Abstract
Yeast Hsp104 and its bacterial homolog, ClpB, are Clp/Hsp100 molecular chaperones and AAA+ ATPases. Hsp104 and ClpB collaborate with the Hsp70 and DnaK chaperone systems, respectively, to retrieve and reactivate stress-denatured proteins from aggregates. The action of Hsp104 and ClpB in promoting cell survival following heat stress is species-specific: Hsp104 cannot function in bacteria and ClpB cannot act in yeast. To determine the regions of Hsp104 and ClpB necessary for this specificity, we tested chimeras of Hsp104 and ClpB in vivo and in vitro. We show that the Hsp104 and ClpB middle domains dictate the species-specificity of Hsp104 and ClpB for cell survival at high temperature. In protein reactivation assays in vitro, chimeras containing the Hsp104 middle domain collaborate with Hsp70 and those with the ClpB middle domain function with DnaK. The region responsible for the specificity is within helix 2 and helix 3 of the middle domain. Additionally, several mutants containing amino acid substitutions in helix 2 of the ClpB middle domain are defective in protein disaggregation in collaboration with DnaK. In a bacterial two-hybrid assay, DnaK interacts with ClpB and with chimeras that have the ClpB middle domain, implying that species-specificity is due to an interaction between DnaK and the middle domain of ClpB. Our results suggest that the interaction between Hsp70/DnaK and helix 2 of the middle domain of Hsp104/ClpB determines the specificity required for protein disaggregation both in vivo and in vitro, as well as for cellular thermotolerance.
Keywords: Hsp40, DnaJ, M-domain, GrpE, nucleotide exchange factor
Hsp104 of yeast and ClpB of bacteria are homologous ATP-dependent molecular chaperones belonging to the Clp/Hsp100 subfamily of the AAA+ (ATPases associated with various cellular activities) superfamily of proteins (1–3). Hsp104 and ClpB are essential for thermotolerance (4, 5) and possess the power to reactivate proteins from insoluble aggregates (6–10). In yeast, during nonstress conditions, Hsp104 is also essential for the propagation of prions (11), which are self-replicating, ordered protein aggregates.
Hsp104 and ClpB consist of four domains: an N-terminal domain (NTD), two AAA nucleotide-binding domains (NBD-1 and NBD-2), and a middle domain (M-domain), which is located within NBD-1 [Fig. 1A; (12)]. Hsp104/ClpB has a hexameric ring structure with a central channel (13–15), resembling other Clp/Hsp100 proteins (16, 17). The current evidence suggests that Hsp104/ClpB uses the energy from ATP hydrolysis to disassemble aggregates by unfolding and translocating the unfolded polypeptides through the Hsp104/ClpB central channel (1, 3, 18, 19).
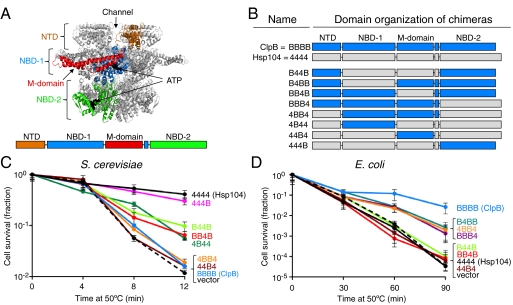
Fig. 1.
Chimeras having the Hsp104 M-domain provide thermotolerance in yeast, while chimeras containing the ClpB M-domain promote cell survival at high temperature in E. coli. (A) ClpB hexamer model of the ClpB protomer was generated (35) based on the T. thermophilus ClpB (pdb:1qvr—chain A) (12) and prepared using PyMOL (www.pymol.org). A diagram of the ClpB/Hsp104 domains is also shown. (B) Domain organization of the ClpB-Hsp104 chimeras with domains from ClpB indicated by “B” and shown in blue, and domains from Hsp104 indicated by “4” and shown in gray. (C) Thermotolerance of S. cerevisiae Δhsp104 cells expressing plasmid-encoded Hsp104, ClpB, or the indicated chimera was determined as described in Methods. (D) Survival of E. coli clpB- cells expressing plasmid-encoded ClpB, Hsp104, or the indicated chimera at elevated temperature was determined as described in Methods. For C and D, data are means ± SEM (n = 3).
