Abstract
Muscle contraction is driven by a change in the structure of the head domain of myosin, the “working stroke” that pulls the actin filaments toward the midpoint of the myosin filaments. This movement of the myosin heads can be measured very precisely in intact muscle cells by X-ray interference, but until now this technique has not been applied to physiological activation and force generation following electrical stimulation of muscle cells. By using this approach, we show that the long axes of the myosin head domains are roughly parallel to the filaments in resting muscle, with their center of mass offset by approximately 7 nm from the C terminus of the head domain. The observed mass distribution matches that seen in electron micrographs of isolated myosin filaments in which the heads are folded back toward the filament midpoint. Following electrical stimulation, the heads move by approximately 10 nm away from the filament midpoint, in the opposite direction to the working stroke. The time course of this motion matches that of force generation, but is slower than the other structural changes in the myosin filaments on activation, including the loss of helical and axial order of the myosin heads and the change in periodicity of the filament backbone. The rate of force development is limited by that of attachment of myosin heads to actin in a conformation that is the same as that during steady-state isometric contraction; force generation in the actin-attached head is fast compared with the attachment step.
Contraction of skeletal muscles is driven by a cyclical interaction between myosin and actin, fueled by the hydrolysis of ATP. The myosin and actin are polymerized into parallel thick and thin filaments, which themselves are organized into a hexagonal array in the muscle cell. The head domains of myosin lie on the surface of the thick filaments and bind to actin in the thin filaments. Filament sliding is driven by a change in conformation of the actin-bound myosin head: its working stroke (1–3). A detailed molecular model for the working stroke has been derived from biochemical and structural studies of isolated myosin head domains and their interaction with actin and ATP (3–6), and the quasi-crystalline organization of myosin and actin in muscle has allowed this model to be tested and elaborated by mechanical and structural studies on muscle cells (1, 2, 7–11).
Many of these cell-based studies used rapid perturbations to synchronize the actions of the myosin heads in a muscle cell. Typically, the length of an active muscle fiber was rapidly decreased, displacing each set of myosin filaments by a few nanometers with respect to the opposing actin filaments (2). Such a shortening step produces an elastic force decrease during the step, followed in the next few milliseconds by rapid force regeneration driven by the working stroke in actin-attached myosin heads (2, 7, 8). This and related protocols have revealed fundamental properties of the working stroke, including its size, speed, and load dependence, and shown how the macroscopic performance of muscle is related to the molecular mechanics and dynamics of the myosin–actin interaction (8–11). However, these perturbations are applied to muscle cells that are already generating force, and do not directly address the question of the physiological mechanism of force generation starting from the resting state in which the myosin heads are detached from actin.
Electrical stimulation of an intact single muscle cell provides an alternative approach to synchronize the action of the myosin heads, in this case starting from the resting detached state, as in an in vivo contraction. The muscle action potential is rapidly conducted into the cell interior via a set of tubular membranes, so that calcium ions are released synchronously throughout the cell volume. The intracellular free calcium concentration peaks at approximately 10 ms after the action potential in the conditions used here (fast-twitch amphibian muscle fibers, 4 °C) (12), much faster than the rate of force development, which has a half-time of approximately 50 ms. Previous studies have used time-resolved X-ray diffraction to investigate the structural changes in the actin and myosin filaments following electrical stimulation (13–18). Here we used the higher resolution technique of X-ray interference (19) to determine the time course of the axial motions of the myosin heads following electrical stimulation and the conformation of the myosin heads in resting muscle.
Results and Discussion
X-Ray Interference Signals from Myosin Heads During Isometric Force Development.
