Abstract
Using genetically modified cyanobacterial strains, we engineered a Green Recovery strategy to convert membrane lipids into fatty acids for economical and environmentally sustainable biofuel production. The Green Recovery strategy utilizes lipolytic enzymes under the control of promoters induced by CO2 limitation. Data indicate that strains of the cyanobacterium Synechocystis sp. PCC6803 engineered for Green Recovery underwent degradation of membrane diacylglycerols upon CO2 limitation, leading to release of fatty acids into the culture medium. Recovered fatty acid yields of 36.1 × 10-12 mg/cell were measured in one of the engineered strains (SD239). Green Recovery can be incorporated into previously constructed fatty-acid-secretion strains, enabling fatty acid recovery from the remaining cyanobacterial biomass that will be generated during fatty acid biofuel production in photobioreactors.
Alternative biofuels are needed to address the issues of climate change and petroleum supply (1). Production of biofuels from photosynthetic microorganisms (which include microalgae and cyanobacteria) has several advantages compared to plant-based biofuel production. Specifically, microalgae and cyanobacteria have higher solar energy efficiency and growth rates compared to plants, and microbial biodiesel production can be based on nonarable land (2). Unlike heterotrophic microbes (e.g., Escherichia coli and yeast), photosynthetic microorganisms such as cyanobacteria do not need costly starch or sugar feed stocks for biofuel production (2).
Cyanobacteria have a robust lipid metabolism. Their lipids are mainly in the form of diacylglycerols as the components of membranes, including monogalactosyl diacylglycerol, digalactosyl diacylglycerol, phosphatidylglycerol, and sulfoquinovosyl diacylglycerol (3). However, the traditional downstream recovery of microbial lipids, requiring physical cell lysis followed by chemical solvent extraction, accounts for 70–80% of the total cost of biofuel production (4). We propose a more cost-effective way to harvest lipids from cyanobacterial biomass for biofuel production that precludes the need for mechanically disrupting the cells. To this end, we developed a Green Recovery system in which lipolytic enzymes degrade the membrane lipids into free fatty acids (FFA) with the collapse of cells. The Green Recovery system controls the synthesis of lipolytic enzymes using CO2-limitation-inducible promoters, which induce expression of the lipolytic genes upon cessation of CO2 aeration.
The lipolytic enzymes (EC 3.1.1), including galactolipase and phospholipase B (5), hydrolyze the carboxylic ester bonds to release the fatty acids from diacylglycerols. Galactolipase (EC 3.1.1.26) catalyzes the hydrolysis of galactolipids by removing one or two fatty acids (6). Phospholipase B is an enzyme with a combination of both phospholipase A1 (EC 3.1.1.32) and phospholipase A2 (EC 3.1.1.4) activities, which can cleave acyl chains from both the sn-1 and sn-2 positions of a phospholipid (7). For the purpose of fatty acid recovery from membrane lipids, we tested the performance of three lipolytic enzymes (from a bacterium, a fungus, and an herbivorous animal) in the cyanobacterium Synechocystis sp. PCC6803 strain SD100 (8). The lipase from Staphylococcus hyicus (Shl) was selected because it has a very broad substrate specificity ranging from triacylglycerol lipids of various chain lengths to phospholipids and lysophospholipids (9). The second candidate was a modified fungal phospholipase from Fusarium oxysporum (Fol) that exhibited galactolipase activity as well as increased phospholipase activity (10). Third, the guinea-pig lipase (Gpl, also called guinea-pig pancreatic lipase-related protein 2) from the digestive juice of guinea pig (11) was also tested. Gpl shows the highest galactolipase activity known to date, and plays a dual role in the digestion of galactolipids and phospholipids, the most abundant lipids occurring in plant thylakoid membranes (11).
