1ar and Camk2d in Cdcs1.3 were more strongly expressed in cells from Cdcs1c/c animals, and these differences (Rwdd3 3.9-fold, P < 0.0001; Ap1ar 3.6-fold, P < 0.0001; Camk2d 1.7-fold, P = 0.009) remained statistically significant after Bonferroni correction for multiple comparisons. Further exploration of the functional outcome of sequence polymorphisms (16) and differential expression of the Cdcs1 locus genes is in progress.
Fig. 5.
Genetic background controls colitis severity through the hematopoietic compartment. (A) Six- to 7-wk-old female BALB/c.TRUC mice (n = 7) were lethally irradiated, followed by intravenous injection of 2 × 106 bone marrow cells (BM) from either BALB/c.TRUC or B6.TRUC donors. Animals were killed at 20 wk of age and colitis severity was scored. Data in this graph are representative of two experiments performed. (B) Dendritic cells were generated in vitro from bone marrow of Cdcs1b/b and Cdcs1c/c mice. Day 6 cells were stimulated with 0, 0.1, or 1 ng/mL LPS for 6 h. CD11c+ cells were purified using magnetic beads and analyzed by qPCR. TNF mRNA levels were normalized to Hprt. Data represent mean and SD from two experiments performed. (C) Unstimulated CD11c+ bone marrow-derived dendritic cells were analyzed by qPCR for expression of Cdcs1 genes. Data were normalized to Hprt and then scaled by dividing over the mean value of the Cdcs1b/b samples (mean and SD of three to five biological samples). (D) Individual data points and group mean for selected genes, as in C.
In summary, we have shown that the Cdcs1 locus on mouse chromosome 3 is the major QTL for colitis severity in the TRUC model of IBD. We were able to map a disease-modulating function of the Cdcs1 locus to the hematopoietic compartment and (by implication) cells of the innate immune system, as TRUC disease, in contrast to the Il10−/− and Gnai2−/− colitis models, is entirely innate immune-mediated. Consistent with that conclusion, we found locus-intrinsic differences of Cdcs1 gene expression in bone marrow-derived dendritic cells serving here as a model innate immune cell. Future research will focus on Cdcs1 effects on the function of additional innate immune-cell subsets, including the recently described subset of innate lymphoid helper cells (17). The human syntenic region of the 120 to 128 Mbp locus on mouse chromosome 3 is located in two parts on human chromosome 1p21-22 and 4q25-26. None of the published CD or UC-associated SNPs map to these regions (2, 3). However, the fact that Cdcs1 plays a role in three rather different IBD models suggests that the biological pathways controlled by Cdcs1 affect inflammatory responses in the intestine in a meaningful way, and that the identification of the disease susceptibility genes in Cdcs1 will have important implications for understanding the pathogenesis of human IBD.
Materials and Methods
Mice.
Cg-Tbx21tm1GlmRag2tm1Fwa (T-bet−/−.Rag2−/−, TRUC) mice have been backcrossed to a BALB/c or C57BL/6 (B6) background for at least 10 generations. BCR3 is the informal designation for an Il10-deficient C57BL/6J congenic stock carrying a C3H/HeJBir-derived chromosome 3 segment between the markers D3Mit11 and D3Mit19 (B6.Cg-Il10tm1CgnCdcs1(D3Mit11-D3Mit19)/JZtm) (16). This segment contains the colitogenic Cdcs1 QTL, herein denoted Cdcs1c. Male BCR3 mice were backcrossed with B6.TRUC females carrying the Cdcs1 resistance QTL (designated Cdcs1b), selecting for the congenic interval and against the targeted Il10 gene to produce Il10 intact TRUC.Cdcs1b/c heterozygous mice. Mice were genotyped for their Cdcs1 genotype using the markers D3Mit316 and D3Mit319 (www.informatics.jax.org). Mice with discordant results indicative of a meiotic recombination event were discarded. All mice were housed in microisolator cages in a specific pathogen-free animal facility at the Harvard School of Public Health and studies were performed according to institutional and National Institutes of Health guidelines for humane animal use. Sulfatrim (Sulfamethoxazole 1 g/L + Trimethoprim 0.2 g/L; Hitech Pharmacal) was added to the drinking water. Mice were weaned between 3 and 4 wk of age.
Cross-Fostering.
Pregnant BALB/c and B6.TRUC females were monitored daily for delivery. Mothers and newborn litters were reciprocally exchanged within 24 h of birth.
Histology.
Animals were killed at the stated age, the colon was isolated in toto, fixed in 4% paraformaldehyde, and embedded in paraffin. Standard H&E-stained sections were examined and scored by an experienced Pathologist (J.G.) in a blinded fashion. The parameters mononuclear cell infiltration, polymorphonuclear cell infiltration, epithelial hyperplasia, and epithelial injury were scored as absent (0), mild (1), moderate (2), or severe (3), giving a total score of 0 to 12 (13).
QTL Analysis.
F1 mice derived from BALB/c × B6.TRUC matings were backcrossed to BALB/c.TRUC males to generate N2 mice. A cohort of 86 animals was killed at 8 wk of age and colitis was scored as described. Tail DNA was prepared using the DNeasy kit (Qiagen). Genotyping and QTL analysis were performed at The Jackson Laboratory.
Antibiotic Treatment.
Starting at weaning, mice were placed continuously on drinking water supplemented with Vancomycin (0.5 g/L; Sigma), Metronidazole (1 g/L; Sigma), or Gentamicin (1 g/L; Cell Gro).
