Abstract
Unique chemical methodology enables the synthesis of innovative and diverse scaffolds and chemotypes and allows access to previously unexplored “chemical space.” Compound collections based on such new synthetic methods can provide small-molecule probes of proteins and/or pathways whose functions are not fully understood. We describe the identification, characterization, and evolution of two such probes. In one example, a pathway-based screen for DNA damage checkpoint inhibitors identified a compound, MARPIN (ATM and ATR pathway inhibitor) that sensitizes p53-deficient cells to DNA-damaging agents. Modification of the small molecule and generation of an immobilized probe were used to selectively bind putative protein target(s) responsible for the observed activity. The second example describes a focused library approach that relied on tandem multicomponent reaction methodologies to afford a series of modulators of the heat shock protein 70 (Hsp70) molecular chaperone. The synthesis of libraries based on the structure of MAL3-101 generated a collection of chemotypes, each modulating Hsp70 function, but exhibiting divergent pharmacological activities. For example, probes that compromise the replication of a disease-associated polyomavirus were identified. These projects highlight the importance of chemical methodology development as a source of small-molecule probes and as a drug discovery starting point.
Keywords: ATPase, diversity oriented synthesis, isosteres, UPCMLD, alpha-methylene cyclopentenone
The discovery of probes for targets or pathways of biological interest often relies on the availability of unique, structurally diverse small molecules. For 8 yr, the National Institute of General Medical Sciences has sponsored Centers for Chemical Methodologies and Library Development (CMLD), including the Center at the University of Pittsburgh (UPCMLD) (1), whose goals are the development of innovative synthetic chemistry methodology, and its application to the synthesis of libraries of unique and complex structures. The availability of such libraries serves a number of purposes. First, application of a new synthetic method for library synthesis not only validates the methodology, but leads to an in depth understanding of its scope and limitations under parallel synthesis conditions or automation, which are becoming routine in synthetic chemistry laboratories. Furthermore, optimization of a synthetic protocol in preparation for library synthesis ensures the availability of a robust and accurate synthetic procedure. Second, the compounds generated during library production, if designed appropriately, have the potential to exhibit unique biological effects (2). To optimize opportunities in this area, the UPCMLD has submitted > 3,000 unique compounds to a variety of screening operations including the National Institutes of Health (NIH) Molecular Libraries Small-Molecule Repository, the Broad Institute, programs sponsored by the National Institute of Allergy and Infectious Diseases, the National Institute of Mental Health, and the National Cancer Institute. We estimate that UPCMLD compounds have been assayed in > 600 biological screens.
Using Pubchem’s data mining tools (3), we recently analyzed the performance of UPCMLD compounds in assays run under the auspices of the NIH’s Molecular Libraries Screening Centers Network and Molecular Libraries Probe Centers Network programs. Based on data available as of August 2010,* > 28% of the compounds submitted by the UPCMLD have been confirmed as “active” in at least one assay, with “active” being determined by criteria set for each individual assay and specified in Pubchem. UPCMLD compounds were confirmed hits in screens spanning virtually all therapeutic areas. These compounds demonstrated activity against diverse protein target classes, including enzymes, transcription factors, ion channels, as well as in protein-protein interactions. Active compounds were also detected in pathway and phenotypic screens. We report herein two examples of chemical probe discoveries that originated from UPCMLD synthetic methodology and library preparation efforts.
In one example, two closely related compounds were found to be DNA damage checkpoint inhibitors when the inhibitory effects of compounds on the Ataxia-Telangiectasia and Rad3-related (ATR) signaling pathway were screened. The compounds also sensitize p53-deficient cells to diverse DNA-damaging agents. We describe herein our efforts to identify the protein target(s) of these compounds. In contrast, the second example exploits the known effects of a class of small molecules on the heat shock protein 70 (Hsp70) molecular chaperone (4). Focused libraries based on tandem multicomponent reactions (MCRs) were used to explore the function of Hsp70s in a variety of organisms and biochemical assay systems. The unique chemical methodology employed to prepare the initial libraries, i.e., tandem Biginelli-Ugi MCRs, allowed the generation of a series of probes of chaperone function and provided starting points for drug discovery.
Results
Replication Checkpoint Inhibitors from a Diversity-Oriented Synthesis Strategy.
Diversity-Oriented Synthesis (DOS), the synthesis of a diverse set of small organic molecules suitable to identify new ligands for a variety of biological targets, has revolutionized how compound libraries are prepared (5). The preparation of subsets of structurally unique compounds typically involves approaches that utilize one or more of the following diversification strategies: building block diversity, appendage diversity, stereochemical diversity, and skeletal diversity (6). Skeletal diversity is the most challenging to achieve (7) and at the same time it is considered the most powerful because it provides molecularly distinct scaffolds that occupy separate regions of chemical space.
