Abstract
The 70-kDa heat shock protein (Hsp70) chaperones perform a wide array of cellular functions that all derive from the ability of their N-terminal nucleotide-binding domains (NBDs) to allosterically regulate the substrate affinity of their C-terminal substrate-binding domains in a nucleotide-dependent mechanism. To explore the structural origins of Hsp70 allostery, we performed NMR analysis on the NBD of DnaK, the Escherichia coli Hsp70, in six different states (ligand-bound or apo) and in two constructs, one that retains the conserved and functionally crucial portion of the interdomain linker (residues  ) and another that lacks the linker. Chemical-shift perturbation patterns identify residues at subdomain interfaces that constitute allosteric networks and enable the NBD to act as a nucleotide-modulated switch. Nucleotide binding results in changes in subdomain orientations and long-range perturbations along subdomain interfaces. In particular, our findings provide structural details for a key mechanism of Hsp70 allostery, by which information is conveyed from the nucleotide-binding site to the interdomain linker. In the presence of ATP, the linker binds to the edge of the IIA β-sheet, which structurally connects the linker and the nucleotide-binding site. Thus, a pathway of allosteric communication leads from the NBD nucleotide-binding site to the substrate-binding domain via the interdomain linker.
) and another that lacks the linker. Chemical-shift perturbation patterns identify residues at subdomain interfaces that constitute allosteric networks and enable the NBD to act as a nucleotide-modulated switch. Nucleotide binding results in changes in subdomain orientations and long-range perturbations along subdomain interfaces. In particular, our findings provide structural details for a key mechanism of Hsp70 allostery, by which information is conveyed from the nucleotide-binding site to the interdomain linker. In the presence of ATP, the linker binds to the edge of the IIA β-sheet, which structurally connects the linker and the nucleotide-binding site. Thus, a pathway of allosteric communication leads from the NBD nucleotide-binding site to the substrate-binding domain via the interdomain linker.
Keywords: conformational ensemble, NMR chemical-shift analysis, subdomain reorientations, actin fold, signal propagation
The 70-kDa heat shock proteins (Hsp70s) compose one of the most well studied and ubiquitously distributed families of allosteric proteins (1). Hsp70s assist in an extraordinarily broad spectrum of cellular processes, including protein folding, disaggregation, and translocation. All chaperone activities of Hsp70s are based on their ability to interact with short hydrophobic peptide segments of protein substrate in an ATP-dependent fashion. Hsp70s contain two domains—a 44-kDa N-terminal nucleotide-binding domain (NBD) and a 15-kDa C-terminal substrate-binding domain (SBD)—connected by a highly conserved hydrophobic linker. The allosteric cycle of Hsp70s involves an alternation between the ATP-bound state with low affinity and fast exchange rates for substrates, and the ADP-bound state with high affinity and low exchange rates for substrates. In turn, substrate binding to the SBD results in about 10-fold stimulation of ATPase activity of the NBD. However, the same ATPase stimulation can be achieved for the isolated NBD in the presence of the conserved interdomain linker sequence motif ( ) (2–4), indicating that the linker plays a key role in NBD function and allostery.
) (2–4), indicating that the linker plays a key role in NBD function and allostery.
The Hsp70 NBD belongs to the Actin/Hexokinase/Hsp70 superfamily, members of which share a number of common features (5, 6). The NBD is composed of two lobes, I and II; each lobe consists, in turn, of two subdomains: IA and IB for lobe I, and IIA and IIB for lobe II. Nucleotide binds at the bottom of the deep central cleft at the interface between subdomains IB and IIB, and all four subdomains are involved in nucleotide coordination. It has been suggested that nucleotide-dependent conformational changes due to subdomain reorientations are an intrinsic property of all NBDs, crucial for their functions (6). Significant subdomain movements have been directly observed between different X-ray structures of hexokinase (6); by contrast, there are no significant conformational changes seen in X-ray structures of Hsp70 NBDs (2, 7) and actin (8). Nonetheless, NMR data (3, 9, 10) and molecular dynamics (MD) calculations (11) for the Hsp70 NBD demonstrated significant conformational flexibility related to subdomain reorientations, which suggests that allosteric coupling in Hsp70s NBD occurs via a network of NBD subdomain motions (10). Despite the progress to date, experimental characterization of the nature of these conformational changes for Hsp70 NBDs and a mechanism by which these subdomain motions are exploited during the allosteric cycle remain elusive.
Some insight into the nature of the conformational landscape of Hsp70 NBDs comes from X-ray structures of their complexes with nucleotide exchange factors (NEFs), which show an opening of the nucleotide-binding pocket relative to other NBD structures conformation (12–15). Additionally, the recently determined X-ray structure of ATP-bound yeast Hsp110, Sse1, a distant relative of the Hsp70s, emerges as a plausible model for the Hsp70 ATP-bound conformation. It shows the interdomain linker and SBD directly interacting with the NBD (16), a likely structural route by which the linker directly influences conformational changes in the Hsp70 NBD. The three structures (i.e., NEF-free and NEF-bound Hsp70 NBD, and Hsp110 NBD) retain similar structural organization within NBD subdomains yet have different relative subdomain orientations. Together they serve as an excellent starting point for modeling the Hsp70 NBD allosteric ensemble.
