Abstract
Affinity reagents that bind to specific molecular targets are an essential tool for both diagnostics and targeted therapeutics. There is a particular need for advanced technologies for the generation of reagents that specifically target cell-surface markers, because transmembrane proteins are notoriously difficult to express in recombinant form. We have previously shown that microfluidics offers many advantages for generating affinity reagents against purified protein targets, and we have now significantly extended this approach to achieve successful in vitro selection of T7 phage-displayed peptides that recognize markers expressed on live, adherent cells within a microfluidic channel. As a model, we have targeted neuropilin-1 (NRP-1), a membrane-bound receptor expressed at the surface of human prostate carcinoma cells that plays central roles in angiogenesis, cell migration, and invasion. We show that, compared to conventional biopanning methods, microfluidic selection enables more efficient discovery of peptides with higher affinity and specificity by providing controllable and reproducible means for applying stringent selection conditions against minimal amounts of target cells without loss. Using our microfluidic system, we isolate peptide sequences with superior binding affinity and specificity relative to the well known NRP-1-binding RPARPAR peptide. As such microfluidic systems can be used with a wide range of biocombinatorial libraries and tissue types, we believe that our method represents an effective approach toward efficient biomarker discovery from patient samples.
Keywords: C-end rule (CendR) peptide, high stringency selection, Phage Display, tumor markers
Since the invention of monoclonal antibodies (1, 2), affinity reagents that specifically bind to molecular targets have become a cornerstone of modern biotechnology (3, 4). In the postgenomic era, there is a pressing need to accelerate the pace of discovery for such reagents, especially as researchers are confronted with an expanding number of targets with posttranslational modifications (5). In vitro evolutionary screening techniques such as phage display (6) offer a powerful alternative for generating polypeptide-based affinity reagents that overcome the limitations inherent to immunoglobulins, including the challenges of uptake and distribution within cells (7–9). This technique has been successfully used to identify affinity reagents for small molecules (10), nucleic acids (11), proteins (12, 13), proteoliposomes (14, 15), and inorganic materials (16). Phage display has proven powerful for use on whole cells (17, 18) and tissues (19, 20) because it allows targeting of transmembrane proteins in their native orientation and conformation with appropriate posttranslational modifications, and does not require a priori knowledge of the targets or their concentrations (21).
The selection of phage-displayed peptides that target a cell-surface marker is typically performed through a biopanning process, either in cell suspension (19, 20, 22) or on a solid support (23, 24). This biopanning process involves four major steps: preparation of phage-displayed peptide libraries, capture of specific phage that bind to the target, washing of low-affinity or nonspecific phage from the cell surface, and recovery of enriched target binders for the next round of selection (25). Though effective, conventional cell-based biopanning approaches suffer from several limitations (26). Each round of biopanning requires a relatively large number of cells (105–107), making the technique difficult to apply to small cell populations harvested from organs and tissues. In addition, fixed-volume manual washing is inefficient and laborious, often yielding irreproducible results and ineffective removal of nonspecifically or weakly bound phage binders. Cells and potential ligands bound to them are often lost during these repeated washes, especially when working with small amount of cells. Thus, there is a need for an alternative selection system capable of handling small numbers of cells without loss, and which can be used to reproducibly apply highly stringent selection conditions and efficient separation technology for the rapid enrichment of peptides that bind to transmembrane proteins with high affinity and specificity.
Towards such a system, we report the successful directed evolution of phage libraries that targets live, adherent cell surfaces using microfluidics technology. The ability of this system to directly target membrane-bound proteins on cell surfaces is significant because most of these proteins are notoriously difficult to produce using recombinant technology. We show that this microfluidic phage selection (MiPS) system offers significant advantages compared to conventional biopanning methods. First, due to the fact that the selection is performed within a microchannel, smaller numbers of target cells are required, which allows us to impose highly stringent mass-action selection pressure (e.g., high molar ratios between the library and target cells) (27), yielding peptides with higher affinity. Second, control of the flow rate of fluids within the microchannel provides a continuous and reproducible means for efficiently removing weakly- or nonspecifically-bound phage (28), resulting in low background binding with minimal cell loss. We demonstrate that continuous washing leads to more efficient enrichment of phage displaying high-affinity peptides to the targeted cell-surface marker. Finally, we have integrated all major components of the selection process—incubation, washing, cell lysis, and lysate collection—within the MiPS device, thereby reducing the risk of contamination and enabling full automation. Importantly, using the MiPS system, we are able to discover unique peptide sequences to an important cancer biomarker neuropilin-1 (NRP-1) with superior affinity and specificity, than the best sequences discovered through conventional biopanning method.
Results and Discussion
Assembly of the Microfluidic Phage Selection Chip.
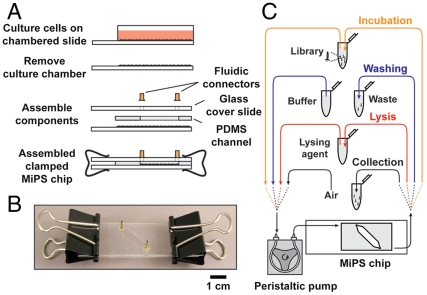
Microfluidic phage selection is performed within the MiPS chip, and the device fabrication scheme is briefly summarized in Fig. 1A. First, we coated a NUNC Lab-Tek culture slide chamber with 10 μg/mL of fibronectin for 12 h at 4 °C to promote cell adhesion. We then seeded the chamber with ∼104 PPC-1 human prostate carcinoma cells in 2 mL of culture medium. PPC-1 cells were selected as a model because they express high levels of NRP-1, a well characterized receptor that binds and internalizes peptides with C-terminal arginine residues, typically within a consensus context of R/KXXR/K (the C-end Rule or CendR motif) (29–31). As a negative control, we used phage expressing hepta-glycine (G7), which exhibits negligible binding to PPC-1 cells (28). Upon reaching 90% confluency, the cells were washed twice with phosphate-buffered saline (PBS). To block exposed surfaces, we incubated the slide with UV-treated, noninfectious G7 phage that can no longer replicate but retain their binding activity (2 × 109 pfu total in 2 mL PBS at 4 °C for 1 h). After aspirating unbound blocking phage, we peeled off the walls of the culture slide chamber to accommodate a thin Polydimethylsiloxane (PDMS) layer, which establishes the microchannel, and a glass layer containing the inlet and outlet for fluidic connections. Finally, the three layers were clamped together by clipping the edges of the device (Fig. 1B).