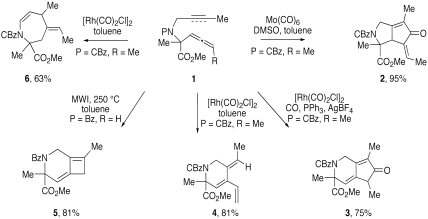
Brummond and coworkers have exploited reactions discovered in their lab with the goal of achieving a skeletal diversification strategy that affords libraries of biologically relevant compounds. Specifically, a reagent-based skeletal diversification strategy has been accomplished by effecting transition metal catalyzed, and thermal carbocyclization and cyclocarbonylation reactions of allene-ynes and ene-allenes (Fig. 1). For example, reaction of allene-yne 1 with molybdenum hexacarbonyl [Mo(CO)6] affords alkylidene cyclopentenone 2 by a selective reaction with the proximal double bond of the allene (8). Alternatively, reaction of the allene-yne 1 with 5 mol% of rhodium biscarbonyl chloride dimer [Rh(CO)2Cl]2 under a carbon monoxide atmosphere gives the 4-alkylidene cyclopentenone 3 via a selective cyclocarbonylation reaction with the distal double bond of the allene. Controlling the reacting double bond of the allene with the transition metal catalyst is unique and was further exploited by exposing allene-yne 1 to the same Rh(I) catalyst but under a nitrogen atmosphere; these conditions produced triene 4 via a formal Alder-ene process. Heating allene-yne 1 in the microwave for 15 min at 250 °C accomplished a rare intramolecular [2 + 2] cycloaddition reaction between an allene and an alkyne to give the bicyclo[4.2.0]octadiene 5 (9). Finally, exchanging the alkyne for an alkene produces the corresponding tetrahydroazepene 6 when reacted with [Rh(CO)2Cl]2 (10).
Fig. 1.
Cyclocarbonylation and cycloisomerization reactions of allene-ynes and ene-allenes.
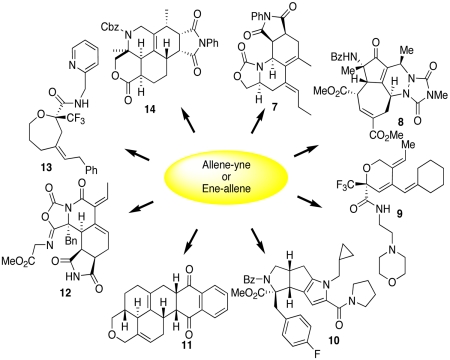
Through one intermediate (1, Fig. 1), skeletal reorganization processes provide diverse and often previously inaccessible substructures; furthermore, it is the different functional groups embedded in these structures (2–6) that are exploited in the preparation of libraries. Moreover, the cyclic scaffolds rigidify the final products, making them more lead-like and appropriate for inclusion in small-molecule screening libraries (11). This reagent-based approach to skeletal diversity has subsequently been demonstrated by the UPCMLD through the preparation of multiple libraries, and specific examples of each are illustrated in Fig. 2 (12). For example, scaffold 2 provides for incorporation of a pyrrole ring using a Stetter/Paal-Knorr condensation reaction sequence to generate compounds such as 10. The library design incorporated diverse substituents at multiple positions of the scaffold to span regions of chemical space that were not typically accessed by natural products or previously known synthetic compounds. In addition, the substituents were chosen to incorporate functional groups that could participate in interactions observed in small-molecule-protein and protein-protein interactions, such as hydrogen-bonding, hydrophobic interactions, and interactions with π systems (13). Screening of these libraries led to the identification of a compound, now called MARPIN (ATM and ATR pathway inhibitor), that sensitizes p53-deficient cells to diverse DNA-damaging agents (Fig. 3A).
Fig. 2.
Representative compounds from libraries based on scaffolds 2–6. Compounds 7–9,11,12,14 originating from chemistries similar to that used to obtain scaffold 4, compound 13 originating from scaffold 6 and compound 10 from scaffold 2.
Fig. 3.
Derivatization of MARPIN. (A) Library design strategy. (B) Structure activity data for a series of MARPIN derivatives. IC50 values indicate the concentrations at which compounds provide 50% inhibition of phosphorylation of Chk1 at Ser345 in cells treated with hydroxyurea.
Screening for DNA Damage Signaling Inhibitors.
Diverse genotoxic stresses are sensed by two major protein kinases called ATM (Ataxia-Telangiectasia Mutated) and ATR (14). These kinases activate DNA damage checkpoints to elicit cell cycle arrest and DNA repair. Inhibition of ATM/ATR pathways in the presence of DNA damage abrogates DNA damage checkpoints and leads to cell death. Because many cancers are resistant to chemotherapeutic agents largely due to defective p53, sensitizing p53-deficient cells to DNA-damaging agents is of great interest in cancer therapy and could be achieved by inhibition of ATM/ATR pathways (15). To discover unique small-molecule inhibitors of ATM/ATR pathways, 9,195 compounds were screened in a cell-based assay for inhibition of phosphorylation of ATR’s key target, Chk1, after induction by replication stress. We identified five compounds that inhibited ATM/ATR signaling pathways. Three compounds were known bioactives, although none had been linked to DNA damage responses, and two compounds, 15 (subsequently renamed MARPIN) and 16, were derived from the UPCMLD DOS library described above (Fig. 3). All inhibitors sensitized p53-deficient cells to DNA-damaging agents. Interestingly, in vitro kinase assays revealed that the inhibitors did not directly suppress ATR’s catalytic activity, implying that the compounds might inhibit mediator proteins involved in ATM/ATR pathways.