To explore the allosteric landscape of the Hsp70 NBD, we have examined the relationship between allosteric modulators and conformational changes in the NBD of the Escherichia coli Hsp70, DnaK. Global chemical-shift analysis of six different states, apo or bound to alternative ligands, of two NBD constructs—one with and one without the conserved  linker sequence—reveals that the NBD exists as an ensemble of several conformations interconverting via rigid-body subdomain motions. Residues located on subdomain interfaces constitute an allosteric network in the NBD, providing multiple pathways for signal transduction. Pairwise comparisons of chemical-shift perturbations in turn reveal how nucleotide binding coordinates subdomain rotations, and consequently enables transmission of signals through the NBD, providing communication with the SBD and Hsp70 cochaperones.
linker sequence—reveals that the NBD exists as an ensemble of several conformations interconverting via rigid-body subdomain motions. Residues located on subdomain interfaces constitute an allosteric network in the NBD, providing multiple pathways for signal transduction. Pairwise comparisons of chemical-shift perturbations in turn reveal how nucleotide binding coordinates subdomain rotations, and consequently enables transmission of signals through the NBD, providing communication with the SBD and Hsp70 cochaperones.
Results
Chemical Shifts as Signposts on the Hsp70 NBD Allosteric Landscape.
The two powerful NMR measurements used most often to monitor protein structural rearrangements—nuclear Overhauser effects and residual dipolar couplings (RDCs)—may not be sensitive in all cases to small to moderate domain reorientations that occur in large proteins with little change in secondary structure, both because experimental data are too sparse and because measurement errors may be comparable to or larger than the overall structural perturbations. By contrast, chemical-shift perturbations, which are also easier to obtain, are extremely accurate and exquisitely sensitive to protein local and global structure. Thus, chemical shifts are excellent reporters to monitor even small changes in a protein conformational ensemble (17–19). To identify residues affected by conformational changes in the isolated 44-kDa NBD of the E. coli Hsp70, DnaK, we compared chemical shifts for backbone amide 1HN, 15N, carbonyl 13C, 13Cα, and 13Cβ atoms in several functionally important states, including the nucleotide-free, ADP-, ADP.Pi(phosphate)-, and ATP-bound states (Table S1). Nucleotide-induced conformational changes in DnaK and other Hsp70s require the correct positioning of the Mg2+ and K+ ions in the nucleotide-binding site. Moreover, the presence of ATP is essential: Functional conformational changes do not occur with several ATP analogs (e.g., ATPγS, AMP-PNP) (1, 20, 21). To understand how these small perturbations in the nucleotide-binding site affect the global NBD conformation, we included in our analysis the ADP.noMg (in the absence of Mg2+ ions) and the ATPγS-bound states. To obtain data for the ATP-bound state over longer experimental times without interference from ATP hydrolysis, we incorporated the T199A mutation, which blocks ATP hydrolysis but still allows functional ATP binding and ATP-induced conformational changes (22).
As noted above, previous biochemical studies revealed the importance of the interdomain linker for NBD conformational changes (2–4). In addition to these data, we found that the presence of the  linker motif changes the energetics of nucleotide binding to the NBD. NMR titrations of the ADP-bound NBD with ATP (Fig. S1) revealed that binding affinities for ATP and ADP.Pi differ significantly for the 1–388 and 1–392 NBD constructs. By NMR, the shorter construct has a higher affinity for ADP than ATP, which agrees with Kd values of 170 and 610 nM for ADP and ATP binding, respectively, as measured by isothermal calorimetry (23). On the contrary, when the
linker motif changes the energetics of nucleotide binding to the NBD. NMR titrations of the ADP-bound NBD with ATP (Fig. S1) revealed that binding affinities for ATP and ADP.Pi differ significantly for the 1–388 and 1–392 NBD constructs. By NMR, the shorter construct has a higher affinity for ADP than ATP, which agrees with Kd values of 170 and 610 nM for ADP and ATP binding, respectively, as measured by isothermal calorimetry (23). On the contrary, when the  motif is present on the isolated NBD, ATP binds more tightly than ADP, mimicking behavior of the full-length protein with Kd values of 250 and 160 nM for ADP and ATP, respectively (23). Moreover, NMR evidence gathered using the NBD392 construct (residues 1–392) pointed to a coupling of the effects of the linker and the nucleotide on NBD conformation (3). To understand the role of the linker
motif is present on the isolated NBD, ATP binds more tightly than ADP, mimicking behavior of the full-length protein with Kd values of 250 and 160 nM for ADP and ATP, respectively (23). Moreover, NMR evidence gathered using the NBD392 construct (residues 1–392) pointed to a coupling of the effects of the linker and the nucleotide on NBD conformation (3). To understand the role of the linker  motif, chemical shifts of NBD were analyzed for two NBD constructs: NBD388 (residues 1–388) and NBD392 (residues 1–392, including the conserved linker
motif, chemical shifts of NBD were analyzed for two NBD constructs: NBD388 (residues 1–388) and NBD392 (residues 1–392, including the conserved linker  motif).
motif).
Backbone amide 1HN, 15N, carbonyl 13C, 13Cα, and 13Cβ assignments for the six different states of NBD388 and NBD392 (a total of 12 NBD states) were obtained for about 85% of all residues. Their chemical shifts were compared pairwise between individual states and as a group.
NBD Nucleotide Cleft Opens by Rotation of Subdomain IIB.