Fig. 1.
MiPS device for the selection of phage-displayed peptides targeting cell-surface markers. (A) The device is fabricated from three layers consisting of glass (top, 1 mm), PDMS (middle, 250 μm), and culture glass (1 mm). The volume of the chamber containing the target cells is ∼25 μL. (B) Photograph of the completed MiPS device. (C) Experimental schematic for phage selection against adherent cells using the assembled MiPS device. All major steps of the selection—library incubation, washing, cell lysis, and lysate collection—are performed within the MiPS chip.
Characterization of Phage Selection in the MiPS Chip.
To ensure that loss of adherent PPC-1 cells was minimal during the selection process, we measured cell recovery by counting the cells before and after the library incubation and washing steps on four different MiPS devices at 90% cell confluency. We first quantified the initial number of cells on a single slide by using 100 μL of cell dissociation buffer to detach the cells from the slide surface and then counting them via flow cytometry (Accuri C6, Accuri Cytometers Inc.). After applying binding and washing conditions to another four slides, the cells were detached, collected, and counted as above. We found that our MiPS selection process results in minimal cell loss, with an average of 93.9% cell recovery for 104 cells (see SI Text, Fig. S1). In contrast, conventional cell suspension biopanning resulted in cell recoveries of 71.7%, 68.1%, and 56.1% for amounts of 106, 105, and 104 cells, respectively, after four fixed-volume centrifuge washes (Fig. S1). These results show that cells are often lost during these repeated centrifuge washes, especially when working with smaller amounts of cells.
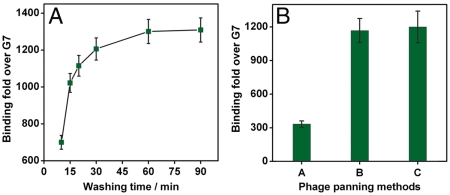
Prior to de novo selection, we performed a trial experiment using the configuration shown in Fig. 1C to screen for phage displaying the RPARPAR peptide sequence, a motif that is known to bind NRP-1 protein expressed on PPC-1 cells. We loaded 2 mL of RPARPAR or G7 phage (2 × 108 pfu/mL in PBS) into the MiPS chip with a peristaltic pump connected via Teflon tubing, and recirculated each sample at a flow rate of 1 mL/ min at 4 °C for 3 h (Fig. 1C, incubation step). We then used the pump to deliver the wash buffer (DMEM supplemented with 1% BSA) at 1 mL/ min to remove nonspecifically- or weakly-bound phage from cell and device surfaces (Fig. 1C, washing step). We lysed the phage-bound cells by pumping 1 mL of lysogeny broth (LB) containing 1% Nonidet P-40 (NP-40) (NP/LB) at 6 mL/ min on ice for 30 min (Fig. 1C, lysis step), and then eluted and quantified the bound phage in the lysate via a titering assay (Fig. 1C, collection step). We observed that relative RPARPAR vs. G7 phage binding increased monotonically with washing time, up to a plateau of ∼1,300-fold enrichment after 60 min of washing (Fig. 2A), suggesting that extended continuous washing favors the selection of peptides with better off-rates.
Fig. 2.
Dependence of washing stringency on selection of phage clones. (A) Binding of phage displaying the RPARPAR sequence relative to G7 control phage as a function of washing time in the MiPS channel. Relative binding of RPARPAR phage increases monotonically with washing time until a plateau (∼1,300-fold enrichment) is reached after 60 min of continuous washing. (B) Enrichment of RPARPAR phage over G7 control phage using three different biopanning methods. Conventional, fixed-volume washing (Method A) yielded a maximum of ∼1,300-fold enrichment, whereas continuous washing in the MiPS device (Methods B and C) resulted in more than 1,000-fold enrichment.
Controlling Washing Efficiency.
Next, we compared the washing efficiency of the fixed-volume washing approach used in conventional biopanning vs. the continuous washing used in the MiPS chip. We performed three different biopanning experiments with 2 × 104 adherent PPC-1 cells incubated with 2 × 108 pfu of RPARPAR or G7 phage. In Method A, incubation was performed in a culture chamber at 4 °C for 3 h, followed by 12 repetitions of a manual, fixed-volume washing step as described in standard protocols (21, 32). Interestingly, application of more washing steps in Method A only improved selection performance to a limited extent; after performing experiments with 2, 4, 6, 9, 12, and 15 washes, we found that the enrichment reached a plateau after 12 washes (Fig. S2). Method B incorporated the same incubation approach, followed by continuous washing in the MiPS chip at 1 mL/ min for 90 min. In Method C, we performed incubation by recirculating the phage in the MiPS chip at 4 °C for 3 h, followed by continuous washing at 1 mL/ min for 90 min. Experimental results showed that Method A yielded a maximum of 300-fold enrichment of RPARPAR phage relative to the G7 phage, whereas Methods B and C yielded ∼1,000-fold enrichment (Fig. 2B). Thus, continuous microfluidic washing appears to be significantly more effective at removing nonspecific phage, presumably by eliminating rebinding events (33–36). This finding is further supported by the work of Yuan et al. which used flow-based phage selection in an surface plasmon resonance (SPR) instrument (37). Since it is well known that shear stress can induce changes in protein expression levels in some cell types, we measured the expression level of NRP-1 on the PPC-1 cells in the MiPS system under nominal operating conditions. Specifically, we labeled the PPC-1 cells with phycoerythrin (PE) conjugated anti-NRP-1 monocolonal antibodies (Miltenyi Biotec) before and after the incubation (3 h, 1 mL/ min) and washing (1.5 h, 1 mL/ min) steps in the MiPS device. Then, the NRP-1 expressions were quantified by measuring the PE intensities of the PPC-1 cells in a flow cytometer. We found that the expression level of NRP-1 is not affected by the incubation and washing process under nominal operating conditions [3 Dyne per square centimeter (dyn/cm2) shear stress] within our device (Fig. S3). We think that the NRP-1 expression was not affected due to the following reasons: First, the shear stress in our microfluidic channel is ∼0.3 Pa (3 dyn/cm2), which is significantly less than that experienced in blood vessels (10–40 dyn/cm2). Second, all of our selection steps were done at 4 °C at which the cells’ response to stimuli is greatly decreased (38–40). However, we recognize other cell types (e.g., endothelial cells) may be more sensitive to changes in shear stress (38) and if selections were to be performed on them, the device operating conditions (i.e., flow rates) and size of device (i.e., the channel width) should be carefully adjusted and tested prior to the selection.