Structure Activity Relationships (SAR) of MARPIN Assessed via Signaling in Live Cells.
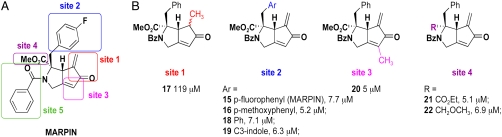
To further characterize the compounds’ mechanisms of action, derivatives of 15 were prepared to establish Structure Activity Relationship (SAR) data and to identify a suitable immobilization/attachment site for affinity matrix experiments. The design strategy (Fig. 3A), based on the structures of the two active compounds 15 (MARPIN) and 16 (IC50 = 7.7 and 5.2 μM, respectively, Fig. S1), maintained the structurally rigid bicyclo[3.3.0] system and incorporated modifications at four sites (Sites 1–4). The benzamide (Site 5) was not modified due to difficulties encountered in removing this group. Thus, a series of derivatives were prepared that were consistent with this skeleton and synthetic accessibility.†
It was hypothesized that the alpha-methylene cyclopentenone at Site 1 is responsible for the biological activity through a covalent modification of the target (16). Thus, cyclopentenone 17 was prepared by selective reduction of the exocyclic double bond of MARPIN (Fig. 3B, reduction not shown; SI Appendix: S1). The resulting diastereomers of 17 were not separated but assayed as a mixture. As an indicator for ATR pathway disruption, we measured inhibition of phosphorylation of Chk1 as induced by hydroxyurea (HU). HU activates the ATR-Chk1 pathway by depleting cellular nucleotides and causing replication stress. MARPIN derivative 17 was considerably less effective in inhibiting HU-induced Chk1 phosphorylation (IC50 = 119 μM), supporting the hypothesis that the alpha-methylene cyclopentenone is critical for MARPIN’s bioactivity (Fig. 4A, Fig. S1). It is noteworthy, however, that other Michael acceptors possessing the alpha-methylene substructure were included in the original screening library, but were not identified as active. This information suggests that while an alpha-methylene unit is important for activity and potency, its mere presence is not sufficient to disrupt the ATR pathway. Structural modifications of the aryl group including incorporation of hydrogen (18) or an electron donating group (16, para-methoxy) at Site 2 had no effect on the bioactivity data when compared to the electron withdrawing substituent (para-fluorine) in MARPIN 15. These data illustrated that electronic effects had no effect on activity. Furthermore, this site was insensitive to changes in size, as replacement of the aromatic ring with a larger indole ring resulted in a compound, 19, that displayed an IC50 of 6.3 μM (Fig. S1). Incorporation of a larger electron releasing methyl group at Site 3 (e.g., 20) also generated a potent inhibitor (Fig. S1). The methyl ester at Site 4 could not be saponified to give the corresponding carboxylic acid, this observation might exclude the possibility that hydrolysis by cellular esterases is required to yield an active derivative. However, the methoxymethyl ether 22 was as potent as MARPIN (Fig. S1), supporting the premise that esterase-catalyzed saponification is not crucial for bioactivity. Together, the SAR allowed us to identify sites 2, 3, and 4 as those that would be appropriate for attachment for immobilization without significantly affecting the inhibitory activity. Site 4 was chosen for pegylation and immobilization due to the synthetic accessibility of these derivatives.
Fig. 4.
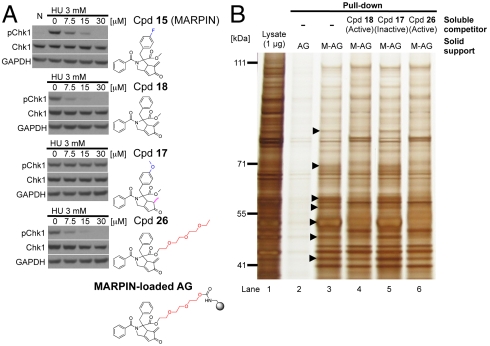
Bioactivities of MARPIN derivatives and their use for detection of MARPIN-binding proteins. (A) Inhibitory effects of MARPIN derivatives on hydroxyurea (HU)-induced phosphorylation of Chk1. 293T cells were treated with 3 mM HU and the indicated concentrations of MARPIN (15) and its derivatives (17, 18, or 26) or with no hydroxyurea (N) for 2 h. Immunoblots quantitated phospho-Chk1 (pChk1) at Ser345, total Chk1, and GAPDH. AffiGel (AG) loaded with pegylated MARPIN was used for pull-down assays. (B) MARPIN affinity matrix studies. Cell lysates of 293T cells were preincubated with vehicle (DMSO) or MARPIN derivatives (active compound 18, 26 or inactive compound 17) as fivefold excess free soluble competitor. Capped AG or MARPIN-loaded AG (M-AG) were then mixed with the preincubated lysates for affinity enrichment. Samples (lanes 2–6) and 1 μg of total protein (lane 1) were analyzed by SDS-PAGE/silver stain. Arrowheads indicate proteins of interest that bound to M-AG (lane 3) and could be competed from binding to M-AG matrix by excess soluble MARPIN derivatives that were active (lanes 4 and 6) but not inactive (lane 5).