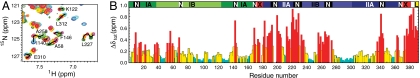
To explore the structural features that enable the NBD to bind and release nucleotide, as required in its allosteric cycle, we analyzed chemical-shift perturbations between the nucleotide-free and ADP-bound states of the NBD; the data are presented as a function of residue position in Fig. 1A. Previous RDC analysis demonstrated that in solution the ADP-bound NBD exists in the “closed” conformation (10). X-ray structures of Hsp70 NBDs bound to their respective NEFs (12–15, 24), which enhance the rate of nucleotide release and binding, show subdomain IIB to be rotated about 10 to 30° with respect to the rest of the NBD and thus opening of the nucleotide-binding cleft (Fig. 2A). For most residues in our analysis, chemical-shift changes between the apo and ADP-bound states were small and moderate (not exceeding 0.3 ppm) (Fig. 1A). As expected, large perturbations (shown in red in Fig. 1A and, on the structure, in Fig. 2B) were observed for residues directly affected by nucleotide binding (i.e., those located in the nucleotide-binding site) and, notably, for some residues relatively distant from the nucleotide-binding site—in particular the two α-helices of subdomain IIB (Fig. 2B, green). These α-helices are located at the interface between subdomains IB and IIB, and consequently would experience large environmental perturbations upon opening of the nucleotide-binding cleft, as seen in the crystal structures of NEF-bound Hsp70 or 70–kDa heat shock cognate proteins (Hsc70s) (Fig. 2A). Thus, our results are consistent with a model where nucleotide dissociation favors rotation of subdomain IIB and opening of the nucleotide-binding cleft. Interestingly, the rest of subdomain IIB (i.e., the double-stranded β-sheet and the loop connecting the two strands, which are distant from the IB–IIB interface) shows almost no chemical-shift perturbation but displays enhanced mobility on the nanosecond timescale, as indicated by the high peak intensities for this region (Fig. S2A). This finding fits remarkably well with elastic network modeling (ENM) predictions showing that the same region enjoys the largest mobility and serves as an adjustable NEF recognition path in the NBD (25). These ENM calculations and previous MD simulations (11) predicted that the open conformation is a state present on the Hsp70 NBD conformational landscape and not induced by NEF binding. Our chemical-shift perturbation data are entirely consistent with this prediction.
Fig. 1.
Nucleotide-induced conformational changes in the DnaK NBD. (A and B) Histograms showing chemical-shift differences,  , for backbone atoms as a function of residue number, where ΔδH, ΔδN, or ΔδCO are 1HN, 15N, and 13CO chemical-shift differences between the apo and ADP-bound states of NBD388 (A), and between the ADP- and ATP-bound states of NBD392 (B). Residues with large Δδtot (> 0.3 ppm) and significant chemical-shift perturbation (at least one ΔδH, ΔδN, or ΔδCO value is larger than two corresponding chemical-shift errors; i.e., 0.06, 0.6, and 0.6 ppm for 1HN and 15N, and 13CO atoms, respectively) are colored red and yellow, respectively; the rest are shown as cyan. The green background highlights regions that are highly affected by nucleotide binding, and the top bar shows NBD subdomains: IA (dark green), IB (light green), IIA (dark blue), IIB (light blue), crossing α-helices (red, X), the
, for backbone atoms as a function of residue number, where ΔδH, ΔδN, or ΔδCO are 1HN, 15N, and 13CO chemical-shift differences between the apo and ADP-bound states of NBD388 (A), and between the ADP- and ATP-bound states of NBD392 (B). Residues with large Δδtot (> 0.3 ppm) and significant chemical-shift perturbation (at least one ΔδH, ΔδN, or ΔδCO value is larger than two corresponding chemical-shift errors; i.e., 0.06, 0.6, and 0.6 ppm for 1HN and 15N, and 13CO atoms, respectively) are colored red and yellow, respectively; the rest are shown as cyan. The green background highlights regions that are highly affected by nucleotide binding, and the top bar shows NBD subdomains: IA (dark green), IB (light green), IIA (dark blue), IIB (light blue), crossing α-helices (red, X), the  linker motif (yellow, L), and the nucleotide-binding site (black, N).
linker motif (yellow, L), and the nucleotide-binding site (black, N).
Fig. 2.
Opening of the nucleotide-binding cleft upon nucleotide dissociation. (A) Comparison of alternative conformations of subdomain IIB in X-ray structures of DnaK homologues: in green, the closed form as seen in the isolated Bos taurus Hsc70 NBD [Protein Data Bank (PDB) ID code 1KAX]; in yellow, the open form as seen in the complex of yeast Sse1 with the Bos taurus Hsc70 NBD (PDB ID code 3C7N); in red, the open form as seen in the complex of yeast Sse1 with human Hsp70 NBD (PDB ID code 3D2F). (B) Mapping of the chemical-shift differences from Fig. 1А onto the structure of the DnaK NBD (PDB ID code 1DKG:D). Residues with large chemical-shift perturbations (red) and the highly affected subdomain IIB α-helices (green) are colored as in Fig. 1А.
ATP and the Interdomain linker Cooperatively Bind the NBD.
To explore the role of the interdomain linker in Hsp70 allostery and understand how the linker and the nucleotide-binding site communicate with each other, we examined how the presence of the  linker motif affects the NBD conformational ensemble by comparing nucleotide-free, ADP-, and ATP-bound states between the two NBD constructs: NBD392 and NBD388 (i.e., with and without the
linker motif affects the NBD conformational ensemble by comparing nucleotide-free, ADP-, and ATP-bound states between the two NBD constructs: NBD392 and NBD388 (i.e., with and without the  motif).
motif).