De Novo Selection of a Random T7 Phage Library.
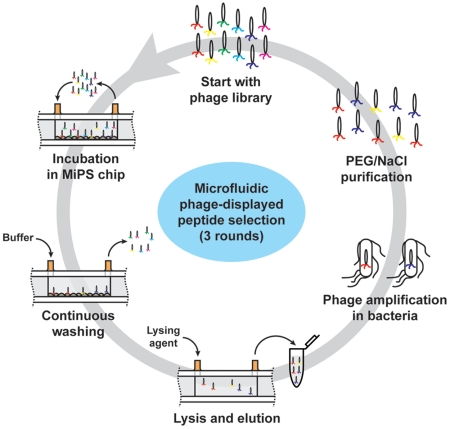
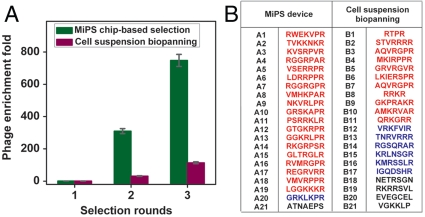
Based on these experimental conditions, we performed de novo selection of a T7 phage library expressing random, linear 7-residue (X7) peptides (diversity approximately 5 × 108) against PPC-1 cells using both conventional biopanning and the MiPS platform (Fig. 3). In both cases, we performed three rounds of selection with a relatively small number of PPC-1 cells (2 × 104). After each round of selection, the recovered phage were amplified in Escherichia coli BLT5403 cells at 37 °C for 2 h, followed by phage precipitation with a polyethylene glycol (PEG)/NaCl solution and purification by CsCl gradient ultracentrifugation (29). We analyzed a small aliquot (100 μL) of the selected phage pool from each round, and found that the enrichment of high-affinity phage was significantly more efficient in the MiPS system; after three rounds of selection in the MiPS chip, the phage from the round 3 pool (R3) demonstrated ∼700-fold higher binding to PPC-1 cells on-chip in comparison to the initial random library (Fig. 4A). In contrast, conventional cell suspension biopanning yielded a 100-fold enrichment, presumably due to a higher level of nonspecific background binding compared to the MiPS system (Fig. 4A). During the selection, we observed that the recovery of phage (number of output phage after a selection round/total number of input phage before a selection round) was lower with MiPS than conventional biopanning (0.001% vs. 0.006%) in the first round, presumably because the highly stringent selection conditions in the MiPS system are more successful at eliminating nonspecifically bound phage. However, phage recovery in the MiPS system was significantly higher in the second (0.48% vs. 0.18%) and third (0.98% vs. 0.50%) rounds in comparison with conventional biopanning, presumably due to the rapid enrichment of high-affinity phage. We believe the capability of the MiPS system in applying highly stringent mass-action selection pressure in combination with its continuous-flow washing within the microchannel, offer an effective means of rapidly enriching phage that display peptides with higher affinity and specificity.
Fig. 3.
Experimental scheme for microfluidic selection of phage-displayed peptides with high affinity and specificity for adherent PPC-1 cells in the MiPS system. 5 × 1010 pfu of phage library (∼100 copies of each unique sequence) were loaded into the device and recirculated at a flow rate of 1 mL/ min for 3 h at 4 °C. The pump then delivered wash buffer at a flow rate of 1 ml/ min for 90 min. Finally, phage-bound cells were lysed with 1 mL of 1% NP/LB solution on ice at a flow rate of 6 mL/ min for 30 min, and the lysate was collected in a tube. The collected phage pool was amplified by infecting BLT5403 bacteria at 37°C for 2 h, followed by phage precipitation with a polyethylene glycol (PEG)/NaCl solution and purification by CsCl gradient ultracentrifugation. The amplified phage pool was used as the starting library pool for the next round of selection.
Fig. 4.
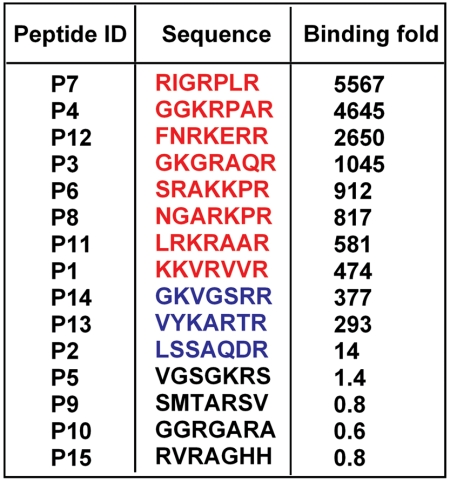
Results from de novo selection with a random linear X7 peptide library and 2 × 104 target cells in the MiPS device. (A) After three rounds of selection using the MiPS chip, the resulting phage pool (R3) demonstrated ∼700-fold higher binding to PPC-1 adherent cells in comparison to the initial random library. In contrast, three rounds of conventional cell suspension-based biopanning yielded ∼100-fold enrichment. (B) Sequences of peptides selected with the MiPS device (left) and conventional biopanning (right). Clones containing the high-affinity (R/K)XXR motif are shown in red, and sequences containing the weak-binding XXXR motif are displayed in blue. Clones that do not exhibit CendR motifs are shown in black. Selection in the MiPS device yielded a significantly higher fraction of phage displaying the R/KXXR motif (92%) compared to conventional biopanning (52%).