In our previous research, we developed a nickel inducible lysis system for SD100 (wild type), which is able to induce phage lysis genes to break down cell walls by adding nickel as an inducer (8). This system showed the feasibility of controlling lethal genes by inducible promoters. However, nickel is toxic to the environment and also adds cost. Therefore, we developed a cyanobacterial CO2-limitation-response mechanism (12) for an inducible transcription system regulated by CO2 rather than nickel. It has been reported that aeration of illuminated SD100 cells with CO2-free air for 30 min depleted the culture CO2 concentration to near zero levels. Under these conditions, transcripts for the three inducible inorganic carbon uptake systems, ndhF3, sbtA, and cmpA, showed near-maximal abundance after 15 min under CO2 limitation (12). By utilizing the promoter sequences for these genes to control the lipase genes, Green Recovery of lipids can be initiated by CO2 limitation resulting from stopping aeration of the culture. Aeration to the photobioreactor is necessary and easy to regulate, thus limiting the CO2 supply is an economical and environmentally friendly method to initiate lipid hydrolysis.
Results
Construction of Green Recovery Strains.
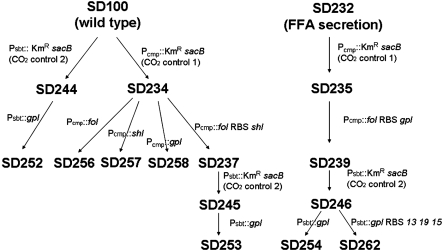
We constructed a series of Sun Devil (SD) strains for testing and optimizing Green Recovery (Fig. 1 and Table S1). By using the KmR/sacB intermediate double cross-over recombination method (13), we inserted synthesized genes fol, shl, gpl, and an artificial operon fol ribosome binding site (RBS) gpl into SD100 after Pcmp and before the start codon of the cmpA gene, resulting in strains SD256, SD257, SD258, and SD237, respectively. The gpl gene was inserted into wild type and SD237 after Psbt and before the ATG of the sbtA gene to result in SD252 and SD253, respectively. To incorporate the Green Recovery system into the FFA-secretion strains (13), we inserted the artificial operon fol RBS shl into an FFA-secretion strain SD232 (13) after Pcmp to result in SD239. To achieve rapid membrane damage, we inserted another artificial operon gpl RBS 13 19 15, in which 13 19 15 are lysis genes from Salmonella phage P22 (8), into SD239 after Psbt to result in SD254 and SD262, respectively. The growth of wild-type based strains (e.g., SD256, SD257, and SD237) was similar to the wild type, with doubling times of about 9 h. The growth of the SD232-based strains (e.g., SD239, SD254, and SD262) was also similar to SD232, with doubling times of about 12 h.
Fig. 1.
Genealogies of the Green Recovery constructions. Three lipolytic genes (fol, shl, and gpl) were inserted into SD100 (6803 wild type) and SD232 (an FFA-secretion 6803 strain), and controlled by two CO2-limitation-inducible promoters (Pcmp and Psbt). Detailed genetic information of the strains is described in Table S1.
Degradation of Membrane Lipids.
Limiting the CO2 supply to the Green Recovery strain cultures (density > 108 cells/mL) resulted in culture discoloration (Fig. 2) and membrane permeability as revealed by SYTOX green staining (8) (Figs. S1 and S2). A comparison between the SYTOX green analysis for membrane permeability and the cfu analysis for cell viability showed that cell death happened before the membrane became noticeably permeable (Fig. S3). When the membrane permeable cell percentage increased to about 50%, the cfu dropped to below 0.1% of the starting cfu. Strains synthesizing the fungal lipase Fol (in SD256) or the bacterial lipase Shl (in SD267) had high levels of permeability when incubated under CO2-limiting conditions compared to wild type (Table 1). The membrane damage rate of SD237 with both fol and shl (45.2% per day) was faster than those of the single lipase synthesizing strains (35.1% per day for SD256 and 33.8% per day for SD257). The guinea-pig lipase gene gpl resulted in a damage rate of 9.4% per day when controlled by Pcmp in SD258, and a faster damage rate of 13.5% per day when controlled by Psbt in SD252 (Table 1). Together these results strongly suggest that the condition of CO2 limitation causes the expression of genes encoding lipases that result in increased membrane permeability.