The last five residues arising from the C-terminus of NBD388 display narrow intense peaks in NMR spectra, and their chemical shifts and peak intensities are identical whether the NBD is bound to ATP or ADP, or no nucleotide is bound (Fig. S2A). This high peak-intensity suggests that these residues are flexible on the nanosecond timescale. We conclude that the NBD388 C-terminus is a flexible, water-exposed segment and that its behavior is not linked to nucleotide-dependent conformational changes of the NBD. In striking contrast, the C-terminal residues of NBD392 experience chemical-shift and mobility changes upon even small changes in the nucleotide-binding site (Fig. 3 A and B). In nucleotide-free NBD392, the C-terminus is highly flexible, and in turn, minimal long-range chemical-shift perturbations were observed when comparing this state to nucleotide-free NBD388 (Fig. 3C). In the ADP- and ATP-bound states, peaks from the C-terminal residues shift and broaden (Fig. 3 A and B), and moreover, the pairwise comparison between NBD388 and NBD392 shows significant effects in the rest of the NBD (Fig. 3C), indicating that there is communication between the linker and the NBD. Previously, it was proposed that a solvent-accessible hydrophobic cleft, which is formed by the crossing helices on the interface between subdomain IA and IIA, is the most probable binding site for the linker (3). Indeed, we found HN-HN NOESY cross-peaks between the linker and residues from the hydrophobic cleft in the ATP-bound state, but not in apo.NBD (Fig. S3A). Upon ADP binding, the  linker motif causes chemical-shift perturbations in the hydrophobic cleft (Fig. S4). These appear to arise from weak binding of the linker to this cleft, as the changes are small and local. Strikingly, ATP binding dramatically enhances the linker effect on the NBD conformational ensemble, causing chemical-shift changes throughout the NBD (Fig. 3C). Our data show that in addition to direct effects of linker binding on the hydrophobic cleft, the entire interface between the NBD lobes is perturbed, suggesting allosteric linker-induced lobe reorientation. In turn, in the ATP-bound state the conformation of the C-terminal segment dramatically changes compared to the flexible solvent-exposed conformation it adopts in the nucleotide-free state.
linker motif causes chemical-shift perturbations in the hydrophobic cleft (Fig. S4). These appear to arise from weak binding of the linker to this cleft, as the changes are small and local. Strikingly, ATP binding dramatically enhances the linker effect on the NBD conformational ensemble, causing chemical-shift changes throughout the NBD (Fig. 3C). Our data show that in addition to direct effects of linker binding on the hydrophobic cleft, the entire interface between the NBD lobes is perturbed, suggesting allosteric linker-induced lobe reorientation. In turn, in the ATP-bound state the conformation of the C-terminal segment dramatically changes compared to the flexible solvent-exposed conformation it adopts in the nucleotide-free state.
Fig. 3.
The cooperative effect of linker and ATP binding. (A) Relative peak intensities (Intrel) of the C-terminal residue for Leu392 for different NBD392 states. The Intrel value is the ratio of the Leu392 peak height to an average peak height in an HNCO spectrum in a corresponding NBD state. (B) Blow-up of the region of the 15N TROSY spectra showing resonances corresponding to the C-terminal Leu392 for different NBD392 states. (C) Histograms showing combined chemical-shift differences (Δδtot) as a function of residue number for backbone 1HN and 15N, and 13CO atoms (as in Fig. 1) between NBD388 and NBD392 in the apo, ADP-, and ATP-bound states. Yellow and cyan colors highlight significant and insignificant chemical-shift perturbations (as defined in Fig. 1), respectively. Gray background highlights the interfaces between two lobes. The top bar is the same as for Fig. 1.
It was previously reported that NBD393, a construct that includes one more residue (D393), had even higher ATPase than NBD392 (4). In our chemical-shift analysis, the addition of D393 to the linker motif (i.e., in the NBD393 construct) results in small long-range perturbations around the nucleotide-binding site and in subdomain IIA (Fig. S5B). However, the overall structure of the ATP-bound state of NBD392 and NBD393 remains very similar. Only a few residues showed significant chemical-shift differences. We suggest that these small perturbations fine-tune the ATPase activity and illustrate how sensitive the catalytic efficiency is to small perturbations.
Taken together, our observations support a two-way coupling mechanism between binding of ATP and the interdomain linker; we conclude that cooperative binding of the  linker sequence and ATP results in an NBD conformation favorable to ATPase hydrolysis and ATP binding and poised for interdomain allostery.
linker sequence and ATP results in an NBD conformation favorable to ATPase hydrolysis and ATP binding and poised for interdomain allostery.
Binding of ATP and Linker Leads to Widespread NBD Structural Reorganization.
A previous RDC analysis of the AMP-PNP–bound state of a construct of Thermus thermophilus DnaK NBD lacking the interdomain linker revealed reorientations of the subdomains IA and IIA relative to each other, such that the hydrophobic cleft became more accessible (9). Our pairwise chemical-shift perturbation analysis of the ADP- and ATP-bound NBD388 construct also showed long-range perturbations of the hydrophobic cleft. Thus, binding of ATP shifts the NBD to a conformation favorable for linker binding.
The consequence of the ATP-induced conformational change in the NBD and resulting linker binding is clearly illustrated by our pairwise chemical-shift perturbation analysis of ADP- and ATP-bound NBD392. In this longer construct, which retains a key structural participant in the allosteric function of Hsp70s, the interfaces between NBD subdomains experience much more extensive perturbations between the ADP- and ATP-bound states than seen for NBD388. The chemical-shift perturbations (shown as a function of residue number in Fig. 1B), when mapped onto the structure (Fig. 4A), revealed the largest changes for the C-terminus, the IIA β-sheet, and the N-terminal crossing α-helix.