We investigated the compositional differences in the peptide motifs enriched by these two selection methods by randomly picking 21 individual phage clones from each R3 pool and comparing their amino acid sequences. Previous studies have shown that peptides with a C-terminal (R/K)XX(R/K) sequence exhibit significantly higher affinity for NRP-1-expressing cells than peptides with a C-terminal arginine alone (XXXR) (29). We found that the MiPS system enables more efficient enrichment of phage displaying this higher-affinity motif (Fig. 4B): 90% of the phage selected using the MiPS contained (R/K)XX(R/K) motifs, while 5% displayed lower affinity XXXR motifs. In contrast, conventional cell suspension-based panning yielded at best 52% (R/K)XX(R/K) phage and 29% XXXR phage.
Affinity of the Phage Pool Selected with the MiPS Chip.
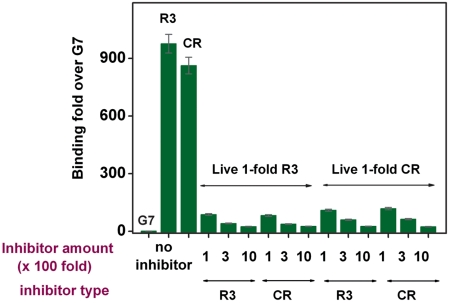
In order to compare the binding affinities between the RPARPAR phage and the R3 phage pool selected by the MiPS chip, we performed a series of inhibitor binding experiments to determine whether the R3 phage pool has sufficient affinity to displace PPC-1-bound RPARPAR phage, and vice versa. We used UV-inactivated noninfectious phage particles expressing about 200 peptides per particle as multivalent inhibitors (29, 30). We incubated PPC-1 cell suspensions (2 × 104 cells/sample) with solutions containing a 100-, 300-, or 1,000-fold excess of noninfectious R3 or RPARPAR phage (inhibitor) at 4 °C for 20 min, after which we added live RPARPAR or R3 phage solutions (1-fold, 3 × 108 pfu). After 1 h incubation, we collected the PPC-1 cells by centrifugation, washed them four times, and then lysed them in order to quantify the bound live phage by titration. For noninhibited samples, we simply incubated target cell suspensions with 3 × 108 pfu of live R3, RPARPAR, or G7 control phage, followed by washing and quantification.
We observed that the phage in the R3 pool bind strongly to PPC-1, and that the average binding affinity of the R3 pool was similar to that of the RPARPAR phage (Fig. 5). For example, using a onefold live R3 phage to compete with a 100-, 300-, or 1,000-fold excess of inhibitors (noninfectious phage particles), we observed 87-, 41-, or 25-fold enrichment of binding against noninfectious R3-pretreated cells and 82-, 38-, or 26-fold enrichment of binding against noninfectious RPARPAR-pretreated cells, respectively, relative to binding of live G7 phage. In equivalent experiments with a onefold live RPARPAR phage, we obtained 110-, 60-, or 26-fold enrichment of binding against noninfectious R3-pretreated cells and 118-, 63-, or 24-fold enrichment of binding against noninfectious RPARPAR-pretreated cells, respectively. In contrast, G7 control exhibited very small inhibition of binding by the R3 pool or RPARPAR (Fig. S4).
Fig. 5.
Inhibitor binding experiments show that the average affinity of the R3 pool, generated through three rounds of MiPS selection, is similar to that of the phage displaying the RPARPAR (CR) sequence. G7 control phage binding was used as a basis for comparison.
Specificity of the Pool Selected with the MiPS Chip.
To further confirm that the enriched R3 phage pool binds specifically to NRP-1, we performed inhibitor binding experiments with an excess of either G7 phage or anti-NRP-1 antibodies. Samples were prepared by incubating PPC-1 cell suspensions with a 100-, 300-, or 1,000-fold excess of G7 control phage, anti-NRP-1 antibody solution (2, 6, or 20 μg/mL), or control IgG antibody solution (20 μg/mL) at 4 °C for 20 min. We then added live R3 or RPARPAR phage (onefold, 3 × 108 pfu total) into these premixed cell suspensions. In the noninhibited samples, cell suspensions were only incubated with live R3, RPARPAR, or G7 phage (onefold, 3 × 108 pfu total). The binding results clearly show that the binding of both R3 and RPARPAR phage to PPC-1 cells was reduced by preincubating the cells with anti-NRP-1 antibodies (Fig. S5), indicating that the specific binding of selected phage to PPC-1 cells was at least partly facilitated by NRP-1 recognition.
Specificity and Affinity of Individual Phage Clones Enriched with the MiPS Chip.
Next, we randomly picked 15 phage clones from the R3 pool, determined their peptide sequences, and measured their binding to PPC-1 relative to M21 melanoma cells (Fig. 6). Of note, in contrast to PPC-1 cells, M21 cells express only minimal amounts of NRP-1 protein (29). As shown in Fig. 6, Fig. S6, clones expressing high-affinity (R/K)XXR motifs exhibited significantly greater specificity for PPC-1 compared to those with XXXR motifs. Interestingly, we found significant differences in specificity among different (R/K)XXR motifs, with phage expressing the RPXR motif (clones P4 and P7) showing the most specific binding to PPC-1 relative to M21 cells (Fig. S7).
Fig. 6.
Peptide sequence displayed by selected phage clones and their relative ratio of binding to PPC-1 cells (expressing NRP-1) compared to M21 cells (negative control). (A) The clones with high-affinity (R/K)XXR motifs are displayed in red, clones with XXXR motifs are shown in blue, and non-CendR sequences are in black.