Preparation of a Water Soluble Competitive Inhibitor and Immobilization of MARPIN for Proteomic Analysis.
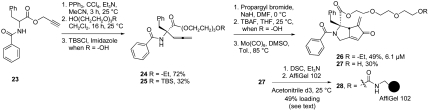
A water soluble competitive inhibitor, 26, was prepared by pegylating an active MARPIN derivative (Fig. 5, SI Appendix: S1). Compound 23 was reacted under standard conditions to effect a dehydrative Claisen rearrangement (vide supra). The intermediate (not shown) was not isolated but reacted with 2-(2-(2-ethoxyethoxy)ethoxy)ethanol to generate allene 24. Treatment of 24 with propargyl bromide and base, and reaction with Molybdenum hexacarbonyl afforded 26. This more water soluble pegylated derivative showed inhibitory effects on HU-induced Chk1 phosphorylation comparable to that of MARPIN (IC50 = 6.1 μM, Fig. 4A).
Fig. 5.
Synthesis of pegylated and immobilized MARPIN derivatives.
With these data in hand, we next immobilized MARPIN. It was reasoned that incorporating a linker between the resin and the functionally dense MARPIN would be advantageous so access to the active site would not be impeded by proximity of the resin. Thus, a bifunctional triethylene glycol was chosen as a suitable linker to maintain distance between the resin and the MARPIN active site. AffiGel (AG) 102 (Bio-Rad) was chosen as the affinity resin, as this matrix is commonly used to identify binding proteins in pull-down assays or via stable isotope labeling using amino acids in cell culture (17, 18). The reaction sequence to attach the resin was performed as described for 26, with the exception that triethylene glycol was used in place of 2-(2-(2-ethoxyethoxy)ethoxy)ethanol, and the resultant product was protected as the tert-butyldimethylsilyl (TBS) ether to give 25. N-Propargylation of the benzoylamide moiety, removal of the TBS protecting group and Pauson-Khand reaction of the allene-yne afforded 27. Next, alcohol 27 was activated with disuccinimide carbonate, and AG 102 was added. The depletion of succinimide activated 27 was monitored by 1H Nuclear Magnetic Resonance (NMR) Spectroscopy NMR and the reaction was stopped after disappearance of 51% of the starting material, corresponding to a 49% loading of 27 on AG 102 to give resin-bound MARPIN 28 (SI Appendix: S1).
Affinity Matrix Data Identify Putative MARPIN-Binding Proteins.
To detect MARPIN-associated proteins, we performed pull-down assays by incubating cell lysates with immobilized MARPIN, and visualized binding proteins on a silver stained gel (Fig. 4B). Minimal protein binding was detected with AG alone (lane 2). In contrast, MARPIN-loaded AG (M-AG) exhibited affinity enrichment of binding proteins (lane 3). We also used excess amounts of the free active derivative 18 and 26 as soluble competitors to detect specifically bound species. In some cases, differences in the abundance of certain proteins were observed (lane 3 vs. lanes 4 and 6). In addition, excess amounts of the free inactive derivative 17 were used (lane 5) and the bound species were similar to those in the M-AG sample (lane 3). Taken together, we have detected specific MARPIN-binding proteins (arrowheads in Fig. 4B) and the identification of these proteins is underway.
Molecular Probes of Heat Shock Protein 70 (Hsp70) Derived from a Focused Library.
Hsp70 molecular chaperones assist the folding of nascent proteins, target misfolded proteins for degradation, and transport proteins across biological membranes. By binding and hydrolyzing ATP, concomitant with the binding and release of aggregation-prone proteins, Hsp70s prevent toxic proteins from accumulating intracellularly. Consequently, Hsp70 function has been linked to many protein conformational diseases, including Alzheimer’s, Parkinson’s, and Huntington’s disease, as well as cystic fibrosis (19, 20). Hsp70s are also antiapoptotic and act at multiple nodes in the apoptotic pathway. Thus, within the stressed tumor cell environment, Hsp70s are critical for cancer cell survival (21). Infectious organisms such as bacteria and viruses also require Hsp70s for survival, replication, and infectivity. Therefore targeting these specialized proteins is a viable approach to antiproliferative and antiinfective drug discovery (22–24). Nevertheless, until recently, relatively few small-molecule modulators of Hsp70 were known (20, 22–26).