Anti-TNF Treatment.
Mice received a total of four weekly injections (15 mg/kg) of either hamster IgG1-anti-mouse TNF (clone TN3-19.12) or control antibody (hamster IgG1 anti-GST) (both Leinco Technologies) starting at weaning.
Adoptive Transfer of Treg Cells.
A single-cell suspension prepared from lymph nodes and spleens of wild-type B6 animals was, after lysis of red blood cells, stained with anti-CD25 PE, anti-CD62L FITC, and anti-CD4 APC (eBioscience), followed by incubation with anti-PE beads (Miltenyi) and enrichment of CD25+ cells using an autoMACS system (Miltenyi). CD4+CD62L+CD25+ Treg cells were FACS sorted to >95% purity on a FACS Aria (BD Biosciences) in the Harvard Institute of Medicine Cell Sorting Facility.
Bone Marrow Chimeras.
Six- to 7-wk-old recipient mice were lethally irradiated with 800 rad. A single-cell suspension was prepared from the bone marrow flushed from femurs and tibias of donor mice. Recipient mice were injected in the lateral tail vein with 2 × 106 donor cells in PBS. Mice were killed at 20 wk of age. Chimerism in the B6.TRUC → BALB/c.TRUC animals determined by FACS analysis of peripheral blood using H2K specific antibodies (BD Biosciences) was > 95% in all animals.
Generation and Stimulation of Bone Marrow Dendritic Cells.
Bone marrow dendritic cells were prepared as previously described (13) by incubation of lineage-depleted bone marrow cells in medium conditioned with supernatant from GM-CSF producing J558L cells for 6 d. On day 6, the medium was replaced with medium containing 0, 0.1, or 1 ng/mL LPS from Salmonella enterica typhimurium (Sigma). Nonadherent cells were harvested after 6 h of stimulation. CD11c+ cells were purified using magnetic beads (Miltenyi) and total RNA was prepared with Qiazol reagent (Qiagen).
Real-Time Quantitative PCR.
Total RNA was reverse-transcribed with the High Capacity cDNA RT Kit (Applied Biosystems) and random primers. Real-time quantitative PCR was performed on a MX3005 cycler (Stratagen) using the SYBR Green method. Data were normalized to Hprt. Primers sequences are provided in Table S1.
Statistical Analysis.
Colitis scores were compared by Mann Whitney U test. Gene-expression data were compared by unpaired t test. All statistical analysis and graphing was done with Prism 5 (Graphpad Software).
Supplementary Material
Acknowledgments
We thank Jacobo Ramirez for excellent animal care, Kirsten Sigrist for help with generating Cdcs1 congenic animals, and Dorothy Zhang for preparing histology slides; and Edward Leiter at The Jackson Laboratory for help with designing the quantitative trait locus study and critical reading of the manuscript. This work was supported by National Institutes of Health Grants CA112663 (to L.H.G.), 1P50CA127003-2 and K08AI078942 (to W.S.G.), a grant from Danone Research (to L.H.G.), a Harvard Digestive Diseases Center (National Institutes of Health Grant P30 DK34845) pilot award (to J.E.), and a Clinical Immunology Society/Talecris fellowship (to J.E.).
Footnotes
Conflict of interest statement: L.H.G. has equity in and is on the corporate board of Bristol-Myers Squibb.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104234108/-/DCSupplemental.
References
- 1.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 5.Budarf ML, Labbé C, David G, Rioux JD. GWA studies: Rewriting the story of IBD. Trends Genet. 2009;25:137–146. doi: 10.1016/j.tig.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Rosenstiel P, Sina C, Franke A, Schreiber S. Towards a molecular risk map—Recent advances on the etiology of inflammatory bowel disease. Semin Immunol. 2009;21:334–345. doi: 10.1016/j.smim.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Beutler B, Du X, Xia Y. Precis on forward genetics in mice. Nat Immunol. 2007;8:659–664. doi: 10.1038/ni0707-659. [DOI] [PubMed] [Google Scholar]
- 8.Farmer MA, et al. A major quantitative trait locus on chromosome 3 controls colitis severity in IL-10–deficient mice. Proc Natl Acad Sci USA. 2001;98:13820–13825. doi: 10.1073/pnas.241258698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borm ME, et al. A major quantitative trait locus on mouse chromosome 3 is involved in disease susceptibility in different colitis models. Gastroenterology. 2005;128:74–85. doi: 10.1053/j.gastro.2004.10.044. [DOI] [PubMed] [Google Scholar]
- 10.Mähler M, et al. Genetics of colitis susceptibility in IL-10–deficient mice: Backcross versus F2 results contrasted by principal component analysis. Genomics. 2002;80:274–282. doi: 10.1006/geno.2002.6840. [DOI] [PubMed] [Google Scholar]
- 11.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrett WS, et al. Colitis-associated colorectal cancer driven by T-bet deficiency in dendritic cells. Cancer Cell. 2009;16:208–219. doi: 10.1016/j.ccr.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benson AK, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ackert-Bicknell CL, et al. Mouse BMD quantitative trait loci show improved concordance with human genome-wide association loci when recalculated on a new, common mouse genetic map. J Bone Miner Res. 2010;25:1808–1820. doi: 10.1002/jbmr.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bleich A, et al. Cdcs1 a major colitis susceptibility locus in mice; subcongenic analysis reveals genetic complexity. Inflamm Bowel Dis. 2010;16:765–775. doi: 10.1002/ibd.21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buonocore S, et al. Innate lymphoid cells drive interleukin-23–dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.