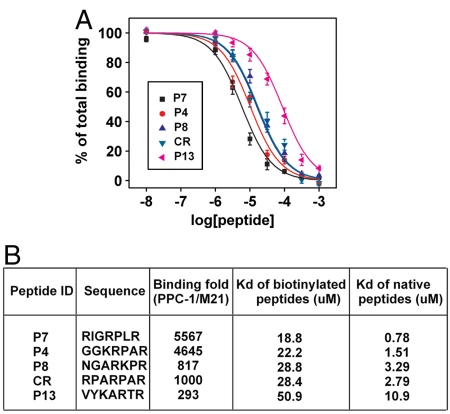
Next, we measured the equilibrium dissociation constants (Kd) of the peptide sequences for binding to the NRP-1 protein. Due to the experimental challenges in directly measuring the Kd values of the native peptides (41, 42), we followed a two-step process wherein we synthesized native and biotinylated versions of each peptide sequence. We first obtained the Kd value of the biotinylated peptide, then performed competitive binding assays to extract the Kd value of the native peptide using standard methods (43, 44). More specifically, to obtain the Kd value of the biotinylated peptide, we incubated NRP-1 protein-coated wells with different concentrations of biotinylated peptide for 1 h, then added streptavidin-conjugated horseradish peroxidase to each well and let it react for 30 min at room temperature. The optical signal from the horseradish peroxidase was fitted to a kinetic model to obtain the Kd values. We confirmed that biotinylated peptides with RXXR motifs (e.g., Kd(RPARPAR) = 28.4 ± 2.9 μM) exhibited higher affinity than those containing XXXR motifs (e.g., Kd(VYKARTR) = 50.9 ± 6.4 μM), and verified that peptides that do not follow the CendR rule, such as GGRGARA and SMTARSV, show minimal affinity for NRP-1. A number of biotinylated sequences showed comparable or superior binding affinities to the biotinylated RPARPAR sequence, including NGARKPR(Kd = 28.8 ± 3.5 μM), GGKRPAR(Kd = 22.2 ± 0.3 μM) and RIGRPLR(Kd = 18.8 ± 3.2 μM). We noticed that sequences with high affinities also showed more specific binding to PPC-1 cells over M21 cells (Fig. 6).
In order to obtain the Kd values of the native peptide sequences, we utilized a competitive binding assay described in the literature (43, 44). We challenged NRP-1-coated microtiter wells with mixtures containing various concentrations of native peptides and a constant concentration of biotinylated peptide for 2 h at room temperature, then added streptavidin-conjugated horseradish peroxidase to each well and let it react for 30 min at room temperature. Finally, we fitted the binding signal using Prism software (Graphpad) (Fig. 7A) to calculate the Kd values for each native peptide sequence (Fig. 7B). We discovered two sequences, GGKRPAR (P4) and RIGRPLR (P7), which show superior affinity and specificity to the well known RPARPAR sequence (CR). We believe that the presence of these peptides explain why R3 pool shows slightly higher enrichment than RPARPAR phage with no inhibitor.
Fig. 7.
Dissociation constants (Kd) of the native peptide sequences measured with competitive binding. (A) Peptides with the RXXR motif [P4, P7, P8, and RPARPAR (CR)] show higher binding affinity for NRP-1 than those with the XXXR motif (P13). (B) Three rounds of selection in MiPS device revealed two peptide sequences (P4 and P7) exhibiting higher affinity and specificity in binding to cells expressing NRP-1 compared with the well known CR.
Conclusion
In this work, we demonstrate a successful microfluidic system for selecting phage-displayed peptides against surface markers from live, adherent mammalian cells. With this system, we discovered two unique peptide sequences within three rounds of selection that bind to NRP-1, an important pleiotropic cell-surface receptor with roles in tumor progression, vascular development, and neurobiology, with superior affinity and specificity to the previously reported RPARPAR sequence (29, 30).
Compared to conventional biopanning methods, our method shows several notable advantages. First, the incorporation of microfluidics technology allows the use of small amounts of target cells with minimal loss, enabling the exertion of stringent mass-action selection pressures that yield peptides with higher affinities. This feature may also provide an important practical advantage for cell populations isolated from primary tissues and biopsy samples where large numbers of cells are not available. The amount of cells that can be harvested through biopsies varies widely depending on the tissue, and extraction processes. The least invasive biopsies (e.g., fine needle aspirate biopsies) usually yield the smallest number of cells in the range of ∼5,000–10,000 cells (45). Given that we obtained recovery performance of our MiPS selection process to be ∼93.9% for 104 cells with the current device, we believe the current design may be sufficient for such applications. In order to explore whether smaller numbers of cells can be processed in the MiPS system, we constructed a number of devices with smaller channel widths: 5 mm, 3.5 mm, 2 mm, and 0.5 mm, which accommodate 9.8 × 103, 6.3 × 103, 3.42 × 103, and 8.6 × 102 cells respectively, and measured the cell recovery. During these experiments, the shear stress was kept constant (3 dyn/cm2) by adjusting the flow rates to be 1 mL/ min (for 5 mm width device), 0.7 mL/ min (for 3.5 mm width device), 0.4 mL/ min (for 2 mm width device), and 0.1 ml/ min (for 0.5 mm width device). As shown in Fig. S8A, the cell recovery remained high (> 87%) even for the smallest device carrying 860 cells. However, if the flow rates were kept constant at 1 mL/ min the cell recovery decreased due to the increased shear stress as shown in Fig. S8B. These results experimentally show that our MiPS system can support smaller number of cells (e.g., ∼1,000 cells) by simply modifying our microchannel dimensions and adjusting the flow rate accordingly.
Second, the capability of the fluidic architecture to continuously wash samples at a reproducible and controlled flow rate allows more efficient removal of background phage compared to conventional, fixed-volume manual washing, leading to more efficient convergence of the phage pool toward phage clones with higher affinities. We tested whether these advantages would hold true even in the smaller devices with ∼1,000 cells, by measuring the binding enrichment of RPARPAR phage over G7 phage in the smallest MiPS chip (1.5 cm × 0.5 mm × 250 μm) at a flow rate of 0.1 mL/ min. This flow rate imposes an average flow velocity 1.3 cm/s and a shear stress 3 dyn/cm2, equivalent to that imposed on the cells in the original device under nominal operating conditions. As shown in Fig. S9, the enrichment reached ∼1,200-fold after 60 min of washing, and this performance is comparable to that of the original device. Thus we postulate that the high washing efficiency can be maintained even with smaller numbers of cells.
Finally, we demonstrate the integration of key selection steps in a monolithic, and disposable format that can be automated to increase reproducibility, minimize labor, eliminate contamination, and prevent loss of target cells and potential ligands. Although some of the process steps were performed outside of the chip, the capability to perform washing, lysis, and phage collection in situ, offers compelling advantages over conventional biopanning assays. Though not shown here, we believe the MiPS system could be configured to perform both positive and negative selection against different cell types in order to simultaneously evolve both affinity and specificity. Because many biocombinatorial libraries and tissue types can be screened with minimal number of cells in our closed, disposable chip, we believe the MiPS system may in principle represent a promising tool for the discovery of biomarkers from patient samples.