Protein disaggregation by Hsp104 and ClpB in vitro requires the collaboration of a second ATP-dependent molecular chaperone, Hsp70/DnaK (6, 7, 9, 10). Hsp70 and DnaK operate with two cochaperones (20, 21); Hsp40 and NEF (nucleotide exchange factor) function with Hsp70 while DnaJ and GrpE, a bacterial nucleotide exchange factor, act with DnaK. Hsp40/DnaJ stimulates Hsp70/DnaK ATPase activity and targets substrates for recognition by Hsp70/DnaK. NEF/GrpE facilitates the exchange of Hsp70/DnaK-bound nucleotide, thereby triggering the release of the substrate from Hsp70/DnaK. The cooperation between Hsp104/ClpB and the Hsp70/DnaK system is specific: Hsp104 acts with the eukaryotic Hsp70 system and ClpB functions with the prokaryotic DnaK system (6, 22). Recently, it was reported that the Hsp104 and ClpB M-domain imparts the specificity between Hsp104/ClpB and Hsp70/DnaK in protein disaggregation in vitro (23). The specificity between Hsp70/DnaK and Hsp104/ClpB implies a direct interaction between the chaperones. Interactions have been observed in vitro between Escherichia coli ClpB and the substrate-binding domain of DnaK (24), between ClpB and DnaK from Thermus thermophilus (25), and between Hsc70 of plants and Hsp101, the Hsp104 homolog in plants (26).
Hsp104 and ClpB have 43% sequence identity and are structural homologs. However, thermotolerance in yeast and bacteria is species-specific (27). “Species-specific” is used here to mean that Saccharomyces cerevisiae Hsp104 cannot function in bacteria or in vitro with prokaryotic DnaK, and E. coli ClpB cannot act in yeast or in vitro with eukaryotic Hsp70. It has been observed that neither the NTD (28, 29) nor NBD-2 is responsible for the species-specificity seen in the thermotolerance process (27). Despite the incompatibility between yeast and bacterial chaperones, functional collaborations of Hsp104 and Hsp70 from distantly related eukaryotic organisms have been shown. For example, mammalian cytoplasmic Hsp70 can cooperate with Hsp104 in providing yeast thermotolerance and in refolding aggregated proteins in vivo and in vitro (6, 18, 30).
To determine the region(s) of Hsp104 and ClpB necessary for the specific action of Hsp104 in S. cerevisiae and ClpB in E. coli, we constructed and tested a series of chimeras in which we exchanged homologous regions of Hsp104 and ClpB. Our results from in vivo and in vitro experiments suggest that species-specific interactions between Hsp70/DnaK and helix 2 of the Hsp104/ClpB M-domain are required for protein disaggregation and cellular thermotolerance.
Results
The M-Domain of Hsp104 and ClpB Confers the Species-Specificity for Thermotolerance.
To identify the region(s) of Hsp104 and ClpB that determine the specific collaborations required for survival during severe heat stress, a series of chimeras were constructed. Based on sequence alignment of E. coli ClpB and S. cerevisiae Hsp104 and the T. thermophilus ClpB crystal structure (12), we exchanged one or more domain(s) from one chaperone with the corresponding domain(s) from the homologous chaperone (Fig. 1 A and B). The chimeras are designated by four characters representing the four domains of Hsp104/ClpB from the N to C terminus of the polypeptide. “B” denotes that the domain is derived from ClpB, and “4” denotes that the domain is from Hsp104. For example, B44B refers to the chimera with the NTD and NBD-2 from ClpB, and NBD-1 and the M-domain from Hsp104. All of the chimeras used in these studies formed hexamers in the presence of ATP and possessed ATPase activity in vitro (SI Text, Fig. S1 and S2).
We first wanted to determine which domain(s) of Hsp104 are required for a chimera to impart thermotolerance in yeast. We expressed plasmid-encoded Hsp104, ClpB, or chimeras under the control of the HSP104 promoter in a S. cerevisiae Δhsp104 strain and measured cell survival after exposure to heat stress at 50 °C. All of the chimeras were expressed in S. cerevisiae, as observed by Western blot analysis (Fig. S3A). The chimera, 444B, provided protection from heat nearly equal to that provided by Hsp104, as previously shown [Fig. 1C, Fig. S3B (27)]. We observed that B44B, BB4B, and 4B44 conferred ∼5-fold more protection from heat than the vector alone, although this level was ∼5-fold less than for 444B and wild type Hsp104 (Fig. 1C, Fig. S3B). All of the chimeras that promoted cell survival contain the Hsp104 M-domain; importantly, in BB4B only the M-domain is derived from Hsp104. Consistent with these findings, 4BB4, 44B4, and ClpB, all of which have the M-domain from E. coli ClpB, provided no additional protection from heat-induced killing over that seen for the vector control. Our observations demonstrate that a chimera requires the M-domain from Hsp104 to function in yeast thermotolerance.