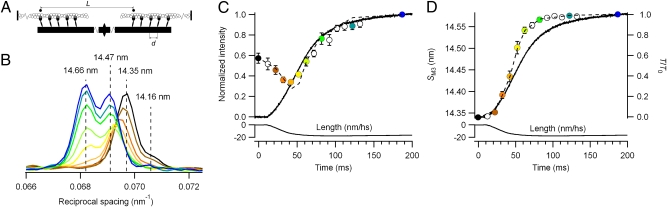
Myosin heads are arranged in a regular array on the surface of the muscle thick filaments (Fig. 1A), with a spacing (d) of approximately 14.5 nm between adjacent layers of heads along the filament (20). This periodic structure gives rise to a strong X-ray reflection called the M3. In resting single fibers from frog muscle, the intensity distribution of the M3 reflection along a line parallel to the fiber axis has a narrow peak at 1/14.35 nm−1 in reciprocal space, with a small but reproducible shoulder at higher diffraction angle, or higher reciprocal spacing (19) (Fig. 1B, black).
Fig. 1.
Changes in the M3 X-ray reflection during isometric force development. (A) Arrangement of thick (black) and thin (white) filaments in the sarcomere showing axial periodicity of myosin heads (d), and interference distance (L) between the two arrays of heads in each thick filament. (B) Axial profiles of the M3 X-ray reflection at different times during force development; colors correspond to symbols in C and D. (C) Intensity of the M3 X-ray reflection (IM3) after width correction, normalized by tetanus plateau value. (D) Spacing of the M3 X-ray reflection (SM3). Thicker line in C and D is force normalized by its plateau value (T0); thinner line is change in half-sarcomere (hs) length. Error bars denote ± SE for five fibers. Dashed lines in C and D were calculated from the model in Fig. 3.
When muscle fibers were activated at fixed length by a train of electrical stimuli to produce an isometric tetanus, force developed with a latency of approximately 10 ms after the first stimulus and reached its half-maximum value at approximately 50 ms (Fig. 1C, Upper, solid line). The sarcomere length in the central region of the fiber illuminated by the X-ray beam (Fig. 1C, Lower) decreased monotonically by approximately 20 nm per half-sarcomere during the initial part of the force development, as the fiber tendons were stretched. The intensity and reciprocal spacing of the M3 X-ray reflection decreased during the first 40 ms of the tetanus (Fig. 1B, brown, dark orange, light orange). The shoulder on the high-angle side of the main peak at a reciprocal spacing of approximately 1/14.16 nm−1 disappeared, and by 40 ms (Fig. 1B, light orange) a shoulder had become evident on the low angle side. As force continued to develop, the reflection resolved into two major peaks with reciprocal spacings of approximately 1/14.47 nm−1 and 1/14.66 nm−1 (Fig. 1B, yellow, light green, dark green). At the tetanus plateau (Fig. 1B, dark blue), the 14.66-nm peak was more intense than the 14.47-nm peak (8, 19). The total intensity of the M3 reflection (IM3), corrected for the decrease in lateral alignment between filaments on activation by multiplying by the radial width of the reflection (13), had a biphasic time course (Fig. 1C), with a minimum at approximately 40 ms. IM3 at the tetanus plateau was almost twice that at rest. The average real-space periodicity reported by the M3 reflection (SM3; Fig. 1D) increased faster than force development, and SM3 was 14.57 nm at the tetanus plateau. These IM3 and SM3 time courses are similar to those observed in previous studies in which the spatial resolution was too low to resolve the individual peaks (13, 15–17).
The multiple peaks of the M3 reflection are a result of X-ray interference between the two arrays of myosin heads in each myosin filament (19) (Fig. 1A). For each head in one half of the filament, there is a head in the other half that is separated from it by a distance L, which is approximately 60 times larger than d. This multiplies the simple diffraction peak corresponding to the head periodicity (d) by an interference fringe pattern, splitting the M3 reflection into two or three component peaks, and allowing the movements of the heads along the filaments to be determined with subnanometer precision (8–11).