Materials and Methods
Materials.
Dulbecco’s Phosphate Buffered Saline solution (DPBS, calcium- and magnesium-free; Thermo Scientific), Dulbecco’s Modified Eagle’s Medium (DMEM; Thermo Scientific), cell dissociation buffer-lysogeny buffer (enzyme-free, PBS-based; Invitrogen), fibronectin (Invitrogen), fetal bovine serum (Invitrogen), Nonidet P-40 (NP-40) (US Biological), and penicillin-streptomycin (Invitrogen) were used as received without further purification. PPC-1 and M21 cell lines were cultured in DMEM supplemented with 10% fetal bovine serum and 1%penicillin/streptomycin. The PDMS layer was purchased from Bisco Silicones, and the PDMS chamber was cut with a Graphtec Cutting Plotter (CE5000-6, Graphtec America, Inc.). The 0.75 mm inlet and outlet holes were drilled through the glass substrates using a computer-controlled CNC mill (Flashcut CNC) and diamond bit (Triple Ripple, Abrasive Technology).
Trial Phage Screening Against Adherent PPC-1 Cells Using the MiPS Device.
PPC-1 cells were grown to nearly 90% confluence in culture slide chambers coated with 10 μg/mL fibronectin. To prevent nonspecific phage binding, the chambers were blocked with UV-treated G7 noninfectious phage. After blocking, the culture slide chamber walls were peeled off the slide surface and the adherent cell-coated slide was assembled into the MiPS device. Phage mixtures were prepared in binding buffer (DMEM supplemented with 1% BSA) containing RPARPAR:G7 ratios of 1∶1 (5 × 108 pfu each), 1∶10 (1 × 108 pfu RPARPAR∶9.9 × 108 pfu G7), 1∶100 (1 × 107 pfu RPARPAR∶9.9 × 108 pfu G7), or 1∶1,000 (1 × 106 pfu RPARPAR∶9.99 × 108 pfu G7) or pure G7 phage (109 pfu total) as a control. These mixtures were loaded into the MiPS device using a peristaltic pump (GE Healthcare) via Teflon tubing, and recirculated at 4 °C at a flow rate of 1 mL/ min for 3 h. To eliminate weak and nonspecific phage binders, washing buffer (DMEM + 1% BSA) was pumped at a flow rate of 1 mL/ min for 90 min. Finally, cells were lysed on ice with 1 mL of 1% NP/LB at a flow rate of 6 mL/ min for 30 min. The lysate was collected in a 2-mL microcentrifuge tube, and a phage titering assay was performed to quantify phage particles bound to the cells in order to calculate the RPARPAR∶G7 binding ratio.
Random Linear X7 Phage Library Screening via MiPS.
The MiPS device was prepared with cultured PPC-1 cells as described above. We applied 4 mL of binding buffer containing 5 × 1010 pfu phage library (∼100 copies of each unique sequence) to the assembled device for phage selection; the library pool was pumped into the device and recirculated with a flow rate of 1 mL/ min for 3 h at 4 °C. The sample was then subjected to washing and lysis under the conditions described above, and the bound phage were quantified by titering. After the first round of selection, we amplified the collected phage pool by infecting BLT5403 bacteria (OD = 0.5) in a bacterial shaker at 37 °C for 2 h until the bacteria were lysed. Amplified phage were mixed with 5 M NaCl (1/10 volume) in a centrifugation tube and centrifuged for 10 min at 13,000 rpm (20,190 × g) at 4 °C and the supernatant was collected in a new centrifugation tube. The amplified phage were precipitated from the collected supernatant with 50% PEG 8000 solution (1/5 volume), and then resuspended in 1.8 mL of PBS for CsCl gradient purification (29) and dialysis. The amplified phage pool was quantified via titering and used as starting library pool for the next round of phage selection. We performed three rounds of phage selection and amplification under these conditions. We used the phage pool selected from the first round as a baseline to determine enrichment of the phage pool selected from the second and third rounds. Individual phage clones were randomly picked from the plated phage pools selected from different rounds and sequenced. The sequences of displayed peptides were determined as described previously (29). The parallel biopanning screen was performed for three rounds with 2 × 104 PPC-1 cells in suspension and 5 × 1010 pfu phage library according to literature procedures (29).
Binding Measurements of the Selected R3 Phage Pool Against PPC-1 Cell Suspensions.
We incubated 2 × 104 PPC-1 cells in 2 mL binding buffer with 3 × 108 pfu of selected R3 phage pool or G7 phage for 1 h at 4 °C. The cells were then manually washed four times with 10 mL of binding buffer followed by centrifugation at 1,700 rpm (350 × g) for 5 min at 4 °C. Finally, the phage-bound cells were lysed with 1 mL of 1% NP/LB solution on ice for 30 min. Titration of the resulting lysate enabled us to determine the relative phage-binding of PPC-1 cell suspensions for selected R3 phage vs. control G7 phage.
Specificity Tests of the Selected R3 Phage Pool Against PPC-1 and M21 Cell Suspensions.
We incubated suspensions of 2 × 104 cells (PPC-1 or M21) in 2 mL binding buffer with 3 × 108 pfu of the selected R3 phage pool for 1 h at 4 °C. The cells were then washed four times by resuspension in 10 mL binding buffer, followed by centrifugation at 1,700 rpm (350 × g) for 5 min and removal of supernatant. The phage-bound cells were lysed with 1 mL of 1% NP/LB solution on ice for 30 min. Titration of the resulting lysate enabled determination of the relative binding of the R3 phage pool against PPC-1 and M21 cells. To further confirm the specificity of the enriched R3 pool, we performed additional inhibitor binding experiments in which 2 × 104 PPC-1 cells suspended in binding buffer were mixed with inhibitors: either a 100-, 300-, or 1,000-fold excess of noninfectious R3, RPARPAR, or G7 phage, or a solution of anti-rat NRP-1 antibody (2, 6, or 20 μg/mL) or IgG antibody (20 μg/mL) at 4 °C for 20 min. 3 × 108 pfu of live R3 or RPARPAR phage were then added into these premixed cell suspensions, which were further incubated at 4 °C for 1 h. The cells were then washed four times by resuspension in 10 mL binding buffer, followed by centrifugation at 1,700 rpm (350 × g) for 5 min and removal of supernatant. The phage-bound cells were lysed with 1 mL of 1% NP/LB solution on ice for 30 min. Titration of the resulting lysates revealed the relative binding affinity of R3 selected phage vs. G7 control in the presence of inhibitors.