Fig. 4.
The linker binds to the hydrophobic cleft between the subdomains IA and IIA. (A) Mapping of the chemical-shift differences from Fig. 1B onto the homology model of the DnaK ATP-bound NBD structure built from the Sse1 structure (PDB ID code 2QXL:B) (Fig. S6). The regions highlighted in Fig. 1B by green background are shown in green on the structure. Residues with large chemical-shift perturbations (highlighted in red in Fig. 1B) are shown as red spheres. (B) Superposition of the ADP- (green) and ATP- (yellow) bound conformations, derived from homology models as described in the text. For the ATP-bound conformation, the C-terminal 12 residues are shown in red, and the interdomain linker ( ) bound to the hydrophobic cleft is shown as red spheres.
) bound to the hydrophobic cleft is shown as red spheres.
To interpret our chemical-shift data structurally, and in so doing describe conformational changes, we generated two model structures representing the ADP- and ATP-bound state of the NBD (SI Text). For the ATP-bound conformation (ATP.NBDM), we built a homology model of the DnaK NBD based on the crystal structure of its distant relative, yeast Hsp110 (Sse1) bound to ATP, which has been suggested to represent the ATP-bound state of Hsp70 (16). The ADP-bound conformation of DnaK (ADP.NBDM) was obtained from the X-ray structure of the NBD of its close homologue, the Bos taurus Hsc70 NBD (26). Whereas the structure within the NBD subdomains for the two models (ADP.NBDM and ATP.NBDM) is quite similar, lobe II is rotated significantly with respect to lobe I, and subdomains IIA and IIB rotate relative to each other (Fig. S6). The resulting rotation of the IA–IIA subdomains against one another exposes binding sites for the interdomain linker along the edge of the IIA β-sheet (Fig. 4B). Moreover, these structural changes correlate well with shear movements of the crossing α-helices relative to each other and to the core IA and IIA β-sheet that have been directly observed for another Hsp70 homolog, hexokinase (6). Consequently, the modeled NBD structures predict that the linker and IIA β-sheet should experience the largest structural perturbations. This prediction is fully confirmed by our experimental data, which indeed reveal the largest chemical-shift changes for these regions (Fig. 4A). In addition, large chemical shifts for the N-terminal crossing α-helix hint at its possible role in domain reorientations, as was predicted from hexokinase structures (6).
In order to obtain more detailed structural information about the ATP-induced NBD conformational changes and specifically the linker conformation, we measured NH RDCs in the apo and ATPγS-bound states of the NBD392 (Fig. S3C). However, the precision of our RDC data did not enable accurate and unambiguous characterization of structural changes between these two states (see SI Text). Nonetheless, RDCs confirmed that the linker undergoes dramatic changes in its conformation upon ATP binding. In the apo state, RDC values for the linker are close to zero, which agrees with its flexible, solvent-exposed conformation, suggested by chemical-shift analysis. On the contrary, in the ATPγS-bound state [which mimics ATP-NBD (Fig. S5A)], the linker displays large positive RDCs, indicative of significantly more structure. Consistent with this observation, Cα and Cβ chemical shifts of the linker show significant β-sheet propensity (Fig. S2B). Moreover, RDC values indicate that NHs of the linker became parallel to those in the IIA β-sheet (Fig. S3C), which agrees well with the homology model derived from the Hsp110 structure (Fig. 4B). We believe these results provide a useful model for the structure of the linker, in general, but nonetheless we caution that the detailed structure of the linker seen for NBD392 may not recapitulate its structure when attached to the rest of the SBD.
An Intramolecular Allosteric Network in the NBD.
In order to gain a holistic picture of the NBD conformational ensemble and in so doing explore the parts of this molecular machine that respond to ligands, we compared chemical shifts of the six different states for both the NBD388 and NBD392 constructs and compiled the greatest shifts, site-by-site. We found that the secondary structure of the protein is largely unchanged: The Cα and Cβ chemical shifts do not vary significantly between different NBD states (Fig. S7); in all cases, these shifts are consistent with the secondary structure derived from the DnaK NBD X-ray structure (Fig. S2B). By contrast, the backbone amide (1HN and 15N) and carbonyl 13C chemical shifts of most residues are modulated by nucleotide binding and/or by the presence of the linker (Fig. 5). Even small changes in the nucleotide-binding site (e.g., between ADP-, ADP.Pi-, and ADP.noMg-bound states), caused long-range perturbations (Fig. S8). We found that about 60% of the NBD residues showed chemical-shift perturbations of backbone amide (1HN and 15N) and carbonyl 13C atoms when all 12 different NBD states were compared; i.e., for these residues chemical shifts were significantly different between at least two states (Fig. 5B). These residues represent “hot spots” in the NBD structure, as they respond to the binding of ligands or presence of the linker. Not surprisingly, the majority of these hot spots map to the interfaces between NBD subdomains (Fig. 6A), whereas the rest of the protein is largely unperturbed. These results fully agree with a model previously proposed based on analysis of RDCs, which showed significant NBD subdomain reorientations in solution compared to X-ray structure and suggested that allosteric coupling in the NBD occurs via a network of subdomain motions (10). Importantly, our chemical-shift analysis explores NBD conformational space more extensively, using NBD388 and NBD392 in six different states, all of which can be explained by an ensemble of conformations with similar overall architecture, the same folds within subdomains, but with different subdomain orientations. A question remains whether nucleotide binding results in a new structure on the NBD energy landscape, or rather shifts a preexisting equilibrium between two or more already populated NBD conformations. Although most NBD residues were represented by single resonances in NMR spectra, significant broadening of a number of peaks (Fig. S2A) suggests that local or global conformational exchange processes are occurring at a rate that is intermediate on the NMR timescale. Moreover, for many residues different chemical shifts were observed for all 12 states examined (Fig. 5A). It is very unlikely that a unique “single structure” exists for each state; instead, the single resonances observed most likely represent averages of multiple conformations rapidly interconverting on the NMR timescale. The “peak walking” pattern observed (Fig. 5A) shows that most resonances do not move in a simple manner in response to ligands, and thus suggests that ligands adjust the populations of more than two NBD conformations. The conformational heterogeneity for the Hsp70 NBD ensemble implied by the chemical-shift behavior is consistent with computational predictions (11) and previous experiments (3, 9, 10, 27). These observations also account for previous results showing that changes in the nucleotide-binding site, such as single point mutations or the absence of inorganic ions, cause disruption of Hsp70 allostery without significant changes in structure by X-ray crystallography (1, 20, 21, 26, 28, 29).