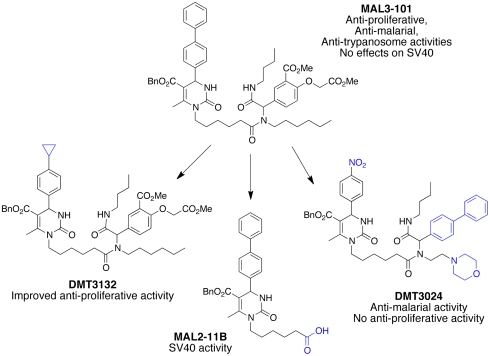
In 2004, the Wipf and Brodsky labs reported on a family of pyrimidinones, exemplified by MAL3-101, which modulated Hsp70 function (4). MAL3-101 was the product of a tandem MCR process under investigation in the Wipf and UPCMLD labs, combining the heterocyclic scaffold of the Biginelli-derived pyrimidinones with the large side-chain diversity accessible by Ugi four-component condensations. The availability of this probe enabled the interrogation of Hsp70’s role in physiological and pathophysiological pathways by numerous laboratories (27–29). Based on these studies, the UPCMLD designed and synthesized second- and third-generation libraries using MAL3-101 as a lead in an effort to improve potency, understand the structural requirements for Hsp70 modulating activity, and improve physical properties. Subsequently, collections of these analogs were submitted for biological evaluations in a variety of Hsp70-dependent assays. These efforts resulted in a suite of MAL3-101 analogs, all modulating Hsp70 function, but exhibiting divergent pharmacological activity (Fig. 6). For example, one analog, DMT3132, maintained Hsp70 modulating activity, but exhibited improved antiproliferative activity in breast cancer cells (IC50 = 3 μM in human breast carcinoma SK-BR-3 cells, compared to IC50 = 27 μM for MAL3-101) and improved physical properties [e.g., molecular weight reduced from 930 to 818; Calculated partition coefficient (clogP) reduced from > 10 to < 8; clogS (calculated log of solubility) reduced from -11.7 to -9.7](30). Other analogs compromised the replication of the causative agent of malaria, Plasmodium falciparum, with ED50’s ranging from 30 nM to ∼2 μM. For example, DMT3024 altered the ATP hydrolyzing activity of P. falciparum Hsp70, inhibited the replication of the parasite in human erythrocyctes, but lacked antiproliferative activity (GI50 SK-BR-3 cells > 50 μM) (31). MAL3-101 itself also inhibited the replication of a Trypanosome species that causes sleeping sickness (29). Finally, MAL2-11B, an intermediate in the synthesis of MAL3-101, inhibited the ATPase activity of Hsp70 as well as the ATPase activity of a chaperone-like protein, T antigen, which is required for polyomavirus (PyV) replication (32). Infection by members of the PyV family contribute to AIDS-related dementias and renal transplant rejection (33).
Fig. 6.
MAL3-101 and three analogs with differential activities in models of cancer, malaria, trypanosome infection, and polyomavirus infection.
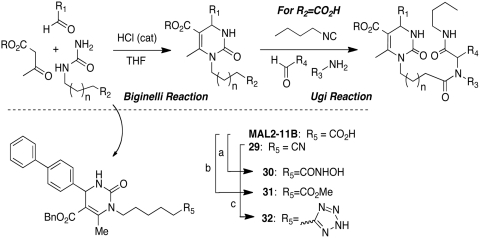
The original libraries based on MAL3-101 were generated using the Biginelli-Ugi multicomponent cascade strategy (Fig. 7) that incorporates at least six points of diversification (34). Initial modifications focused on the C4-pyrimidine substituent (R1), as well as substitutions on the amide side-chain (R3 and R4). Subsequent libraries also added changes in the linker (n) and the ester (R) groups. To date, six libraries based on MAL3-101 and an earlier design have been obtained, resulting in ∼500 analogs. We have found distinct, and often independent, SARs for each pharmacological activity (4, 30, 31, 32).
Fig. 7.
General synthesis of MAL3-101 libraries, MAL2-11B, and MAL2-11B isosteres. (a) (i) TEA, ClCO2Et; (ii) NH2OH; (b) SOCl2, MeOH; (c) TMSN3, TBAF, microwave irradiation.
Hsp70 activity is enhanced by interaction with Hsp40 chaperones, which contain a “J domain” that binds directly to the ATPase domain in Hsp70 (35). Thus, the divergent pharmacological activities of closely related structures might be explained by differential effects on J domain interaction. Moreover, because there are significantly more Hsp40 homologs than Hsp70s present in cells, the interaction of Hsp70 with a J domain protein might be inhibited by one analog, while Hsp70’s interaction with a distinct cochaperone is more effectively inhibited by another. Furthermore, nonmammalian Hsp70s (e.g., those found in P. falciparum and trypanosomes) may exhibit unique preferences of Hsp40s. Recent biochemical and structural studies support the notion that closely related small molecules can exhibit opposing effects on Hsp70 function. For example, MAL2-11B suppressed J domain-enhanced Hsp70 ATPase activity, but a structural analog stimulated ATPase activity (36). These results are in-line with our model that chemically related structures can exhibit differential or even opposing effects on Hsp70, and that the resulting biological outcomes may depend on the Hsp70 and Hsp40 repertoire in the cell.
Optimization of Pyrimidinone Probes of SV40 T-Antigen.