The axial profile of the M3 reflection at each time point was accurately fitted by two or three Gaussian peaks of the same width (Fig. 2). Three peaks were required to fit the M3 profile in the resting state (Fig. 2A) and for the first 40 ms of stimulation, and two peaks at later times and at the tetanus plateau (Fig. 2B). Because the total intensity of the M3 reflection (IM3; Fig. 1C) is influenced by variable changes in lateral alignment between myosin filaments, as noted earlier, we used the fractional intensity of the component peaks (Fig. 2C) for detailed analysis. The lowest angle peak (LA; Fig. 2A, red) increased from a small shoulder at rest to become the dominant peak at the tetanus plateau. Despite its long latency, the change in the intensity of this peak was half complete by approximately 50 ms (Fig. 2C), similar to the half-time for force, and most of the fractional intensity change took place between 40 and 60 ms. The intensity of the central peak (MA; Fig. 2A, blue), which dominates the resting M3 reflection, decreased with essentially the same time course. The intensity of the high angle peak (HA; Fig. 2A, green) decreased in the first 40 ms and was subsequently undetectable. The real-space periodicities corresponding to the three peaks (Fig. 2D) increased together, with time courses similar to that of SM3 (Fig. 1D).
Fig. 2.
Analysis of the component peaks of the M3 reflection. (A and B) Black lines are axial profiles of the M3 reflection at rest (A) and at the tetanus plateau (B). Red, blue, and green lines are Gaussian functions fitted to the lower, middle, and higher angle peaks, respectively. (C) Fractional intensity and (D) axial spacing of the three peaks (mean ± SE, n = 5 fibers); continuous lines show mean calculated from the model described in the text.
Conformation of Myosin Heads in Resting Muscle.
To calculate the axial movements of the myosin heads from these X-ray interference data, we started from the conformation at the tetanus plateau that had been determined previously by imposing small length or force changes (8–11, 21). Approximately 30% of the myosin heads are attached to actin at the tetanus plateau, and their axial mass distribution can be represented by using the crystallographic structure of the myosin head with its catalytic domain bound to actin as in the absence of nucleotide (4, 5), and its light-chain domain tilted at 60° to the filament axis (Fig. 3A, Top, red), with a ± 17° uniform angular dispersion (9–11). The axial mass distribution of the actin-attached heads has a peak (zA) displaced by +3.27 nm from the myosin head–tail junction (Fig. 3B, red), where positive zA denotes a position further from the midpoint of the myosin filament. The other 70% of the heads are detached from actin (Fig. 3A, Top, orange), and were represented by a Gaussian mass distribution with peak displacement (zD) 2.07 nm and standard deviation (SD; σD) 3.6 nm (Fig. 3B, orange) (9, 10). The combined axial mass distributions of these attached and “active detached” heads gives a good fit to the profile of the M3 reflection at the tetanus plateau (Fig. 3C, thin red line).
Fig. 3.
Three populations of myosin heads during isometric force development: (A) Force-generating (Top, red), active detached (Top, orange), and two possible resting conformations (Middle and Lower, green). The globular catalytic domain of the head binds to actin, and the more elongated light chain domain connects it to the myosin tail in the filament backbone. (B) Axial mass distributions of these three conformations, and from isolated thick filaments (22) (black). (C) Comparison of experimental and model M3 axial profiles at rest (black and thick red continuous lines, respectively) and at the tetanus plateau (blue and thin red line). (D) Fraction of heads in the resting (fR, green), active detached (fD, orange), and force-generating (fA, red) conformations, and mean center of mass (CoM) of the heads (violet).
Knowing the axial mass distribution of the myosin heads at the tetanus plateau, their distribution in resting muscle can be calculated from the difference between the axial profiles and intensities of the M3 reflection in the two states. The only free parameters are the peak position (zR) and dispersion (σR) of the resting distribution. A good fit to the resting M3 profile (Fig. 3C, thick red line) was obtained with a Gaussian mass distribution with a zR of −0.80 + 7.17n nm (where n is an integer) and SD (σR) of 3.5 nm, close to the σD value of 3.6 nm for the active detached heads. A more detailed analysis, taking into account the nonuniform spacing of the three layers of myosin heads in each helical repeat of approximately 43 nm, gave a slightly smaller estimate of σR of 3.2 nm (SI Text). The large transient decrease in IM3 during force development was not reproduced by the n = 0 (zR = −0.80 nm) solution, and the size of the myosin head excludes n > 1 and n < −1. Thus, zR is either +6.37 or −7.97 nm (Fig. 3B, green).