Fig. 5.
An intramolecular allosteric network in the NBD. (A) Representative regions from overlaid 1H-15N TROSY spectra of the 12 NBD states. Several examples of nonoverlapping resonances for residues with significant conformational changes are labeled to highlight their peak-walking patterns. The color code for the 12 spectra is: red (ATP.NBD392), magenta (ATPγS.NBD392), green (ADP/ADP.Pi/ADP.noMg.NBD392), light blue (apo.NBD392), orange (ATP.NBD388), yellow (ATPγS.NBD388), light green (ADP/ADP.Pi/ADP.noMg.NBD388), blue (apo.NBD388). (B) Histograms showing chemical-shift differences,  , for backbone atoms as a function of residue number; where ΔδH, ΔδN, or ΔδCO are the largest 1HN, 15N, and 13CO chemical-shift differences between any 2 of the 12 states of NBD388 and NBD392. Coloring and the top bar are the same as for Fig. 1.
, for backbone atoms as a function of residue number; where ΔδH, ΔδN, or ΔδCO are the largest 1HN, 15N, and 13CO chemical-shift differences between any 2 of the 12 states of NBD388 and NBD392. Coloring and the top bar are the same as for Fig. 1.
Fig. 6.
Mechanism of intramolecular allostery in the Hsp70 NBD. (A) Mapping of allosteric hot spots onto the structure of the DnaK NBD (PDB ID code 1DKG:D): Residues with large and significant chemical-shift differences from Fig. 5B are shown in red and yellow, respectively. Cyan and gray indicate insignificant changes and residues with no data, respectively. (B) Structural model for two-way coupling pathway between the nucleotide-binding site and the interdomain linker: superposition of the IIA β-sheet and the crossing α-helices for the ATP- (yellow) and ADP- (green) bound conformations of the NBD. The ATP γ-phosphate and the linker are in red, and residues involved in nucleotide binding are shown as spheres.
Discussion
Our results provide detailed molecular insights into intramolecular signal transduction in Hsp70 molecular chaperones. Our chemical-shift data lead us to conclude that binding of a nucleotide ligand and the docking of the interdomain linker result in changes in subdomain and lobe orientations in the Hsp70 NBD, providing long-range perturbations along lobe and subdomain interfaces (Fig. 6A). Importantly, by interacting with all four subdomains, nucleotide binding selects preferred features on the protein landscape and consequently coordinates further allosteric events. An allosteric signal can propagate bidirectionally. Therefore, not only do perturbations in the nucleotide-binding site affect subdomain interfaces, but also a signal can propagate from subdomain interfaces to the nucleotide-binding site, and in so doing regulate nucleotide binding and ATPase activity. Our results are entirely consistent with a previous hypothesis that allosteric signal transduction occurs via a network of motions of protein modules (e.g., subdomains) (30, 31). According to this model, residues at the interfaces between modules form an allosteric network between protein active sites. More specifically, our results argue that nucleotide dissociation favors rotation of subdomain IIB and opening of the nucleotide-binding cleft, which had been predicted early by ENM (25) and MD (11) calculations. Additionally, NEF binding, which accelerates nucleotide exchange, appears to act by selection of preexisting conformations on the landscape (25).
Because our study included a comparison of NBD constructs with and without the functionally critical first segment of the interdomain linker, we can provide a structural model for two-way coupling pathway between binding of ATP and the linker, which helps understanding of NBD to SBD communication. Via subdomain reorientations, ATP binding results in exposure of a binding surface for the interdomain linker. Linker binding, in turn, stabilizes the ATPase-active conformation of the NBD, which is similar to that seen in the Hsp110 NBD crystal structure (16). As a result, the β-sheet of subdomain IIA structurally connects the interdomain linker and the nucleotide-binding site (Fig. 6B). Indeed, linker binding to an edge of the IIA β-sheet results in rotation of subdomain IIA relative to subdomain IA and the crossing α-helices, and consequently changes the orientation of the β1–β2 turn in subdomain IIA that is responsible for coordination of the ATP γ-phosphate (6). Consequently, the presence of the ATP γ-phosphate and the linker is essential for the ATPase active conformation. The activation of the ATPase activity upon extension of the construct [e.g., in NBD393 (4)] is evidence that additional structural adjustments can occur and enable further regulation of ATPase activity by Hsp70 cochaperones and the SBD.