We next tested the chimeras for the ability to support the growth of an E. coli clpB- strain at high temperature. Hsp104, ClpB, or chimeras were expressed from plasmids under the control of an arabinose-inducible promoter and cell survival at 50 °C was measured. Western blot analysis showed that the chimeras were expressed in E. coli (Fig. S3C). B4BB, 4BB4, and BBB4 increased cell survival by ∼30-fold in comparison to the vector control (Fig. 1D, Fig. S3D). Notably, these three chimeras all contain the M-domain of ClpB, however ClpB provided ∼10-fold greater protection from heat stress than these chimeras. Hsp104 and chimeras containing the M-domain of Hsp104, B44B, and BB4B, did not enable E. coli clpB- cells to survive at high temperature (Fig. 1D, Fig. S3D). The chimera with only the M-domain derived from ClpB, 44B4, did not provide protection from heat; however, this chimera is not active in protein reactivation in vitro, shown below. These observations imply that for a chimera to confer protection against heat-induced killing in E. coli, the ClpB M-domain is necessary. Taken together, the results suggest that the M-domain of Hsp104 and ClpB determines the species-specificity for the thermotolerance process in S. cerevisiae and E. coli.
The Hsp104 M-Domain Is Required for Chimeras to Function in Protein Reactivation in Yeast.
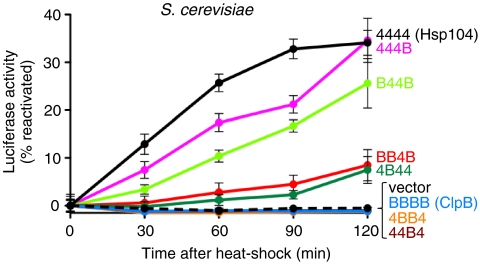
We tested whether the chimeras could reactivate a specific heat-inactivated substrate, luciferase, in yeast. Plasmids expressing Hsp104, ClpB, or chimeras under the control of the HSP104 promoter were introduced into S. cerevisiae Δhsp104 cells harboring a second plasmid encoding firefly luciferase. After induction of the chimeras at 37 °C, the temperature was shifted to 44 °C for 1 h to inactivate luciferase. Then the temperature was decreased to 25 °C and luciferase activity was monitored (Fig. 2). Hsp104 and the chimeras possessing the M-domain of Hsp104, 444B, B44B, BB4B, and 4B44, promoted luciferase reactivation (Fig. 2). Conversely, ClpB and the chimeras tested that contained the ClpB M-domain, 4BB4, and 44B4, were unable to reactivate luciferase. These results demonstrate the importance of the Hsp104 M-domain for compatibility with the yeast protein disaggregation machinery.
Fig. 2.
Chimeras possessing the Hsp104 M-domain reactivate heat-inactivated luciferase in yeast. In vivo luciferase reactivation was determined in Δhsp104 yeast cells expressing firefly luciferase and Hsp104, ClpB, or the indicated chimera following heat inactivation as described in Methods. Data are averages of three independent experiments  .
.
The M-Domain of Hsp104 and ClpB Provides the Specificity for the Hsp70 and DnaK System, Respectively, in Protein Reactivation In Vitro.
We next tested whether the M-domain acts as a specificity determinant in vitro using purified proteins. It has been shown that both Hsp104 and ClpB are able to reactivate aggregated proteins in vitro (6, 10). Moreover, Hsp104 disaggregates protein aggregates in conjunction with the eukaryotic Hsp70 chaperone system [composed of human or yeast Hsp70 and yeast Hsp40 (6, 18)] and is unable to function with the bacterial DnaK system [(6), Fig. 3]. Similarly, E. coli ClpB functions with the E. coli DnaK system and is inactive with the eukaryotic cytoplasmic Hsp70 system [(22), Fig. 3].
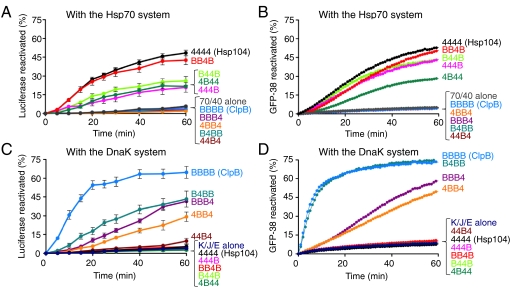
Fig. 3.
The M-domain of Hsp104 and ClpB provides the specificity for the Hsp70 and DnaK system, respectively, in protein disaggregation in vitro. (A, C) Reactivation of urea-denaturated luciferase by Hsp104, ClpB, or the indicated chimera in the presence of the Hsp70 (A) or DnaK (C) system was measured as described in Methods. Data are means ± SEM (n = 3). (B, D) Reactivation of heat-aggregated GFP-38 by Hsp104, ClpB, or the indicated chimera in the presence of the Hsp70 (B) or DnaK (D) system was measured as described in Methods. A representative experiment of three replicates is shown.