This 7-nm offset of the center of mass of the myosin head from its head–tail junction can only be achieved if the light chain domain of the head is roughly parallel to the filament axis (Fig. 3A, green). Such a conformation has been observed in isolated thick filaments by EM (22, 23), and the axial mass distribution of the head conformations relative to the head–tail junction reported in those studies (Fig. 3B, black) is close to that for the −7.97 nm solution. A Gaussian fit to the mass distribution in isolated filaments gave a zR of −7.4 nm and σR of 3.05 nm, close to the parameters determined here (zR = −7.97 nm; σR = 3.2 or 3.5 nm; Fig. 3B, thicker green line). The zR = −7.97 nm solution, in which the heads are folded back along the myosin tail toward the M line of the sarcomere (Fig. 3A, Lower), is therefore likely to represent the conformation of myosin heads in resting muscle. It follows that, during muscle activation, the center of mass of the heads moves by approximately 10 nm (from the green peak at left to the red and orange peaks in Fig. 3B) away from the midpoint of the sarcomere, i.e., in the opposite direction to the working stroke in the actin-attached myosin heads. This general conclusion is not affected by our simplifying assumption that the mass distribution of the resting heads has a Gaussian shape, although we cannot exclude the possibility that a small fraction of the heads in resting muscle take up a markedly different conformation. For example, the present results are consistent with as many as 10% of the heads in resting muscle being in the active detached conformation.
Time Course of Axial Motion of Myosin Heads During Force Development.
A similar analysis was applied to the X-ray data collected at each time point during force development. As stiffness measurements indicate that the time course of myosin head attachment to actin is almost the same as that of force development (17), we constrained the fraction of attached heads (fA) to be proportional to the force at each time point, leaving the resting fraction (fR) as the only free parameter to be determined by fitting the M3 profile. The detached fraction (fD) is given by (1− fA − fR). The axial periodicity of the heads, assumed to be the same for the three populations at each time point, was also determined from the fit. This model reproduced the shape of the observed M3 profiles (Fig. S1), the relative intensities and spacings of the three M3 components at each time point (Fig. 2 C and D, lines), and the total intensity and spacing of the M3 reflection (Fig. 1 C and D, dashed line).
The time course of the decrease in the fraction of resting heads (fR) was sigmoidal (Fig. 3D, green), with a half-time (t1/2) of 48.6 ± 1.4 ms (mean ± SE; n = 5), the same as that of the increase in the fraction of active detached heads (fD; Fig. 3D, orange), 47.2 ± 1.7 ms, and slightly faster than that of the attached heads (fA; Fig. 3D, red) and force development, 53.8 ± 1.8 ms. The motion of the mean center of mass of the myosin heads (Fig. 3D, violet), with a t1/2 of 48.8 ± 1.4 ms, also led force development by approximately 5 ms, and this difference was marginally significant at the 5% level in a paired t test (P ∼ 0.03; n = 5). Thus, the changes in the axial profile of the M3 reflection during force development can be quantitatively explained by the progressive loss of heads in the resting conformation and their replacement by a combination of attached and active detached heads with the same conformation as at the tetanus plateau. Although the present results provide no direct evidence for a transient population of heads in a “prepower stroke” state during isometric force development, the axial movement of the heads seems to lead force development by a few milliseconds.
Sequence of Structural Changes in Myosin and Actin Filaments During Isometric Force Development.
The earliest structural changes following electrical stimulation of skeletal muscle are in the thin filaments. The intracellular free calcium transient peaks approximately 10 ms after the first stimulus (12), at approximately the same time as latency relaxation and the loss of the viscoelasticity characteristic of resting muscle (17). The intensity of the T1 meridional X-ray reflection (IT1) associated with the approximately 38-nm repeat of troponin along the thin filaments increases during latency relaxation (Fig. 4A), signaling a change in the structure of troponin on binding calcium (18, 24, 25). However, this IT1 increase is transient, and IT1 has already fallen to near its tetanus plateau value by approximately 50 ms, when force is only half-maximum. The azimuthal motion of tropomyosin around the thin filaments, indicated by the increase in intensity of the second actin-based layer line, is approximately 90% complete when force is only approximately 10% of its maximum value (14, 18), approximately 20 ms after the first stimulus in the present conditions.