Thus, we offer an explanation for how the interdomain linker works as a switch: By binding to the hydrophobic cleft in a nucleotide-dependent fashion, the linker directly affects the NBD conformation and regulates interdomain communication, bringing the SBD close to the NBD. Indeed, single point mutations of the linker and hydrophobic cleft residues perturb the NBD ATPase activity and disturb its regulation by the SBD and cochaperones (2, 16, 32, 33). The fact that the hydrophobic cleft is the binding site for the Hsp70 cochaperone DnaJ, which regulates the rate of ATP hydrolysis (2, 34, 35), indicates that cochaperone regulation of ATPase activity most likely occurs through the same allosteric networks (i.e., via the linker and the hydrophobic cleft). Intriguingly, the hydrophobic cleft comprises a sector of coevolved residues in the NBD, which is important for stabilizing the interdomain interfaces and mediating allosteric communication between the NBD and SBD (36).
Evolution has capitalized on the ability of the Actin/Hexokinase/Hsp70 family NBD structure to utilize ATP as a signal for downstream functions, and the present study using chemical-shift perturbation has shed light on the structural origins of this conserved allosteric machine. Chemical-shift perturbation analysis has proven itself to be a rich source of information in a complex conformational landscape with shifting populations.
Materials and Methods
Protein Expression and Purification.
Plasmids encoding DnaK NBD(1–388)T199A, NBD(1–392)T199A, and NBD(1–393)T199A were prepared previously in our laboratory; they were expressed in E. coli and purified as described (3, 37). 2H-, 13C-, and 15N-labeled NMR samples were prepared using established protocols (38), as described previously (37) (see SI Text).
NMR Spectroscopy.
To obtain backbone and Cβ resonance assignments, we recorded standard sets of transverse-relaxation optimized spectroscopy (TROSY)-modified triple-resonance experiments, developed for 15N-, 13C-, and 2H-labeled proteins. Details about NMR experiments and sample conditions are summarized in Table S1. All spectra were obtained at 26 °C on a 600-MHz Bruker Avance spectrometer using a TXI cryoprobe, processed using nmrPipe (39), and analyzed using Cara (40).
Chemical-Shift Analysis.
Backbone 1HN, 15N,  , 13Cα, and 13Cβ chemical shifts for individual states were obtained with the program AutoAssign (41) and/or manually (see SI Text).
, 13Cα, and 13Cβ chemical shifts for individual states were obtained with the program AutoAssign (41) and/or manually (see SI Text).
The 1HN/15N/13CO combined chemical-shift change of a particular residue between different conformations was calculated as described previously (42):
 |
where 0.154 and 0.341 are the weighting factors for 15N and 13CO, respectively. Errors in chemical-shift values, 0.03, 0.3, 0.3, 0.3, and 0.5 ppm for 1HN, 15N, 13CO, 13Cα, and Cβ atoms, respectively, were obtained as average line-widths for 1H dimensions and spectral resolution in corresponding 3D spectra and 2D 15N-TROSY for 13C and 15N dimensions, respectively.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grant GM027616.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: 1HN, 15N, 13CO, 13Cα, and 13Cβ chemical shifts for ADP.NBD388, apo.NBD388, and ATP.NBD392 have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession nos. 17208, 17209, and 17210).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014448108/-/DCSupplemental.
References
- 1.Mayer MP, Brehmer D, Gassler CS, Bukau B. Hsp70 chaperone machines. Adv Protein Chem. 2001;59:1–44. doi: 10.1016/s0065-3233(01)59001-4. [DOI] [PubMed] [Google Scholar]
- 2.Jiang JW, et al. Structural basis of J cochaperone binding and regulation of Hsp70. Mol Cell. 2007;28:422–433. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swain JF, et al. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell. 2007;26:27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogel M, Mayer MP, Bukau B. Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J Biol Chem. 2006;281:38705–38711. doi: 10.1074/jbc.M609020200. [DOI] [PubMed] [Google Scholar]
- 5.Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurley JH. The sugar kinase heat shock protein 70 actin super family: Implications of conserved structure for mechanism. Annu Rev Biophys Biomol Struct. 1996;25:137–162. doi: 10.1146/annurev.bb.25.060196.001033. [DOI] [PubMed] [Google Scholar]
- 7.Flaherty KM, Wilbanks SM, DeLuca-Flaherty C, McKay DB. Structural basis of the 70-kilodalton heat-shock cognate protein ATP hydrolytic activity. II. Structure of the active-site with ADP or ATP bound to wild-type and mutant ATPase fragment. J Biol Chem. 1994;269:12899–12907. [PubMed] [Google Scholar]
- 8.Reisler E, Egelman EH. Actin structure and function: What we still do not understand. J Biol Chem. 2007;282:36133–36137. doi: 10.1074/jbc.R700030200. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharya A, et al. Allostery in Hsp70 chaperones is transduced by subdomain rotations. J Mol Biol. 2009;388:475–490. doi: 10.1016/j.jmb.2009.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang YB, Zuiderweg ERP. The 70-kDa heat shock protein chaperone nucleotide-binding domain in solution unveiled as a molecular machine that can reorient its functional subdomains. Proc Natl Acad Sci USA. 2004;101:10272–10277. doi: 10.1073/pnas.0401313101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woo HJ, Jiang JW, Lafer EM, Sousa R. ATP-induced conformational changes in Hsp70: Molecular dynamics and experimental validation of an in silico predicted conformation. Biochemistry. 2009;48:11470–11477. doi: 10.1021/bi901256y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arakawa A, et al. The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange. Structure. 