We first measured reactivation of urea-denatured luciferase by our chimeras in combination with a eukaryotic cytoplasmic Hsp70 system, comprised of human Hsp70 and yeast Hsp40. We found that all of the chimeras containing the M-domain of Hsp104 reactivated luciferase, while chimeras possessing the M-domain of ClpB were unable to cooperate with Hsp70 and Hsp40 (Fig. 3A). However, the chimeras exhibited different relative luciferase reactivation activities in vitro (Fig. 3A) and in vivo (Fig. 2), suggesting that different conditions in vivo and in vitro might affect the activity of the chimera.
We also tested the ability of the chimeras to collaborate with the Hsp70 system in the reactivation of another substrate, heat-aggregated GFP-38, which is a GFP fusion protein containing a 38 amino acid C-terminal polypeptide. We observed that chimeras with the M-domain of Hsp104 were able to reactivate heat-inactivated GFP-38 in the presence of the Hsp70 system, while those containing the ClpB M-domain were not (Fig. 3B). These results corroborate the luciferase reactivation experiments showing that the Hsp104 M-domain provides the specificity for the Hsp70 system.
Reactivation of urea-denatured luciferase and heat-inactivated GFP-38 by the chimeras was also examined in combination with the E. coli DnaK system. We observed that both substrates were reactivated by the chimeras that contained the ClpB M-domain when incubated with DnaK, DnaJ, and GrpE (Fig. 3 C and D). An exception was 44B4, which was not active with either the DnaK or Hsp70 system*. As expected, chimeras containing the Hsp104 M-domain were unable to reactivate either substrate with the DnaK system (Fig. 3 C and D).
Our data suggest that the M-domain determines the specific collaboration between Hsp104/ClpB and the Hsp70/DnaK system in protein reactivation in vitro. Taken together with the in vivo luciferase reactivation data, these results imply that the cooperation between the M-domain of Hsp104/ClpB and the Hsp70/DnaK system is required for protein disaggregation in vivo. Due to the requirement for protein disaggregation for thermotolerance (8), it can be inferred that this same collaboration is important for thermotolerance.
The M-Domain of ClpB Interacts Specifically with DnaK In Vivo.
The in vitro protein reactivation results suggest that the Hsp70/DnaK system collaborates with Hsp104/ClpB through contacts between the Hsp104/ClpB M-domain and Hsp70/DnaK or Hsp40/DnaJ. A previous study demonstrated that Hsp104 is able to reactivate protein aggregates in vitro in combination with S. cerevisiae Hsp70 and E. coli DnaJ (6). Therefore, it appeared likely that the component of the Hsp70/DnaK system that interacts with the Hsp104/ClpB M-domain is Hsp70/DnaK. To test this possibility, we used a bacterial two-hybrid system (31). DnaK was fused to the C terminus of one fragment of Bordetella pertussis adenylate cyclase, T25. ClpB, Hsp104, or a chimera was fused to the C terminus of the other fragment, T18. If DnaK and the chimera being tested interact, the T18 and T25 fragments reconstitute an active adenylate cyclase and a cAMP-dependent reporter gene, β-galactosidase, is expressed. T25-DnaK and the various T18-chimeras were cotransformed into an E. coli Δcya ΔclpB strain. Expression of fusion proteins was confirmed by Western blot analysis (Fig. S4A). In control experiments, when the T18 and T25 fragments alone were coexpressed, colonies appeared white on indicator plates and expressed basal levels of β-galactosidase (Fig. 4 A and B). When a leucine zipper domain was fused to both the T18 and T25 fragments (31) and coexpressed, colonies appeared red on indicator plates and expressed high levels of β-galactosidase (Fig. 4A, Fig. S4B). When ClpB was fused to each of the cyclase fragments and coexpressed, colonies expressed high levels of β-galactosidase, as expected for a multimeric protein (Fig. S4B).
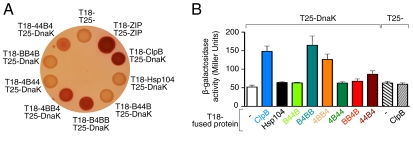
Fig. 4.
Chimeras containing the ClpB M-domain interact with DnaK in vivo. A bacterial two-hybrid system was used to detect interactions between chimeras and DnaK in vivo, as described in Methods. Interaction between DnaK and the target protein was monitored by the expression of β-galactosidase on MacConkey indicator plates (A) and in liquid assays (B). A representative plate from three independent experiments is shown in A and data in B are means ± SEM (n = 9).