Fig. 4.
Changes in the intensities of other X-ray reflections during isometric force development. (A) Meridional associated with troponin (T1). (B) First myosin layer line (ML1). (C) First myosin-based meridional (M1). (D) Second myosin-based meridional (M2). Line is force normalized by its plateau value. Error bars denote ± SE for the five fibers in Fig. 1.
The next set of structural changes take place in the thick filaments (13–17). The intensity of the first myosin layer line (IML1), associated with the quasi-helical organization of the myosin heads on the surface of the resting thick filaments, decreased with a t1/2 of 30.5 ± 2.2 ms in these fibers (Fig. 4B). The intensities of the M1 X-ray reflection (IM1; Fig. 4C) associated with the 44-nm periodicity of the thick filaments and with myosin binding protein C, and its second order, the M2 reflection (IM2; Fig. 4D), decreased with t1/2 values of 23.7 ± 0.9 ms and 23.8 ± 1.2 ms, respectively. These t1/2 values may underestimate those of the underlying structural changes because the fraction of diffractors in a given state is more closely related to the reflection amplitude, i.e., the square root of the intensity. However, t1/2 values for the amplitudes of the ML1, M1, and M2 reflections—39.4, 27.3, and 30.6 ms, respectively—are still significantly smaller than that of force development. The axial periodicity of the backbone of the thick filament, reported by the spacing of the M6 axial reflection (9, 11), increases by approximately 1.6% during force development with a t1/2 of 32 ms after the first stimulus (17), also much faster than force development.
The signaling pathway leading from activation of the thin filaments to these structural changes in the thick filaments is not understood. The widely accepted paradigm for regulation of striated muscle contraction assumes that the azimuthal motion of tropomyosin in the thin filament is sufficient to allow myosin head binding and force generation, implying that the structural changes in the thick filament are triggered in some way by myosin head binding to actin. However, this hypothesis, at least in its simplest form, seems difficult to reconcile with the finding that nearly all the myosin heads in resting muscle are folded back against the filament backbone (Fig. 3A, green), and that the structural changes in the thick filament are significantly faster than both the strong binding of myosin to actin as determined by stiffness measurements (17) and force generation itself. These considerations suggest the existence of a fast signaling pathway between the thin and thick filaments. Such signaling might be mediated by the detachment of the mechanical links between the thick and thin filaments that are responsible for the resting viscoelasticity, a switch-like process that is synchronous with the calcium-dependent change in troponin structure (26). The identity and function of such “resting links” remain to be established, although a recent X-ray study on insect flight muscle (27) implicated myosin–troponin connections in a related context. The present results do not exclude the possible involvement in the signaling pathway of a small fraction (10% or less) of myosin heads that are not folded back against the thick filament in resting muscle, and the conventional view would be that such heads might detect the activation state of the thin filament by weak binding to actin. However, such a mechanism would not readily explain the switch-like loss of the resting viscoelasticity, suggesting that, as in insect flight muscle, other protein–protein interactions may be involved in the signaling between thin and thick filaments in vertebrate skeletal muscle.
The slowest structural change during force development is the net axial movement of the myosin heads, which leads force generation by only approximately 5 ms. Within the temporal resolution of the current structural and previous mechanical (17, 28) measurements, i.e., a few milliseconds, the net axial movement of the heads is approximately synchronous with their strong binding to actin, but slower than the disordering of the myosin heads accompanying the earlier structural changes in the thick filaments described earlier. Moreover, at this temporal resolution, the attached head conformation seen during force development is the same as that at the tetanus plateau; no intermediate conformations related to de novo force generation at the start of the working stroke were detected. The working stroke in the myosin heads during physiological force development following electrical stimulation occurs on the millisecond timescale, like that accompanying force recovery after imposition of length steps at the tetanus plateau (2, 8, 21). The time course of force generation in physiological conditions is not limited by the speed of the working stroke, or by that of structural changes in the thin or thick filaments, but by the rate of strong myosin head attachment to actin.