Dissociation Constant (Kd) Measurements for Nonlabeled Peptides.
We used a competitive ELISA-based immunochemical assay to determine the dissociation constants of the unmodified peptides. We coated the wells of a microtiter plate with 50 μL of purified NRP-1 (R and D Systems) at 5 μg/mL overnight at 4 °C. After washing with PBST (PBS + 0.01% Tween 20), the wells were incubated for 2 h at room temperature with 50 μL of DPBS containing various concentrations of native test peptides and a constant concentration of biotinylated reporter peptide (Bio-RIGRPLR, 100 μM), and washed with the same washing buffer. We then added streptavidin-conjugated horseradish peroxidase (Vector Laboratories) to each well and incubated for 30 min at room temperature. Finally, we quantified the amount of bound horseradish peroxidase using 2,2-azino-bis (3-ethylbenzthiazoline-6-sulfonic acid) substrate. Using Prism software, we obtained the dissociation constants (Kd) of the native test peptides by fitting the dependence of the OD405 value on the concentration of native test peptides to the one-site competition equations (43).
SI Text Available: Phage Display; Cell recovery experiments for conventional suspension biopanning; Dissociation constant (Kd) measurements of biotinylated peptides; Trial phage screening against adherent PPC-1 cells using the MiPS device; Random linear X7 phage library screening via MiPS; Binding measurements of the selected R3 phage pool against PPC-1 cell suspensions; Specificity tests of the selected R3 phage pool against PPC-1 and M21 cell suspensions are available free of charge via Internet at http://www.pnas.org.
Supplementary Material
Acknowledgments.
We thank Joshua A. Olson for technical help and Dr. David Cheresh for the M21 cell line. We are grateful for the financial support of the ARO Institute for Collaborative Biotechnologies, National Institutes of Health, California Institute for Regenerative Medicine (CIRM), Midwestern Progenitor Cell Consortium, Armed Forces Institute for Regenerative Medicine, and Otis Williams Foundation.
Footnotes
The authors declare no conflict of interest..
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014753108/-/DCSupplemental.
References
- 1.Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 2.Riechmann L, Clark M, Waldmann H, Winter G. Reshaping human antibodies for therapy. Nature. 1988;332:323–327. doi: 10.1038/332323a0. [DOI] [PubMed] [Google Scholar]
- 3.Mori T. Cancer-specific ligands identified from screening of peptide-display libraries. Curr Pharm Design. 2004;10:2335–2343. doi: 10.2174/1381612043383944. [DOI] [PubMed] [Google Scholar]
- 4.Neri D, et al. Targeting by affinity-matured recombinant antibody fragments of an angiogenesis associated fibronectin isoform. Nat Biotechnol. 1997;15:1271–1275. doi: 10.1038/nbt1197-1271. [DOI] [PubMed] [Google Scholar]
- 5.MacBeath G. Protein microarrays and proteomics. Nat Genet. 2002;32(Suppl):526–532. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- 6.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 7.Reilly RM, et al. Problems of delivery of monoclonal antibodies. Pharmaceutical and pharmacokinetic solutions. Clin Pharmacokinet. 1995;28:126–142. doi: 10.2165/00003088-199528020-00004. [DOI] [PubMed] [Google Scholar]
- 8.Aina OH, Sroka TC, Chen ML, Lam KS. Therapeutic cancer targeting peptides. Biopolymers. 2002;66:184–199. doi: 10.1002/bip.10257. [DOI] [PubMed] [Google Scholar]
- 9.Nilsson F, Tarli L, Viti F, Neri D. The use of phage display for the development of tumour targeting agents. Adv Drug Deliv Rev. 2000;43:165–196. doi: 10.1016/s0169-409x(00)00068-5. [DOI] [PubMed] [Google Scholar]
- 10.Rozinov MN, Nolan GP. Evolution of peptides that modulate the spectral qualities of bound, small-molecule fluorophores. Chem Biol. 1998;5:713–728. doi: 10.1016/s1074-5521(98)90664-0. [DOI] [PubMed] [Google Scholar]
- 11.Wölcke J, Weinhold E. A DNA-binding peptide from a phage display library. Nucleos Nucleot Nucl. 2001;20:1239–1241. doi: 10.1081/NCN-100002526. [DOI] [PubMed] [Google Scholar]
- 12.Brown KC. New approaches for cell-specific targeting: identification of cell-selective peptides from combinatorial libraries. Curr Opin Chem Biol. 2000;4:16–21. doi: 10.1016/s1367-5931(99)00045-9. [DOI] [PubMed] [Google Scholar]
- 13.Hartley O. The use of phage display in the study of receptors and their ligands. J Recept Signal Tr R. 2002;22:373–392. doi: 10.1081/rrs-120014608. [DOI] [PubMed] [Google Scholar]
- 14.Babcock GJ, Mirzabekov T, Wojtowicz W, Sodroski J. Ligand binding characteristics of CXCR4 incorporated into paramagnetic proteoliposomes. J Biol Chem. 2001;276:38433–38440. doi: 10.1074/jbc.M106229200. [DOI] [PubMed] [Google Scholar]
- 15.Mirzabekov T, Kontos H, Farzan M, Marasco W, Sodroski J. Paramagnetic proteoliposomes containing a pure, native, and oriented seven-transmembrane segment protein, CCR5. Nat Biotechnol. 2000;18:649–654. doi: 10.1038/76501. [DOI] [PubMed] [Google Scholar]
- 16.Whaley SR, English DS, Hu EL, Barbara PF, Belcher AM. Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly. Nature. 2000;405:665–668. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]
- 17.Shadidi M, Sioud M. Identification of novel carrier peptides for the specific delivery of therapeutics into cancer cells. FASEB J. 2003;17:256–258. doi: 10.1096/fj.02-0280fje. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Spring H, Schwab M. Neuroblastoma tumor cell-binding peptides identified through random peptide phage display. Cancer Lett. 2001;171:153–164. doi: 10.1016/s0304-3835(01)00575-4. [DOI] [PubMed] [Google Scholar]