We observed that cells coexpressing ClpB and DnaK formed red colonies on indicator plates and exhibited 3-fold higher β-galactosidase activity than the cells expressing DnaK and the T18 fragment alone (Fig. 4 A and B). These results suggest that ClpB and DnaK interact in vivo. Conversely, there was no detectable interaction between Hsp104 and DnaK (Fig. 4 A and B). Cells coexpressing DnaK and either B4BB or 4BB4 formed colonies that appeared red on indicator plates and had higher β-galactosidase activity than the control (3.2- and 2.5-fold, respectively), suggesting that there is a specific interaction between DnaK and these chimeras (Fig. 4 A and B). When 44B4, which contains only the M-domain of ClpB, was coexpressed with DnaK, colonies appeared pink on indicator plates and showed slightly higher β-galactosidase activity than the control (1.7-fold), suggestive of an interaction with DnaK (Fig. 4 A and B). There was no detectable interaction between DnaK and chimeras containing the Hsp104 M-domain, B44B, 4B44, and BB4B (Fig. 4 A and B). Taken together, these results suggest that DnaK interacts specifically with E. coli ClpB via the ClpB M-domain.
Helix 2 and Helix 3 of the Hsp104/ClpB M-Domain Are Implicated in the Specificity for the Hsp70/DnaK System.
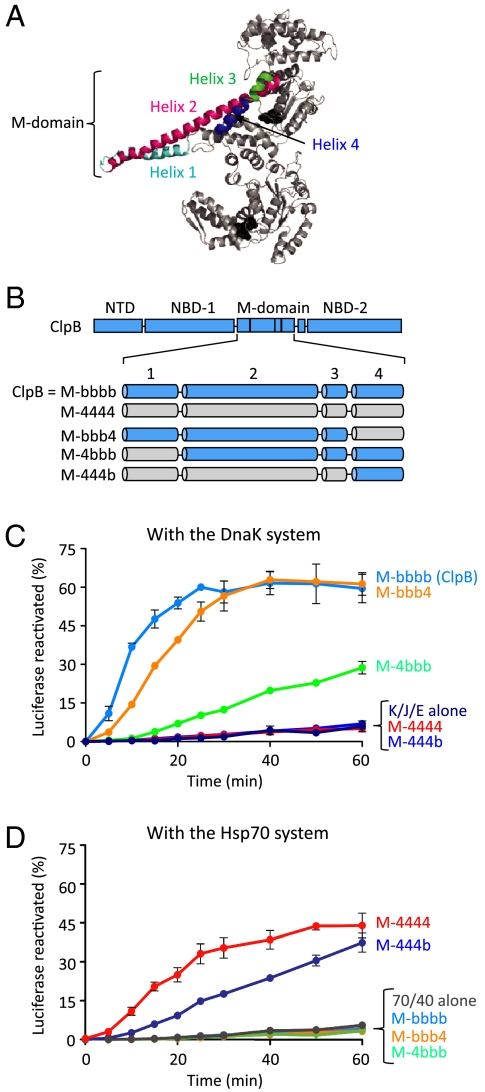
We next wanted to more precisely identify the region within the M-domain that is involved in the collaboration between Hsp104/ClpB and Hsp70/DnaK. We constructed several chimeras of ClpB in which one or more of the four M-domain helices were replaced with the corresponding Hsp104 helices (Fig. 5 A and B). To designate these ClpB chimeras we use “M-”, for M-domain chimera, followed by lowercase “b” to designate that the helix is from ClpB, and “4” to indicate that it is from Hsp104. For example, M-bbb4 denotes ClpB with M-domain helices 1, 2, and 3 from ClpB and helix 4 from Hsp104.
Fig. 5.
Helix 2 and helix 3 of the Hsp104/ClpB M-domain are implicated in the ClpB/Hsp104 specificity for the DnaK/Hsp70 system in vitro. (A) Monomer model of E. coli ClpB generated using the T. thermophilus ClpB structure (pdb:1qvr—chain B) showing M-domain helices. (B) Diagram of ClpB M-domain chimeras with M-domain helices originating from ClpB indicated by “b” and solid colors and helices originating from Hsp104 designated by “4” and hatched colors. (C, D) In vitro reactivation of urea-denaturated luciferase by ClpB or the ClpB M-domain chimeras was measured as described in Methods in the presence of the DnaK system (C) or the Hsp70 system (D). Data in C and D are means ± SEM (n = 3).
When we tested the ClpB M-domain chimeras in luciferase reactivation assays in vitro with the DnaK system, we observed that M-4bbb and M-bbb4 reactivated luciferase, while M-444b was unable to reactivate luciferase (Fig. 5C). These results show that helix 1 and helix 4 are not responsible for the specificity of ClpB for DnaK, suggesting that helix 2 and helix 3 are required for the collaboration. M-444b was able to reactivate luciferase with the Hsp70 system, while M-4bbb and M-bbb4 were inactive with the Hsp70 system (Fig. 5D), showing that helices 1–3 from Hsp104 are sufficient to promote the collaboration with the Hsp70 system. Taken together, the results imply that helices 2 and 3 of the Hsp104 and ClpB M-domain participate in the collaboration with the Hsp70 and DnaK systems, respectively.