Fig. 2.
The cultures of 6803 wild type, SD256 and SD257 in sealed flasks for four days for CO2 limitation after being grown with vigorous aeration.
Table 1.
Membrane damage and FFA yields of SD strains for Green Recovery
| Strain | Genetic description * | Starting cell density, cfu/mL | Membrane damage,† %/d | Recovered FFA,‡ mg/L | FFA yield, 10-12 mg/cell |
| SD100 | Wild type | 3.2 × 109 | 8.9 | 16.5 ± 2.5 | 5.1 ± 0.8 |
| SD200 | ΔlipA∷sacB KmR | 3.4 × 109 | 2.5 | 2.7 ± 1.1 | 0.8 ± 0.3 |
| SD256 | Pcmp∷fol | 1.6 × 109 | 35.1 | 19.3 ± 1.0 | 12.4 ± 0.6 |
| 3.9 × 108(1/4) § | 94.7 | 5.5 ± 0.7 | 14.0 ± 1.8 | ||
| SD257 | Pcmp∷shl | 2.9 × 109 | 33.8 | 22.7 ± 1.0 | 7.9 ± 0.4 |
| 7.2 × 108(1/4) § | 83.4 | 5.7 ± 1.2 | 8.0 ± 1.7 | ||
| SD237 | Pcmp∷fol RBS shl | 1.5 × 109 | 45.2 | 23.6 ± 1.1 | 15.7 ± 0.7 |
| 3.8 × 108(1/4) § | 91.3 | 10.6 ± 0.1 | 28.0 ± 0.4 | ||
| SD258 | Pcmp∷gpl | 3.0 × 108 | 9.4 | 5.2 ± 0.6 | 17.4 ± 2.0 |
| SD252 | Psbt∷gpl | 3.5 × 108 | 13.5 | 3.1 ± 1.4 | 8.8 ± 4.1 |
| SD239 | Δslr1609∷PpsbA2 ‘tesA Δ(slr1993-slr1994)∷ PcpcaccBC PrbcaccDA Δsll1951∷*PpsbA2 Uc fatB1 Prbc Ch fatB2 Pcmp∷fol RBS shl | 5.8 × 108 | 56.7 | 23.6 ± 0.02 ¶ | 40.7 ± 0.03 ¶ |
| 44.6 ± 2.8 ∥ | 76.8 ± 4.7 ∥ | ||||
| 21.0 ± 2.8 ** | 36.1 ± 4.7 ** | ||||
| SD254 | Δslr1609∷PpsbA2 ‘tesA Δ(slr1993-slr1994)∷ PcpcaccBC PrbcaccDA Δsll1951∷*PpsbA2 Uc fatB1 Prbc Ch fatB2 Pcmp∷fol RBS shl Psbt∷gpl | 7.3 × 108 | 34.5 | 45.1 ± 0.4 ¶ | 61.7 ± 0.5 ¶ |
| 63.6 ± 0.4 ∥ | 87.2 ± 0.5 ∥ | ||||
| 18.6 ± 0.4 ** | 25.4 ± 0.5 ** | ||||
| SD262 | Δslr1609∷PpsbA2 ‘tesA Δ(slr1993-slr1994)∷ PcpcaccBC PrbcaccDA Δsll1951∷*PpsbA2 Uc fatB1 Prbc Ch fatB2 Pcmp∷fol RBS shl Psbt∷gpl RBS 13 19 15 | 8.9 × 108 | 67.3 | 47.0 ± 0.7 ¶ | 52.8 ± 0.8 ¶ |
| 73.4 ± 1.4 ∥ | 82.5 ± 1.5 ∥ | ||||
| 26.5 ± 1.4 ** | 29.7 ± 1.5 ** |
*Detailed genetic information is described in Table S1.
†Membrane damage was detected by SYTOX staining. The damage rates (percent per day) were estimated from starting time point to the 50% permeable cell time point, when over 99.9% cells were dead. If the permeable cell percentage did not drop to 50%, the damage rates were estimated over the entire experimental period.