Materials and Methods
Muscle Fibers and Experimental Protocol.
General methods for muscle fiber preparation, attachment to transducers, electrical stimulation, measurement of force, sarcomere length, fiber length, and cross-sectional area, and the adaptations of these methods for the study of vertically mounted muscle fibers at synchrotron beam lines have been described previously (17, 28). Frogs (Rana temporaria) were killed by decapitation and destruction of the brain and the spinal cord according to European Community Directive 86/609/EEC and the United Kingdom Animals Scientific Procedures Act of 1986. Single fibers dissected from the tibialis anterior muscle were mounted in Ringer solution (115 mM NaCl, 2.5 mM KCl, 1.8 mM CaCl2, 3 mM phosphate buffer, pH 7.1). Resting sarcomere length was set to 2.14 ± 0.02 μm. Fibers were selected for low tendon compliance and homogeneous sarcomere length; in the central two thirds of the fiber length, where X-ray measurements were made, the average sarcomere length dispersion was 0.6% (SD). Force was measured with a capacitance transducer. Changes in mean sarcomere length of central fiber segments during force development were measured with a striation follower. Fibers were electrically stimulated for 390 to 550 ms by 20- to 24-Hz trains of alternating polarity stimuli delivered every 4 min at 4 °C.
X-Ray Data Collection.
Fibers were mounted vertically at beam line ID02 of the European Synchrotron Radiation Facility (ESRF; Grenoble, France). The full width at half-maximum of the X-ray beam at the fiber was approximately 100 μm vertically and 300 μm horizontally. The beam provided up to 2 × 1013 photons s−1 at 0.1 nm wavelength, and was attenuated for fiber alignment. X-ray exposure was controlled by using a fast electromagnetic shutter. The fiber was moved vertically between tetani to spread the radiation dose. There was no sign of radiation damage after the 40 to 120 5-ms exposures made in each fiber. X-ray data were collected using a FReLoN CCD-based detector with active area 200 × 200 mm2. The recorded sequence of time frames was smoothed with a 1:2:1 algorithm, and results are presented at 10-ms intervals. The camera length was 10 m, allowing resolution of the interference fine structure in the M3 axial reflection. The 2,048 × 2,048 pixels of the CCD were binned by 16 in the horizontal direction and by two in the vertical direction before readout to increase signal-to-noise ratio. X-ray data are presented from five fibers with cross-sectional area 20,400 ± 7,900 μm2 (mean ± SD) and isometric plateau force (T0) 194 ± 16 kPa.
Data Analysis.
X-ray diffraction data were analyzed using the SAXS package (P. Boesecke, ESRF), Fit2D (A. Hammersley, ESRF), and IgorPro (WaveMetrix). Diffraction patterns were centered and aligned using the equatorial 1,1 reflections, and mirrored horizontally and vertically. The intensity distribution along the meridian of the X-ray pattern was calculated by integrating radially to 0.012 nm−1 on each side of the meridian for the M3 reflection, and to 0.0046 nm−1 for the M1 and M2 reflections. The first myosin layer line (ML1) was integrated radially from 0.037 to 0.064 nm−1. Background intensity was fitted by using a convex hull algorithm or straight-line fit and subtracted; the small residual background was removed using the intensity from a nearby region containing no reflections. Integration along the meridian for the M3, M2, M1, T1, and ML1 reflections was from 0.067 to 0.072 nm−1, 0.046 to 0.048 nm−1, 0.021 to 0.024 nm−1, 0.025 to 0.027 nm−1, and 0.019 to 0.023 nm−1, respectively. The radial width of the M3 reflection was determined by Gaussian fitting. Reduction in intensities of the M1, M2, and M3 reflections caused by axial misalignment between neighboring filaments was corrected by multiplying by the radial width of the M3 reflection. No correction was applied for the T1 reflection. The axial profile of the M3 reflection was fitted by multiple Gaussian peaks with the same axial width, and its spacing calculated as the weighted mean of these peaks, assuming a resting spacing of 14.34 nm. The combined instrumental point spread function of the beam and detector was negligible compared with the radial width of the reflections. Half-times were estimated by sigmoidal fitting data from seven time points at 10-ms intervals around the half-maximum change.