Mutations in Helix 2 of the ClpB M-Domain Affect Cooperation with the DnaK Chaperone System.
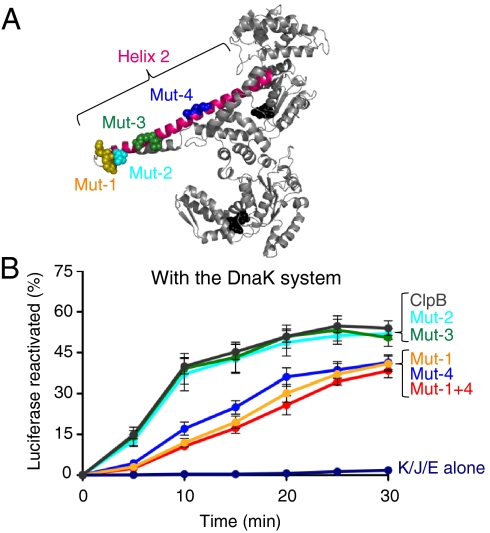
We wanted to identify residues within the E. coli ClpB M-domain that are important for collaboration with the DnaK system. Because it has been shown that helix 3 interacts with NBD-1 and modulates ClpB ATPase activity (32), it seemed more likely that helix 2 would be involved in the collaboration with DnaK. We made site-directed mutations in groups of one to three residues in helix 2 that were predicted to be surface exposed (Fig. 6A).
Fig. 6.
Amino acid substitutions within helix 2 of the ClpB M-domain alter collaboration between ClpB and the DnaK system in vitro. (A) Monomer model of E. coli ClpB showing Mut-1 (K438E,K439E,R440E) in yellow, Mut-2 (L444A) in aqua, Mut-3 (R453A,Q454A) in green, and Mut-4 (A467H,S470E,G471E) in blue. (B) In vitro reactivation of urea-denatured luciferase by ClpB M-domain mutants in the presence of the DnaK chaperone system was measured as described in Methods. Data are means ± SEM (n = 3).
The mutants were tested in vitro for their ability to reactivate urea-denatured luciferase in conjunction with the DnaK system. Mut-1 (ClpBK438E,K439E,R440E) and Mut-4 (ClpBA467H,S470E,G471E) reactivated luciferase at ∼30% of the rate of wild type ClpB in combination with DnaK, DnaJ, and GrpE (Fig. 6B, Table S1). A double mutant containing the substitutions of both Mut-1 and Mut-4 (Mut-1+4) was also defective in luciferase reactivation (Fig. 6B, Table S1). A similar defect in protein disaggregation activity by Mut-1, Mut-4, and Mut-1+4 in combination with the DnaK system was observed using two other model substrates, heat-aggregated malate dehydrogenase, and GFP (Fig. S5 A and B). Two additional M-domain mutants, Mut-2 (ClpBL444A) and Mut-3 (ClpBR453A,Q454A), which are located in helix 2 between Mut-1 and Mut-4, were indistinguishable from wild type ClpB in their ability to reactivate luciferase and other model substrates with the DnaK system (Fig. 6 A and B, Fig. S5 A and B). In control experiments, all of the helix 2 mutants had ATPase activity similar to wild type ClpB (Fig. S5C), formed hexamers in the presence of ATP (Fig. S1), and were similar to wild type in their ability to perform protein remodeling independent of the DnaK system (Fig. S5D). Taken together these results suggest that certain residues located in helix 2 of the M-domain are important for the collaboration between ClpB and the DnaK system.
Discussion
Our results show that the unique M-domain of Hsp104/ClpB determines species-specific tolerance to heat stress and protein reactivation in vivo and in vitro. Moreover, the results suggest that this specificity is likely derived from a region within helix 2 of the M-domain.
The bacterial two-hybrid experiments point to an interaction between DnaK and the ClpB M-domain; intermolecular interactions were detected between DnaK and ClpB and also between DnaK and chimeras possessing the ClpB M-domain. Moreover, no interaction was detected between DnaK and chimeras containing the M-domain of Hsp104, showing that the interaction is specific and likely biologically relevant. However, these experiments do not resolve whether the interaction between the M-domain of ClpB and DnaK is direct or mediated through another protein, such as the cochaperone DnaJ or a protein substrate. Previous studies have shown that the Hsp40/DnaJ component of the Hsp70/DnaK system is exchangeable between various organisms (6, 22). Thus, it is likely that Hsp70/DnaK is providing the interaction that determines species-specificity. Interestingly, while mammalian cells lack cytoplasmic Clp/Hsp100 proteins, mammalian cytoplasmic Hsp70 is able to function with yeast Hsp104 (30). These studies suggest that Hsp70 has retained the ability to interact with Hsp104 and that the site of interaction may have been conserved as part of another function of Hsp70.