‡Recovered FFAs were extracted by 10 mL hexane from 16-mL cultures after 4 d of CO2 limitation.
§The data for 1/16 dilutions were similar to the data for one-fourth dilutions.
¶The secreted FFAs before CO2 limitation.
∥Total FFAs after CO2 limitation, including secreted FFAs and released FFAs.
**For combination strains, released FFAs were calculated by subtracting the total amount from the secreted amount.
SD100 also showed membrane damage at high cell density under CO2-limitation conditions (Fig. S2). We therefore constructed a lipA deficient strain SD200 by interrupting the putative lipase gene (lipA, sll1969) with the KmR/sacB cassette. Compared with wild type, SD200 showed much less background autolysis under these conditions (Table 1 and Fig. S2), supporting the conclusion that the lipA gene is responsible for background autolysis, and that the presence of the lipA gene contributes to the efficiency of Green Recovery. We assume that the Green Recovery system can be more tightly controlled by exchanging the native promoter of lipA with a CO2-limitation-inducible promoter.
Conditions for Green Recovery.
We tested several optimal conditions for Green Recovery, such as cell density of the cultures (Fig. S4), intensity of illumination (Fig. S5), and agitation of the cultures (Fig. S5). It was observed that the membrane damage was faster and FFA release was higher after one-fourth dilution (about 108 cells/mL) than at the original cell density (about 109 cells/mL) (Table 1 and Fig. S4). Light is essential for Green Recovery, which is negligible in the dark (less than 2 μmol photons m-2 s-1), and much slower in low light (20 μmol photons m-2 s-1) than in higher light levels (140 μmol photons m-2 s-1) (Fig. S5). A probable explanation for these observations is that energy from photosynthesis is required for gene expression, synthesis, and function of lipase enzymes. Because of self-shading, photosynthetic cells have a higher metabolic activity at low cell density than at high cell density, consistent with the higher FFA yields that we observed at low cell concentrations (Table 1). The intensity of agitation did not affect the rate of lysis; for example, continuous rotation at 60 rpm/ min and intermittent shaking (once per day) produced similar membrane damage curves (Fig. S5).
Green Recovery for FFAs.
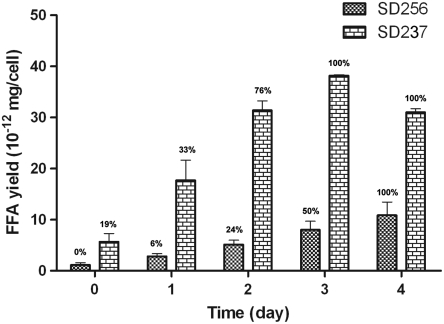
GC (14) showed that a significant amount of FFAs were released by lipolytic degradation of the membrane lipids (Table 1 and Fig. S6). Green Recovery FFA yield of 36.1 × 10-12 mg/cell was measured in SD239, compared to 200 × 10-12 mg FFA/cell secreted by an FFA-secretion strain SD277 (13) grown in the same conditions. The FFA yields of Green Recovery were closely related to the membrane damage even under different conditions (Table 1). About 40% of the total membrane diacylglycerol lipid is recovered as FFA at the cell density of 4 × 108 cells/mL (Table S2). GC also showed that the profile of the released FFA was close to the fatty acid profile of the SD100 membrane lipids with abundant unsaturated fatty acids (Fig. 3), suggesting that the released FFAs were degraded from membrane lipids. FFA release occurred concomitantly with membrane damage during Green Recovery, and the released FFA amount reached the maximum when most cells became permeable to the SYTOX stain (Fig. 4).
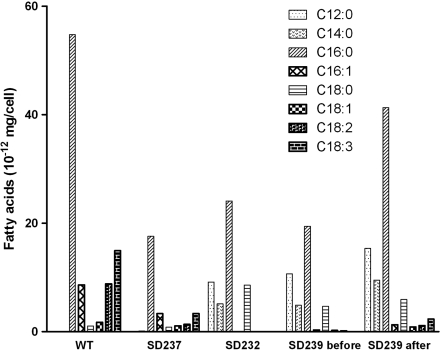
Fig. 3.