- 19.Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 20.Rajotte D, et al. Molecular heterogeneity of the vascular endothelium revealed by in vivo phage display. J Clin Invest. 1998;102:430–437. doi: 10.1172/JCI3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barry MA, Dower WJ, Johnston SA. Toward cell-targeting gene therapy vectors: selection of cell-binding peptides from random peptide-presenting phage libraries. Nat Med. 1996;2:299–305. doi: 10.1038/nm0396-299. [DOI] [PubMed] [Google Scholar]
- 22.Robert R, et al. Identification of human scFvs targeting atherosclerotic lesions: selection by single round in vivo phage display. J Biol Chem. 2006;281:40135–40143. doi: 10.1074/jbc.M609344200. [DOI] [PubMed] [Google Scholar]
- 23.Barbas CF, Kang AS, Lerner RA, Benkovic SJ. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hust M, Maiss E, Jacobsen HJ, Reinard T. The production of a genus-specific recombinant antibody (scFv) using a recombinant potyvirus protease. J Virol Methods. 2002;106:225–233. doi: 10.1016/s0166-0934(02)00166-0. [DOI] [PubMed] [Google Scholar]
- 25.Mandecki W, Chen YC, Grihalde N. A mathematical model for biopanning (affinity selection) using peptide libraries on filamentous phage. J Theor Biol. 1995;176:523–530. doi: 10.1006/jtbi.1995.0218. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman JA, Laakkonen P, Porkka K, Bernasconi M, Ruoslahti E. In: Phage Display: A Practical Approach. Clackson T, Lowman H, editors. Oxford, United Kingdom: Oxford University Press; 2004. pp. 171–192. [Google Scholar]
- 27.Dennis MS, Lowman HB. In: Phage Display: A Practical Approach. Clackson T, Lowman H, editors. Oxford, United Kingdom: Oxford University Press; 2004. pp. 61–82. [Google Scholar]
- 28.Liu Y, et al. Controlling the selection stringency of phage display using a microfluidic device. Lab Chip. 2009;9:1033–1036. doi: 10.1039/b820985e. [DOI] [PubMed] [Google Scholar]
- 29.Teesalu T, Sugahara KN, Kotamraju VR, Ruoslahti E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc Natl Acad Sci USA. 2009;106:16157–16162. doi: 10.1073/pnas.0908201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugahara KN, et al. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell. 2009;16:510–520. doi: 10.1016/j.ccr.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starzec A, et al. Antiangiogenic and antitumor activities of peptide inhibiting the vascular endothelial growth factor binding to neuropilin-1. Life Sci. 2006;79:2370–2381. doi: 10.1016/j.lfs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Fong S, Doyle LV, Devlin JJ, Doyle MV. Scanning whole cells with phage-display libraries: identification of peptide ligands that modulate cell function. Drug Develop Res. 1994;33:64–70. [Google Scholar]
- 33.Myszka DG, Jonsen MD, Graves BJ. Equilibrium analysis of high affinity interactions using BIACORE. Anal Biochem. 1998;265:326–330. doi: 10.1006/abio.1998.2937. [DOI] [PubMed] [Google Scholar]
- 34.Mulvaney SP, et al. Rapid, femtomolar bioassays in complex matrices combining microfluidics and magnetoelectronics. Biosens Bioelectron. 2007;23:191–200. doi: 10.1016/j.bios.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 35.Levitan B. Stochastic modeling and optimization of phage display. J Mol Biol. 1998;277:893–916. doi: 10.1006/jmbi.1997.1555. [DOI] [PubMed] [Google Scholar]
- 36.Warrick J, Meyvantsson I, Ju J, Beebe DJ. High-throughput microfluidics: improved sample treatment and washing over standard wells. Lab Chip. 2007;7:316–321. doi: 10.1039/b613350a. [DOI] [PubMed] [Google Scholar]
- 37.Yuan B, Schulz P, Liu R, Sierks MR. Improved affinity selection using phage display technology and off-rate based selection. Electron J Biotechn. 2006;9:171–175. [Google Scholar]
- 38.Nagel T, Resnick N, Atkinson WJ, Dewey CF, Jr, Gimbrone MA., Jr Shear stress selectively upregulates intercellular adhesion molecule-1 expression in cultured human vascular endothelial cells. J Clin Invest. 1994;94:885–891. doi: 10.1172/JCI117410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fisher AB, Chien S, Barakat AI, Nerem RM. Endothelial cellular response to altered shear stress. Am J Physiol Lung C. 2001;281:L529–533. doi: 10.1152/ajplung.2001.281.3.L529. [DOI] [PubMed] [Google Scholar]
- 40.Silverstein SC, Steinman RM, Cohn ZA. Endocytosis. Annu Rev Biochem. 1977;46:669–722. doi: 10.1146/annurev.bi.46.070177.003321. [DOI] [PubMed] [Google Scholar]
- 41.Ng EW, et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2001;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 42.Baldrich E, Restrepo A, O’Sullivan CK. Aptasensor development: elucidation of critical parameters for optimal aptamer performance. Anal Chem. 2004;76:7053–7063. doi: 10.1021/ac049258o. [DOI] [PubMed] [Google Scholar]
- 43.Motulsky HJ, Neubig RR. Analyzing binding data. Current Protocols in Neuroscience. 2010;52:7.5.1–7.5.65. doi: 10.1002/0471142301.ns0705s52. [DOI] [PubMed] [Google Scholar]
- 44.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 percent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 45.Stomper PC, Nava ME, Budnick RM, Stewart CC. Specimen mammography-guided fine-needle aspirates of clinically occult benign and malignant lesions. Analysis of cell number and type. Invest Radiol. 1997;32:277–281. doi: 10.1097/00004424-199705000-00004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.