Calculation of M3 Intensity Profiles from Structural Models.
A previous structural model for the thick filament during maximal activation (10) was modified by adding a “resting” (R) population of myosin heads to the “attached” (A) and “detached” (D) populations considered before, as described earlier. Essentially the same results were obtained with a more complicated structural model in which the two heads of each myosin molecule were treated explicitly (29), and with a model in which the light-chain domains of the A heads had a Gaussian rather than a uniform angular distribution. The A, D, and R populations have the same axial periodicity and diffract coherently. The approximately 1.6% increase in the axial periodicity of the thick filament observed at maximal activation was assumed to be confined to the region of the filament containing myosin heads; the periodicity of the bare zone was assumed to increase by 0.5%, corresponding to the thick filament compliance (9). The conclusions were not significantly different when the entire filament was assumed to undergo the approximately 1.6% periodicity change, except that the calculated transient decrease in IM3 approximately 50 ms after stimulation became larger than the observed decrease. The calculated M3 meridional intensity profiles were convoluted with the instrumental point-spread function (full width at half maximum, 290 μm), then analyzed as for the experimental profiles.
Supplementary Material
Acknowledgments
We thank M. Dolfi and J. Gorini for electronic and mechanical engineering support. This study was supported by Fondo per gli Investimenti della Ricerca di Base Futuro in Ricerca Project RBFR08JAMZ_001; and by Ministero dell'Istruzione, dell'Università e della Ricerca; Ente Cassa di Risparmio di Firenze; National Institutes of Health; Medical Research Council; Royal Society (United Kingdom); European Molecular Biology Laboratory; and European Synchrotron Radiation Facility.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018330108/-/DCSupplemental.
References
- 1.Huxley HE. The mechanism of muscular contraction. Science. 1969;164:1356–1365. doi: 10.1126/science.164.3886.1356. [DOI] [PubMed] [Google Scholar]
- 2.Huxley AF, Simmons RM. Proposed mechanism of force generation in striated muscle. Nature. 1971;233:533–538. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- 3.Lymn RW, Taylor EW. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry. 1971;10:4617–4624. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- 4.Rayment I, et al. Three-dimensional structure of myosin subfragment-1: A molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 5.Rayment I, et al. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 6.Geeves MA, Holmes KC. The molecular mechanism of muscle contraction. Adv Protein Chem. 2005;71:161–193. doi: 10.1016/S0065-3233(04)71005-0. [DOI] [PubMed] [Google Scholar]
- 7.Irving M, Lombardi V, Piazzesi G, Ferenczi MA. Myosin head movements are synchronous with the elementary force-generating process in muscle. Nature. 1992;357:156–158. doi: 10.1038/357156a0. [DOI] [PubMed] [Google Scholar]
- 8.Piazzesi G, et al. Mechanism of force generation by myosin heads in skeletal muscle. Nature. 2002;415:659–662. doi: 10.1038/415659a. [DOI] [PubMed] [Google Scholar]
- 9.Reconditi M, et al. The myosin motor in muscle generates a smaller and slower working stroke at higher load. Nature. 2004;428:578–581. doi: 10.1038/nature02380. [DOI] [PubMed] [Google Scholar]
- 10.Piazzesi G, et al. Skeletal muscle performance determined by modulation of number of myosin motors rather than motor force or stroke size. Cell. 2007;131:784–795. doi: 10.1016/j.cell.2007.09.045. [DOI] [PubMed] [Google Scholar]
- 11.Huxley HE, Reconditi M, Stewart A, Irving TC. X-ray interference studies of crossbridge action in muscle contraction: Evidence from quick releases. J Mol Biol. 2006;363:743–761. doi: 10.1016/j.jmb.2006.08.075. [DOI] [PubMed] [Google Scholar]