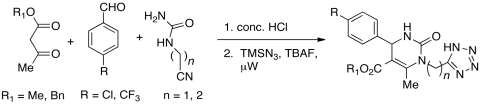
As noted above, MAL2-11B inhibited the endogenous ATPase activity and T-antigen mediated activation of Hsp70. This effect was observed at 100 μM. MAL2-11B also reduced the replication of a PyV, simian virus 40 (SV40), in cell culture by inhibiting viral DNA synthesis. Interestingly, MAL3-101 had little effect on PyV replication. MAL2-11B also reduced the growth of a human polyomavirus (BK virus) in kidney cells, as measured by assaying the levels of viral DNA, with no apparent effect on cell viability (32). To optimize the antiviral activity and properties of the MAL2-11B series of pyrimidinones, and to develop a SAR, we incorporated isosteric replacements for the carboxylic acid groups. Our initial hypothesis was that by replacing the charged carboxylate with bioisosteres, such as tetrazoles and hydroxamic acids, we could maintain the compound’s biological properties but improve cellular permeability. A small set of target compounds was assembled using the one-pot, three-component Biginelli sequence (Fig. 7, SI Appendix: S2). This sequence directly led to the acid MAL2-11B and the nitrile 29, by use of the appropriate urea in the Biginelli reaction (34). The carboxylic acid derivative, MAL2-11B was converted to the hydroxamic acid and methyl ester analogs (30 and 31, respectively). The hydroxamic acid was formed via the mixed anhydride, and the methyl ester was synthesized under standard conditions using thionyl chloride and methanol (37). Tetrazole 32 was generated via a microwave facilitated [3 + 2] cycloaddition of 29 with trimethylsilyl azide using tetrabutylammonium fluoride as catalyst under solventless conditions (38).
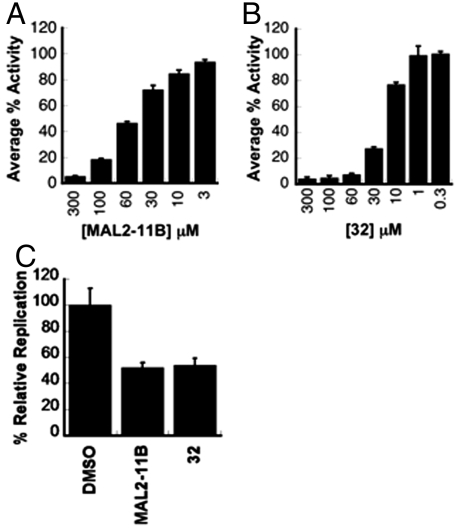
Each agent was examined for its effects on the ATP hydrolyzing activity of T-antigen (SI Appendix: S2). As shown in Fig. 8 A and B, the tetrazole analog (32) inhibited the ATPase activity of purified T-antigen somewhat more potently than MAL2-11B. For example, at a final concentration of 60 μM, the residual activity in the presence of MAL2-11B was 47%, while in the presence of tetrazole 32 only 7.5% of the initial activity remained. Neither the nitrile (29), the hydroxamic acid (30), nor the ester (31) were active in this assay. We then measured the replication of BK virus in a mammalian kidney cell culture system (SI Appendix: S2). When examined at a final concentration where both agents reduced the ATPase activity to < 20% (100 μM), the compounds inhibited viral replication to the same degree (Fig. 8C). Of note, there were no adverse effects on cell growth or viability at this concentration.
Fig. 8.
Effect of MAL2-11B and 32 on T-antigen and viral replication. Steady-state ATPase assays with purified T-antigen were performed as described (32, SI Appendix: S2) in the presence of the indicated concentrations of (A) MAL2-11B or (B) 32. Data represent the means of at least three determinations ± SD. (C) The replication of the BK virus and viral DNA content were assessed in the presence of the indicated supplements as outlined (SI Appendix: S2). MAL2-11B and 32 were used at a final concentration of 100 μM. Data represent the means of two independent infections, each performed in triplicate, ± SEM; p ≤ 0.007 for DMSO vs. the compound treated samples.
Based on the promising activity of the tetrazole 32 and its potential for improved permeability, a small library of tetrazole analogs was prepared. This library was designed to further probe the SAR while maintaining or improving physical properties. Modifications in the phenyl ring (R), the ester (R1), and the carbon linker (n) were made (Fig. 9), and a total of eight compounds have so far been prepared in this preliminary work. These compounds are currently undergoing biological evaluation in PyV replication assays.
Fig. 9.
Synthesis of tetrazole library.
Discussion
The two examples presented herein highlight the power of synthetic chemistry methodology and the impact it can have on biomedical research. In the first example, a DOS strategy afforded an array of scaffolds, one of which (exemplified by MARPIN) exhibited activity in a pathway screen for inhibitors of ATM/ATR signaling. Next, SAR data were obtained, sites for immobilization were identified, and potential cellular targets were uncovered. The use of competitors (both active and inactive variants) confirmed that a subset of small-molecule-binding proteins most likely represent relevant biological targets. Although further investigation of these binding proteins is required, this approach is one key step toward target identification of small-molecule modulators of key protein(s) in ATM/ATR signaling. Knowledge of this protein target may provide a unique target for cancer therapy.