Results from the characterization of Hsp104/ClpB M-domain chimeras indicated that helix 2 and helix 3 likely contain the region of collaboration with the Hsp70/DnaK system. Further evidence for the involvement of helix 2 in the specificity between ClpB and the DnaK system was provided by the ClpB helix 2 mutants that are defective in collaborating with the DnaK system. Previous work of Bukau and colleagues found that mutants in helix 3 of the M-domain are also defective in DnaK-dependent disaggregation (32). However, due to the proximity of helix 3 to NBD-1 and observed alterations in ClpB ATPase activity, it was suggested that helix 3 is involved in coupling the activity of DnaK with the protein threading function of ClpB (32). Together these observations suggest a role of helix 2 in interacting with DnaK and transmitting signals through helix 3, to NBD-1 of ClpB.
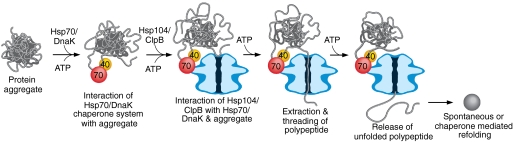
Building upon the current models for protein disaggregation by Hsp104/ClpB and the Hsp70/DnaK system (1–3), our data are most consistent with a model in which Hsp70/DnaK functions directly with Hsp104/ClpB in the disaggregation process (Fig. 7). The Hsp70/DnaK system interacts with the aggregate, likely performing some loosening and partial remodeling of the aggregate. Interactions between Hsp70/DnaK and a small region of the Hsp104/ClpB M-domain promote and stabilize Hsp104/ClpB binding to the aggregate. In addition, the interactions between Hsp70/DnaK and the M-domain of Hsp104/ClpB trigger signals that are propagated through the M-domain to NBD-1 of Hsp104/ClpB. These signals activate the coupling of ATP hydrolysis by Hsp104/ClpB to polypeptide unfolding and translocation. After the polypeptide is unfolded and translocated through the central channel of Hsp104/ClpB, it is released in an unfolded conformation. The polypeptide either spontaneously refolds or is refolded with the aid of cellular chaperones. Establishing the precise functions of the Hsp70/DnaK system in disaggregation with Hsp104/ClpB remains an important subject for future investigation.
Fig. 7.
Model illustrating the collaboration between Hsp104/ClpB and the Hsp70/DnaK system in protein disaggregation. The Hsp70/DnaK system binds the aggregate and may carry out some loosening and remodeling of the aggregate. Hsp104/ClpB directly interacts with the Hsp70/DnaK system through a small region of the Hsp104/ClpB M-domain, which promotes interaction of Hsp104/ClpB with the aggregate. The interaction with Hsp70/DnaK and Hsp104/ClpB causes transmission of signals through the Hsp104/ClpB M-domain to NBD-1, thereby coupling ATP hydrolysis to polypeptide unfolding and translocation. Following translocation, the unfolded polypeptide is released from Hsp104/ClpB and may spontaneously refold or is refolded with the assistance of cellular chaperones.
Methods
Strains, Plasmids, and Proteins.
E. coli Rosetta(DE3) ΔclpB and BTH101 ΔclpB were constructed as described (SI Text). SG22100 (E. coli MC4100 clpB∷kan) was from S. Gottesman (National Institutes of Health) (4). S. cerevisiae strain YKT52 Δhsp104 was from J. Weissman (University of California, San Francisco) (27). Plasmids used for in vivo assays, for protein purification and for the two-hybrid assay were constructed as described in SI Text. Proteins were prepared as described in SI Text. The chimeras and ClpB mutants used had properties similar to wild type ClpB and Hsp104 (Fig. S1, S2, S5C, S6). Protein concentrations given are for the protein oligomer.
Assays.
In vivo assays for induced thermotolerance in S. cerevisiae (5, 27), luciferase reactivation in S. cerevisiae (27), cell survival at high temperature in E. coli (33), and the two-hybrid assay (31) were performed as described with slight modifications (SI Text). Assays for reactivation of urea-denaturated luciferase (22) and heat-inactivated GFP-38 (34) were performed as described with some modifications (SI Text).
Supplementary Material
Acknowledgments.
We thank Susan Gottesman (NCI), Len Neckers (NCI), and Jonathan Weissman (UCSF) for strains and plasmids; A. Battesti for help with the two-hybrid system; and Jodi Camberg and Susan Gottesman for reading of the manuscript and discussions. This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, Center for Cancer Research, and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102828108/-/DCSupplemental.
*A chimera similar to 44B4, but differing by several amino acids in the domain transitions has recently been observed to reactivate denatured luciferase with the DnaK system ∼10% above the level seen with the DnaK system alone, but only after several hours of incubation (23).