2010;18:309–319. doi: 10.1016/j.str.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Polier S, Dragovic Z, Hartl FU, Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133:1068–1079. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Schuermann JP, et al. Structure of the Hsp110 : Hsc70 nucleotide exchange machine. Mol Cell. 2008;31:232–243. doi: 10.1016/j.molcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sondermann H, et al. Structure of a Bag/Hsc70 complex: Convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291:1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- 16.Liu QL, Hendrickson WA. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131:106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavalli A, Salvatella X, Dobson CM, Vendruscolo M. Protein structure determination from NMR chemical shifts. Proc Natl Acad Sci USA. 2007;104:9615–9620. doi: 10.1073/pnas.0610313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulder FAA, Filatov M. NMR chemical shift data and ab initio shielding calculations: Emerging tools for protein structure determination. Chem Soc Rev. 2010;39:578–590. doi: 10.1039/b811366c. [DOI] [PubMed] [Google Scholar]
- 19.Shen Y, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 21.Palleros DR, Reid KL, Shi L, Welch WJ, Fink AL. ATP-induced protein Hsp70 complex dissociation requires K+ but not ATP hydrolysis. Nature. 1993;365:664–666. doi: 10.1038/365664a0. [DOI] [PubMed] [Google Scholar]
- 22.McCarty JS, Walker GC. DnaK as a thermometer: Threonine-199 is site of autophosphorylation and is critical for ATPase activity. Proc Natl Acad Sci USA. 1991;88:9513–9517. doi: 10.1073/pnas.88.21.9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taneva SG, Moro F, Velazquez-Campoy A, Muga A. Energetics of nucleotide-Induced DnaK conformational states. Biochemistry. 2010;49:1338–1345. doi: 10.1021/bi901847q. [DOI] [PubMed] [Google Scholar]
- 24.Harrison CJ, HayerHartl M, DiLiberto M, Hartl FU, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Gierasch LM, Bahar I. Role of the Hsp70 ATPase domain intrinsic dynamics and sequence evolution in enabling its function interactions with NEFs. PLoS Comput Biol. 2010;6:1–15. doi: 10.1371/journal.pcbi.1000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Brien MC, Flaherty KM, McKay DB. Lysine 71 of the chaperone protein Hsc70 is essential for ATP hydrolysis. J Biol Chem. 1996;271:15874–15878. doi: 10.1074/jbc.271.27.15874. [DOI] [PubMed] [Google Scholar]
- 27.Mapa K, et al. The conformational dynamics of the mitochondrial Hsp70 chaperone. Mol Cell. 2010;38:89–100. doi: 10.1016/j.molcel.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Johnson ER, McKay DB. Mapping the role of active site residues for transducing an ATP-induced conformational change in the bovine 70-kDa heat shock cognate protein. Biochemistry. 1999;38:10823–10830. doi: 10.1021/bi990816g. [DOI] [PubMed] [Google Scholar]
- 29.Sousa MC, McKay DB. The hydroxyl of threonine 13 of the bovine 70-kDa heat shock cognate protein is essential for transducing the ATP-induced conformational change. Biochemistry. 1998;37:15392–15399. doi: 10.1021/bi981510x. [DOI] [PubMed] [Google Scholar]
- 30.Daily MD, Gray JJ. Allosteric communication occurs via networks of tertiary and quaternary motions in proteins. PloS Comput Biol. 2009;5:1–14. doi: 10.1371/journal.pcbi.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.del Sol A, Tsai CJ, Ma BY, Nussinov R. The origin of allosteric functional modulation: Multiple pre-existing pathways. Structure. 2009;17:1042–1050. doi: 10.1016/j.str.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis JE, Voisine C, Craig EA. Intragenic suppressors of Hsp70 mutants: Interplay between the ATPase- and peptide-binding domains. Proc Natl Acad Sci USA. 1999;96:9269–9276. doi: 10.1073/pnas.96.16.9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogel M, Bukau B, Mayer MP. Allosteric regulation of Hsp70 chaperones by a proline switch. Mol Cell. 2006;21:359–367. doi: 10.1016/j.molcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 34.Gassler CS, et al. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc Natl Acad Sci USA. 1998;95:15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suh WC, et al. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc Natl Acad Sci USA. 1998;95:15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smock RG, et al. An interdomain sector mediating allostery in Hsp70 molecular chaperones. Mol Syst Biol. 2010;6:414. doi: 10.1038/msb.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clerico EM, Zhuravleva A, Smock RG, Gierasch LM. Segmental isotopic labeling of the Hsp70 molecular chaperone DnaK using expressed protein ligation. Biopolymers. 2010;94:742–752. doi: 10.1002/bip.21426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 39.Delaglio F, et al. NMRpipe—A multidimensional spectral processing system based on Unix pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 40.Keller R. Zurich, Switzerland: Eidgenössiche Technische Hochschule; 2004. Optimizing the process of nuclear magnetic resonance spectrum analysis and computer aided resonance assignment. Doctoral thesis. [Google Scholar]
- 41.Zimmerman DE, et al. Automated analysis of protein NMR assignments using methods from artificial intelligence. J Mol Biol. 1997;269:592–610. doi: 10.1006/jmbi.1997.1052. [DOI] [PubMed] [Google Scholar]
- 42.Evenas J, et al. Ligand-induced structural changes to maltodextrin-binding protein as studied by solution NMR spectroscopy. J Mol Biol. 2001;309:961–974. doi: 10.1006/jmbi.2001.4695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.