In the second example, access to an efficient chemical methodology allowed the rapid synthesis of libraries of diverse analogs of a tandem MCR-derived scaffold. The resulting building blocks provided easy access to diverse analogs, all impacting Hsp70 chaperone activity, but with divergent pharmacological activities. Specifically, pyrimidinones have been identified with potential applications in cancer, malaria, sleeping sickness, and PyV-associated disease. Further synthetic manipulations to optimize the activity, selectivity, and physical properties of this class of Hsp modulators provide excellent starting points for drug discovery efforts in various therapeutic areas.
Materials and Methods
Synthetic procedures (SI Appendix: S1 and S2), compound characterization (Dataset S1), and assay protocols (SI Appendix: S1 and S2) can be found in SI Appendices: S1 and S2.
Supplementary Material
Acknowledgments.
We thank Jim Pipas and Michael Imperiale for reagents and advice on polyomavirus propagation. D.M.H. thanks Jay Kostman for technical assistance during manuscript preparation. This work was supported in part by National Institutes of Health grants P50GM067082 (the University of Pittsburgh CMLD), P41GM081275, DK79307 (the University of Pittsburgh George O’Brien Kidney Research Core Center) and R01-AR049832. A.W.I. acknowledges support from the Beckman Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
*Data available in August 2010 reflects approximately 1,000 compounds submitted to the MLSMR prior to 2009. There is a lag time between when UPCMLD compounds are submitted to the MLSMR and their distribution to MLSCN and MLPCN screening centers for inclusion in screening campaigns.
†The syntheses of these second generation analogs were carried out as previously reported by the Brummond labs and synthetic details can be found in Text S1 and Dataset S1. All compounds were obtained as a racemates.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015251108/-/DCSupplemental.
References
- 1.Center for Chemical Methodologies & Library Development (UPCMLD) 2002. http://ccc.chem.pitt.edu/
- 2.Dancik V, Seiler KP, Young DW, Schreiber SL, Clemons PA. Distinct biological network properties between the targets of natural products and disease genes. J Am Chem Soc. 2010;132:9259–9261. doi: 10.1021/ja102798t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.NCBI Pubchem Structure Search: 2010. http://pubchem.ncbi.nlm.nih.gov/
- 4.Fewell SW, et al. Small molecule modulators of endogenous and co-chaperon-stimulated Hsp70 ATPase activity. J Biol Chem. 2004;279:51131–51140. doi: 10.1074/jbc.M404857200. [DOI] [PubMed] [Google Scholar]
- 5.Tan DS. Diversity-oriented synthesis: exploring the intersections between chemistry and biology. Nat Chem Biol. 2005;1:74–84. doi: 10.1038/nchembio0705-74. [DOI] [PubMed] [Google Scholar]
- 6.Dandapani S, Marcaurelle LA. Current strategies for diversity-oriented synthesis. Curr Opin Chem Biol. 2010;14:362–370. doi: 10.1016/j.cbpa.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 7.Lipkus AH, et al. A scaffold analysis of the 24 million compounds in the Chemical Abstracts (CAS) database reveals that half of the compounds can be described by a mere 143 molecular frameworks. J Org Chem. 2008;73:4443–4451. doi: 10.1021/jo8001276. [DOI] [PubMed] [Google Scholar]
- 8.Brummond KM, Mitasev B. Allenes and transition metals: a diverging approach to heterocycles. Org Lett. 2004;6:2245–2248. doi: 10.1021/ol0492391. [DOI] [PubMed] [Google Scholar]
- 9.Brummond KM, Chen D. Microwave-assisted intramolecular [2 + 2] allenic cycloaddition reaction for the rapid assembly of bicyclo[4.2.0]octa-1,6,-dienes and bicyclo[5.2.0]nona-1,7-dienes. Org Lett. 2005;7:3473–3475. doi: 10.1021/ol051115g. [DOI] [PubMed] [Google Scholar]
- 10.Brummond KM, Chen H, Mitasev B, Casarez AD. A Rhodium(I)-catalyzed ene-allene carbocyclization strategy for the formation of azepines and oxepines. Org Lett. 2004;6:2161–2163. doi: 10.1021/ol049390a. [DOI] [PubMed] [Google Scholar]
- 11.Hann MM, Oprea TI. Pursuing the lead likeness concept in pharmaceutical research. Curr Opin Chem Biol. 2004;8:255–263. doi: 10.1016/j.cbpa.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Werner S, et al. Solution-phase synthesis of a tricyclic pyrrole-2-carboxamide discovery library applying a Stetter-Paal-Knorr reaction sequence. J Comb Chem. 2006;8:368–380. doi: 10.1021/cc050160c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bizzantz C, Kuhn B, Stahl M. A medicinal chemist’s guide to molecular interactions. J Med Chem. 2010;53:5061–5084. doi: 10.1021/jm100112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou BB, Bartek J. Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nat Rev Cancer. 2004;4:216–225. doi: 10.1038/nrc1296. [DOI] [PubMed] [Google Scholar]