The fatty acid profiles of SD strains. All the cultures were grown to about 4 × 108 cells/mL at 30 °C. For wild type, the columns show the fatty acid profile of total membrane lipids. For SD237, the columns show the released FFA profile by Green Recovery, which is similar to that of wild type with abundant unsaturated fatty acids. For SD232, the columns show the profile of secreted FFAs, which are highly saturated with significant amounts of C12∶0 and C14∶0. For SD239 before (Green Recovery), the columns show the profile of secreted FFAs before CO2 limitation, which is similar to that of the FFA-secretion strain SD232. For SD239 after (Green Recovery), the columns show the profile of all the FFAs contributed by SD239 after CO2 limitation, which is a mixture of secreted FFAs (e.g., C12∶0 and C14∶0) and released FFAs (e.g., C18∶2 and C18∶3).
Fig. 4.
FFA yields of SD256 and SD237 during Green Recovery. Ten CO2-limiting flasks with 16-mL cultures were set in the same conditions on day zero for each strain. Every day, the whole cultures in duplicate flasks were extracted by hexane for FFA yields. The cell membrane damage was observed after SYTOX staining, and the permeable cell percentages are indicated above the columns.
Green Recovery with FFA Secretion.
Recently, we reported our results in constructing SD strains that secrete FFAs into the culture media by introducing acyl–acyl carrier protein (acyl-ACP) thioesterase genes into SD100 and other genetic modifications (13). The FFA-secretion strains that harbor the Green Recovery system (SD239, SD254, and SD262 in Table 1) are still able to release membrane lipids as FFAs at rates faster than non-FFA-secretion strains following CO2 limitation. GC analysis showed that the profile of the FFAs overproduced by acyl-ACP thioesterases by FFA-secretion strains was different from the profile of the membrane-released lipids. The FFAs recovered from overproducing strains are highly saturated and rich in C12∶0 and C14∶0, whereas FFAs obtained via the Green Recovery system contained substantial amounts of unsaturated fatty acids and only a small portion of C12∶0 and C14∶0, which is the same composition observed in membrane lipids (Fig. 3). The released FFA amount from membrane lipids is similar to the amount of secreted FFA from thioesterases (Table 1, SD239). As anticipated, the FFAs recovered from the combination strains (e.g., SD239 and SD262) after CO2 limitation was a mixture of the overproduced FFAs and the released membrane FFAs.
Discussion
The Green Recovery system was designed for production of scalable and cost-effective renewable biofuels in photobioreators. Productive photobioreators require aeration systems to supply the photosynthetic microorganisms with CO2. Lipid recovery from biomass by limiting CO2 supply is clearly an efficient and effective method. The system we describe here does not require traditional biomass processes (4), such as cell harvesting, dewatering, cell disruption, solvent extraction, or inducer molecules like those employed in our previous inducible cyanobacteria lysis system (8), thus considerably reducing the cost of lipid recovery. Because continuous agitation is not required for Green Recovery (Fig. S5), this system only needs sunlight and possibly intermittent agitation to convert biomass into FFAs. Another advantage of Green Recovery is that lipolytic enzymes convert diacylglycerols in the membranes into FFAs, which, due to their low density and low solubility in water, are easier to harvest and refine than the diacylglycerol lipids. Some unstaturated species can be saturated by hydrogenation if the FFA products will undergo a hydrogen decarboxylation to yield alkanes. Although it was reported that the guinea-pig lipase Gpl had the strongest galactolipase activities ever identified (10), our experiment showed that Gpl was not as effective as the others in our SD strains (Table 1). We speculate that the compromised performance of Gpl in our SD strains was due to different lipid substrates in cyanobacteria and plants or improper protein folding of an animal protein in a cyanobacterial protein synthesis system. We anticipate that Green Recovery would provide the same advantages when applied in other microbial bioreactors such as algae, E. coli, and yeast systems (maybe using different inducible promoters).