- 12.Caputo C, Edman KA, Lou F, Sun Y-B. Variation in myoplasmic Ca2+ concentration during contraction and relaxation studied by the indicator fluo-3 in frog muscle fibres. J Physiol. 1994;478:137–148. doi: 10.1113/jphysiol.1994.sp020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huxley HE, Faruqi AR, Kress M, Bordas J, Koch MHJ. Time-resolved X-ray diffraction studies of the myosin layer-line reflections during muscle contraction. J Mol Biol. 1982;158:637–684. doi: 10.1016/0022-2836(82)90253-4. [DOI] [PubMed] [Google Scholar]
- 14.Kress M, Huxley HE, Faruqi AR, Hendrix J. Structural changes during activation of frog muscle studied by time-resolved X-ray diffraction. J Mol Biol. 1986;188:325–342. doi: 10.1016/0022-2836(86)90158-0. [DOI] [PubMed] [Google Scholar]
- 15.Martin-Fernandez ML, et al. Time-resolved X-ray diffraction studies of myosin head movements in live frog sartorius muscle during isometric and isotonic contractions. J Muscle Res Cell Motil. 1994;15:319–348. doi: 10.1007/BF00123484. [DOI] [PubMed] [Google Scholar]
- 16.Piazzesi G, et al. Changes in conformation of myosin heads during the development of isometric contraction and rapid shortening in single frog muscle fibres. J Physiol. 1999;514:305–312. doi: 10.1111/j.1469-7793.1999.305ae.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunello E, et al. Structural changes in the myosin filament and cross-bridge formation during the development of the isometric force in single fibres from frog muscle. J Physiol. 2006;577:971–984. doi: 10.1113/jphysiol.2006.115394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuo T, Yagi N. Structural changes in the muscle thin filament during contractions caused by single and double electrical pulses. J Mol Biol. 2008;383:1019–1036. doi: 10.1016/j.jmb.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Linari M, et al. Interference fine structure and sarcomere length dependence of the axial x-ray pattern from active single muscle fibers. Proc Natl Acad Sci USA. 2000;97:7226–7231. doi: 10.1073/pnas.97.13.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huxley HE, Brown W. The low-angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J Mol Biol. 1967;30:383–434. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- 21.Irving M, et al. Conformation of the myosin motor during force generation in skeletal muscle. Nat Struct Biol. 2000;7:482–485. doi: 10.1038/75890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodhead JL, et al. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436:1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 23.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci USA. 2008;105:2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugimoto Y, et al. Structural changes of the regulatory proteins bound to the thin filaments in skeletal muscle contraction by X-ray fiber diffraction. Biochem Biophys Res Commun. 2008;369:100–108. doi: 10.1016/j.bbrc.2007.11.088. [DOI] [PubMed] [Google Scholar]
- 25.Matsuo T, Iwamoto H, Yagi N. Monitoring the structural behavior of troponin and myoplasmic free Ca2+ concentration during twitch of frog skeletal muscle. Biophys J. 2010;99:193–200. doi: 10.1016/j.bpj.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yagi N. An x-ray diffraction study on early structural changes in skeletal muscle contraction. Biophys J. 2003;84:1093–1102. doi: 10.1016/S0006-3495(03)74925-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perz-Edwards RJ, et al. X-ray diffraction evidence for myosin-troponin connections and tropomyosin movement during stretch activation of insect flight muscle. Proc Natl Acad Sci USA. 2011;108:120–125. doi: 10.1073/pnas.1014599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunello E, et al. Structural changes in myosin motors and filaments during relaxation of skeletal muscle. J Physiol. 2009;587:4509–4521. doi: 10.1113/jphysiol.2009.176222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brunello E, et al. Skeletal muscle resists stretch by rapid binding of the second motor domain of myosin to actin. Proc Natl Acad Sci USA. 2007;104:20114–20119. doi: 10.1073/pnas.0707626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.