- 15.Nghiem P, Park PK, Kim YS, Vaziri C, Schreiber SL. ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc Natl Acad Sci USA. 2001;98:9092–9097. doi: 10.1073/pnas.161281798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potashman MH, Duggan ME. Covalent modifiers: an orthogonal approach to drug design. J Med Chem. 2009;52:1231–1246. doi: 10.1021/jm8008597. [DOI] [PubMed] [Google Scholar]
- 17.Godl K, et al. An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc Natl Acad Sci USA. 2003;100:15434–15439. doi: 10.1073/pnas.2535024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ong SE, et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci USA. 2009;106:4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherman MY, Goldberg AL. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 20.Brodsky J, Chiosis G. Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr Top Med Chem. 2006;6:1215–1225. doi: 10.2174/156802606777811997. [DOI] [PubMed] [Google Scholar]
- 21.Didelot C, et al. Anti-cancer therapeutic approaches based on intracellular and extracellular heat shock proteins. Curr Med Chem. 2007;14:2839–2847. doi: 10.2174/092986707782360079. [DOI] [PubMed] [Google Scholar]
- 22.Evans CG, Chang L, Gestwicki JE. Heat shock protein 70 (Hsp70) as an emerging drug target. J Med Chem. 2010;53:4585–4602. doi: 10.1021/jm100054f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shonhai A. Plasmodial heat shock proteins: targets for chemotherapy. FEMS Immunol Med Microbiol. 2010;58:61–74. doi: 10.1111/j.1574-695X.2009.00639.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang YP, et al. Heat stress cognate 70 host protein as a potential drug target against drug resistance in hepatitis B virus. Antimicrob Agents Chemother. 2010;54:2070–2077. doi: 10.1128/AAC.01764-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powers MV, et al. Targeting Hsp70: the second potentially druggable heat shock protein and molecular chaperone? Cell Cycle. 2010;9:1542–1550. doi: 10.4161/cc.9.8.11204. [DOI] [PubMed] [Google Scholar]
- 26.Patury S, Miytata Y, Gestwicki JE. Pharmacological targeting of the Hsp70 chaperone. Curr Top Med Chem. 2009;9:1337–1351. doi: 10.2174/156802609789895674. and references cited within. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodina A, et al. Selective compounds define Hsp90 as a major inhibitor of apoptosis in small-cell lung cancer. Nat Chem Biol. 2007;3:498–507. doi: 10.1038/nchembio.2007.10. [DOI] [PubMed] [Google Scholar]
- 28.Rabu C, Wipf P, Brodsky J, High S. A precursor-specific rold for Hsp40/hsc70 during tail-anchored protein integration at the endoplasmic reticulum. J Biol Chem. 2008;283:27504–27513. doi: 10.1074/jbc.M804591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patham B, et al. Post-translational import of protein into the endoplasmic reticulum of a trypanosome: an in vitro system for discovery of anti-trypansomal chemical entities. Biochem J. 2009;419:507–517. doi: 10.1042/BJ20081787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright CM, et al. Pyrimidinone-peptoid hybrid molecules with distinct effects on molecular chaperone function and cell proliferation. Bioorg Med Chem. 2008;16:3291–3301. doi: 10.1016/j.bmc.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiang AN, et al. Select pyrimidinones inhibit the propagation of the malarial parasite, Plasmodium falciparum. Bioorg Med Chem. 2009;17:1527–1533. doi: 10.1016/j.bmc.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright CM, et al. Inhibition of simian virus 40 replication by targeting the molecular chaperone function and ATPase activity of T antigen. Virus Res. 2009;141:71–80. doi: 10.1016/j.virusres.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang M, Abend JR, Johnson SF, Imperiale MJ. The role of polyomaviruses in human disease. Virology. 2009;384:266–273. doi: 10.1016/j.virol.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Werner S, Turner DM, Lyon MA, Huryn DM, Wipf P. A focused library of tetrahydropyrimidinone amides via a tandem Biginelli-Ugi multi-component process. Synlett. 2006;14:2334–2338. [Google Scholar]
- 35.Kampinga HH, Craig EA. The Hsp70 chaperone machinery: J proteins as drivers of functional specificity. Nature Rev Mol Cell Bio. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wisen S, et al. Binding of a small molecule at a protein-protein interface regulates the chaperone activity of Hsp70-Hsp40. ACS Chem Biol. 2010;5:611–622. doi: 10.1021/cb1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy AS, Kumar MS, Reddy GR. A convenient method for the preparation of hydroxamic acids. Tetrahedron Lett. 2000;41:6285–6288. [Google Scholar]
- 38.Amantini D, Beleggia R, Fringuelli F, Pizzo F, Vaccaro L. TBAF-catalyzed synthesis of 5-substituted 1H-tetrazoles under solventless conditions. J Org Chem. 2004;69:2896–2898. doi: 10.1021/jo0499468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.