Green Recovery exhibits other advantages when combined with the previously described cyanobacterial FFA-secretion system (13). The FFA-secretion system avoids the energy-intensive biomass processes such as concentration and extraction by directly recovering the secreted FFA from the culture medium. However, the FFA-secretion system still requires substantial biomass to achieve cost-effective FFA production, which means a significant amount of fixed carbon has to be converted and stored as lipid membranes. It is expected that the Green Recovery system will recover the membrane lipids in the potential remaining cyanobacterial biomass generated by the FFA-secretion system, and will also cause cell lysis and release of the unsecreted intracellular FFAs. To our surprise, incorporation of the Green Recovery system into the FFA-secretion strains resulted in an increased damage rate upon CO2 limitation (Table 1, SD239 and SD262). We postulate that secretion of FFAs through the cytoplamstic membranes creates some lesions in the membranes that facilitate the contact of lipolytic enzymes to the acyl glycerol ester bonds. These findings demonstrate the practical combination of the FFA-secretion system and Green Recovery in photobioreactors, where the old cultures or the spent biomass can be utilized for extra FFA yield. This Green Recovery is also applicable in the SD100 strains we generated for producing other biofuel molecules, such as triacylglycerols, fatty acyl methyl, or ethyl esters (biodiesel), and alkanes. However, the yields of the other biofuel molecules are lower than would be needed for a productive process. We believe that cyanobacterial biofuel will be instrumental in developing a carbon neutral source of sustainable fuels.
Materials and Methods
Bacterial Strain Construction, Media, and Growth Conditions.
All SD strains are derived from Synechocystis sp. PCC6803 strain SD100 (8). Gene modifications are applied into SD strains by using a KmR/sacB cassette (13). Methods for DNA manipulation are standard (15). The primers for constructions and genotype verifications are listed in Table S3. Optimized gene segments are synthesized at GenScript, Inc. and listed in Table S4. SD strains were grown at 30 °C in BG-11 medium (16) under continuous illumination (140 μmol photons m-2 s-1) and bubbled with air or 1% CO2-enriched air. The details for preservation of SD cells and growing an SD culture from a colony are described previously (13).
Growth and Cell Membrane Damage Measurement.
Bacterial growth in liquid culture was monitored spectrophotometrically and/or by dilution and plating. The cell membrane damage was detected by staining with 5 μM SYTOX green nucleic acid stain (Invitrogen Molecular Probes, Inc.) for 5 min with observation with an Axioskop 40 fluorescence microscope (Zeiss) (8).
CO2 Limitation and FFA Extraction.
After the cultures achieved above 109 cells/mL, they were limited for CO2 supply by stopping aeration and transferring to sealed flasks for Green Recovery. Briefly, 16-mL cultures or dilutions were transferred from bubbling flasks to triplicate or duplicate 25-mL flasks (Pyrex), sealed with plastic wrap (SaranWrap) and Parafilm, and rotated at 100 rpm under continuous illumination (140 μmol photons m-2 s-1). At the time points during or after Green Recovery, the whole 16-mL culture would be mixed with 0.3 g NaCl, 0.3 mL 1M H3PO4, and 10 mL hexane, and rotated in a 50-mL centrifuge tube at 200 rpm at 37 °C for 30 min. After a centrifugation ((6,000 × g) × 10 min), 5 mL of the upper organic layer was collected for FFA analysis. The composition and total amount of FFA samples were analyzed by GC (17). The total membrane lipids were extracted by Folch method (18) and transesterified into fatty acid methyl ester by methanolic HCl (Supelco). The fatty acid profile of the total lipids in 6803 wild type was analyzed by GC (19).
Statistical Analysis.
Most data were expressed as means ± standard deviation. Student’s t test was used for pairwise comparisons. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments.