References
- 1.Doyle SM, Wickner S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem Sci. 2009;34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Wendler P, Saibil HR. Cryo electron microscopy structures of Hsp100 proteins: crowbars in or out? Biochem Cell Biol. 2010;88:89–96. doi: 10.1139/o09-164. [DOI] [PubMed] [Google Scholar]
- 3.Haslberger T, Bukau B, Mogk A. Towards a unifying mechanism for ClpB/Hsp104-mediated protein disaggregation and prion propagation. Biochem Cell Biol. 2010;88:63–75. doi: 10.1139/o09-118. [DOI] [PubMed] [Google Scholar]
- 4.Squires CL, Pedersen S, Ross BM, Squires C. ClpB is the Escherichia coli heat shock protein F84.1. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 6.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 7.Motohashi K, Watanabe Y, Yohda M, Yoshida M. Heat-inactivated proteins are rescued by the DnaK-J-GrpE set and ClpB chaperones. Proc Natl Acad Sci USA. 1999;96:7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 9.Goloubinoff P, Mogk A, Zvi AP, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci USA. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zolkiewski M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J Biol Chem. 1999;274:28083–28086. doi: 10.1074/jbc.274.40.28083. [DOI] [PubMed] [Google Scholar]
- 11.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion- like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 12.Lee S, et al. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 13.Lee S, Choi JM, Tsai FT. Visualizing the ATPase cycle in a protein disaggregating machine: structural basis for substrate binding by ClpB. Mol Cell. 2007;25:261–271. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wendler P, et al. Motor mechanism for protein threading through Hsp104. Mol Cell. 2009;34:81–92. doi: 10.1016/j.molcel.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S, Sielaff B, Lee J, Tsai FT. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc Natl Acad Sci USA. 2010;107:8135–8140. doi: 10.1073/pnas.1003572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White SR, Lauring B. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic. 2007;8:1657–1667. doi: 10.1111/j.1600-0854.2007.00642.x. [DOI] [PubMed] [Google Scholar]
- 17.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 18.Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem. 2004;279:29139–29146. doi: 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- 19.Weibezahn J, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 20.Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol Microbiol. 2007;66:840–857. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- 21.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krzewska J, Langer T, Liberek K. Mitochondrial Hsp78, a member of the Clp/Hsp100 family in Saccharomyces cerevisiae, cooperates with Hsp70 in protein refolding. FEBS Lett. 2001;489:92–96. doi: 10.1016/s0014-5793(00)02423-6. [DOI] [PubMed] [Google Scholar]
- 23.Sielaff B, Tsai FT. The M-domain controls Hsp104 protein remodeling activity in an Hsp70/Hsp40-dependent manner. J Mol Biol. 2010;402:30–37. doi: 10.1016/j.jmb.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kedzierska S, Chesnokova LS, Witt SN, Zolkiewski M. Interactions within the ClpB/DnaK bi-chaperone system from Escherichia coli. Arch Biochem Biophys. 2005;444:61–65. doi: 10.1016/j.abb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Schlee S, Beinker P, Akhrymuk A, Reinstein J. A chaperone network for the resolubilization of protein aggregates: direct interaction of ClpB and DnaK. J Mol Biol. 2004;336:275–285. doi: 10.1016/j.jmb.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C, Guy CL. Co-immunoprecipitation of Hsp101 with cytosolic Hsc70. Plant Physiol Biochem. 2005;43:13–18. doi: 10.1016/j.plaphy.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell. 2008;32:584–591. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chow IT, Barnett ME, Zolkiewski M, Baneyx F. The N-terminal domain of Escherichia coli ClpB enhances chaperone function. FEBS Lett. 2005;579:4242–4248. doi: 10.1016/j.febslet.2005.06.055. [DOI] [PubMed] [Google Scholar]
- 29.Hung GC, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics. 2006;173:611–620. doi: 10.1534/genetics.106.056820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tutar Y, Song Y, Masison DC. Primate chaperones Hsc70 (constitutive) and Hsp70 (induced) differ functionally in supporting growth and prion propagation in Saccharomyces cerevisiae. Genetics. 2006;172:851–861. doi: 10.1534/genetics.105.048926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haslberger T, et al. M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol Cell. 2007;25:247–260. doi: 10.1016/j.molcel.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 33.Chow IT, Baneyx F. Coordinated synthesis of the two ClpB isoforms improves the ability of Escherichia coli to survive thermal stress. FEBS Lett. 2005;579:4235–4241. doi: 10.1016/j.febslet.2005.06.054. [DOI] [PubMed] [Google Scholar]
- 34.Hoskins JR, Doyle SM, Wickner S. Coupling ATP utilization to protein remodeling by ClpB, a hexameric AAA+ protein. Proc Natl Acad Sci USA. 2009;106:22233–22238. doi: 10.1073/pnas.0911937106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.