The authors thank Michael Fisher, Rebecca Allen, and Greg Golden for valuable comments on the manuscript. This work was supported by Arizona State University Startup Funding to R.C. and US Department of Energy Advanced Research Projects Agency–Energy Grant (XKS0012).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103016108/-/DCSupplemental.
References
- 1.Peralta-Yahya PP, Keasling JD. Advanced biofuel production in microbes. Biotechnol J. 2010;5:147–162. doi: 10.1002/biot.200900220. [DOI] [PubMed] [Google Scholar]
- 2.Dismukes GC, Carrieri D, Bennette N, Ananyev GM, Posewitz MC. Aquatic phototrophs: Efficient alternatives to land-based crops for biofuels. Curr Opin Biotechnol. 2008;19:235–240. doi: 10.1016/j.copbio.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Wada H, Murata N. Temperature-induced changes in the fatty acid composition of the cyanobacterium, Synechocystis PCC6803. Plant Physiol. 1990;92:1062–1069. doi: 10.1104/pp.92.4.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molina Grima E, Belarbi EH, Acien Fernandez FG, Robles Medina A, Chisti Y. Recovery of microalgal biomass and metabolites: Process options and economics. Biotechnol Adv. 2003;20:491–515. doi: 10.1016/s0734-9750(02)00050-2. [DOI] [PubMed] [Google Scholar]
- 5.Svendsen A. Lipase protein engineering. Biochim Biophys Acta. 2000;1543:223–238. doi: 10.1016/s0167-4838(00)00239-9. [DOI] [PubMed] [Google Scholar]
- 6.Helmsing PJ. Purification and properties of galactolipase. Biochim Biophys Acta. 1969;178:519–533. doi: 10.1016/0005-2744(69)90221-6. [DOI] [PubMed] [Google Scholar]
- 7.Kohler GA, et al. Phospholipase A2 and phospholipase B activities in fungi. Biochim Biophys Acta. 2006;1761:1391–1399. doi: 10.1016/j.bbalip.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, Curtiss R., 3rd Nickel-inducible lysis system in Synechocystis sp. PCC 6803. Proc Natl Acad Sci USA. 2009;106:21550–21554. doi: 10.1073/pnas.0911953106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenstein R, Gotz F. Staphylococcal lipases: Biochemical and molecular characterization. Biochimie. 2000;82:1005–1014. doi: 10.1016/s0300-9084(00)01180-9. [DOI] [PubMed] [Google Scholar]
- 10.Rapp P. Production, regulation, and some properties of lipase activity from Fusarium oxysporum f. sp. vasinfectum. Enzyme Microb Technol. 1995;17:832–838. [Google Scholar]
- 11.Andersson L, Carriere F, Lowe ME, Nilsson A, Verger R. Pancreatic lipase-related protein 2 but not classical pancreatic lipase hydrolyzes galactolipids. Biochim Biophys Acta. 1996;1302:236–240. doi: 10.1016/0005-2760(96)00068-9. [DOI] [PubMed] [Google Scholar]
- 12.McGinn PJ, Price GD, Maleszka R, Badger MR. Inorganic carbon limitation and light control the expression of transcripts related to the CO2-concentrating mechanism in the cyanobacterium Synechocystis sp. strain PCC6803. Plant Physiol. 2003;132:218–229. doi: 10.1104/pp.019349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Jie S, Curtiss R., 3rd Fatty acid production in genetically modified cyanobacteria. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1103014108. 10.1073/pnas.1103014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lalman JA, Bagley DM. Extracting long-chain fatty acids from a fermentation medium. J Am Oil Chem Soc. 2004;81:105–110. [Google Scholar]
- 15.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 16.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 17.Lalman JA, Bagley DM. Extracting long-chain fatty acids from a fermentation medium. J Am Oil Chem Soc. 2004;81:105–110. [Google Scholar]
- 18.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 19.Sheng J, Vannela R, Rittmann BE. Evaluation of methods to extract and quantify lipids from Synechocystis PCC 6803. Bioresour Technol. 2011;102:1697–1703. doi: 10.1016/j.biortech.2010.08.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.