Abstract
Quantitative resistance to plant pathogens, controlled by multiple loci of small effect, is important for food production, food security, and food safety but is poorly understood. To gain insights into the genetic architecture of quantitative resistance in maize, we evaluated a 5,000-inbred-line nested association mapping population for resistance to northern leaf blight, a maize disease of global economic importance. Twenty-nine quantitative trait loci were identified, and most had multiple alleles. The large variation in resistance phenotypes could be attributed to the accumulation of numerous loci of small additive effects. Genome-wide nested association mapping, using 1.6 million SNPs, identified multiple candidate genes related to plant defense, including receptor-like kinase genes similar to those involved in basal defense. These results are consistent with the hypothesis that quantitative disease resistance in plants is conditioned by a range of mechanisms and could have considerable mechanistic overlap with basal resistance.
Keywords: Zea mays, Setosphaeria turcica, Exserohilum turcicum, quantitative trait loci mapping
Resistance to diseases in many plant and animal systems is based on complex inheritance. Complex or quantitative disease resistance in plants (QDR), conditioned by numerous genes of small effect, is of practical importance in agriculture because it is less readily overcome by the evolution of pathogen populations than simply inherited forms of resistance (1). Resistance-gene alleles providing complete or near-complete resistance are typically involved in direct or indirect recognition of the pathogen (2). Mutation or loss of the recognition target in the pathogen renders one or more resistance alleles at these genes ineffective. On the other hand, QDR tends to be associated with more durable resistance, as a pathogen strain that overcomes a single allele of small effect does not gain a large selective advantage, and loss of effectiveness of a single gene does not leave the host completely susceptible (1, 3–5). For some important plant diseases, including most caused by necrotrophic pathogens, resistance genes of large effect are unknown and QDR is the only available form of resistance.
Race-specific resistance genes have been found to largely fall into few classes of genes, the most common being the group containing nucleotide binding and leucine-rich repeat (NB-LRR) motifs (2). In maize, major race-specific resistance genes are limited mainly to the rust diseases (6) and to a lesser extent northern leaf blight (7). Although it appears that only a limited number of mechanisms provide plants with potentially complete resistance against pathogens, it is likely that a wider range of mechanisms allow plants to reduce the success of their pathogens. Indeed, several types of novel defense-related genes conditioning quantitative resistance have been recently identified (8–11), supporting the proposition that a range of genes and mechanisms are involved in QDR (3).
Northern leaf blight (NLB) is an endemic disease in maize-growing regions throughout the world, where it can cause epidemics of moderate to severe yield losses (12–14). Caused by the hemibiotrophic fungal pathogen Setosphaeria turcica (anamorph Exserohilum turcicum), NLB is of particular concern in the tropical highlands, where conditions favor disease development and subsistence farming and food insecurity are prevalent. Previous research on resistance to NLB has suggested a complex genetic architecture, with quantitative trait loci (QTL) dispersed throughout the genome (6, 15, 16). Three genes conferring race-specific, although incomplete, resistance have been mapped for S. turcica: Ht1, located in maize bin 2.08 (17), and Ht2 (18–20) and Htn1 (21), located in maize bin 8.06. Due to low resolution of previous QTL mapping studies, it has been difficult to use this information in breeding programs, to identify positional candidate genes or to make strong inferences on the linkage relationships among these and other QTL. In light of the economic importance of resistance to NLB and the availability of advanced genetic resources in maize, we have used this pathosystem as a model for understanding the genetic architecture of QDR in plants while gaining valuable information for maize resistance breeding.
A large nested association mapping (NAM) population consisting of 25 recombinant inbred-line (RIL) populations has been developed in maize for dissection of complex traits (22–24). The 25 NAM founder lines were selected to maximize diversity from a larger panel of diverse maize inbreds (25), and each was crossed in a half-sib design to the common reference parent B73. The half-sib design enables joint analysis of the full population and narrows the physiological maturity range, which is beneficial for disease resistance evaluations. RILs from the intermated B73 x Mo17 population (IBM) were included as a 26th family (26). The NAM RILs are genotyped with 1,106 SNP markers (23) and are publicly available (http://www.panzea.org).
Results and Discussion
We evaluated 4,630 of the NAM RILs, along with each of the founder inbred lines, over three seasons for resistance to NLB (SI Appendix, Table S1) in nurseries artificially inoculated with a single isolate of S. turcica race 1. Lines were scored at three time points each season, and RIL best linear unbiased predictions (BLUPs) were predicted from a multivariate mixed model for each rating and an NLB index was calculated by averaging the three BLUPs for each line. The founder lines showed a wide range of phenotypic variation for resistance to NLB (Fig. 1A and SI Appendix, Table S1 and Fig. S1). The common reference parent, B73, was moderately susceptible, with 34% diseased leaf area (DLA) at the third disease rating (rating 3). At rating 3, the NAM founders varied over a 10-fold range (5–60% DLA), whereas the RIL progeny varied over nearly a 40-fold range (2.5–80% DLA).
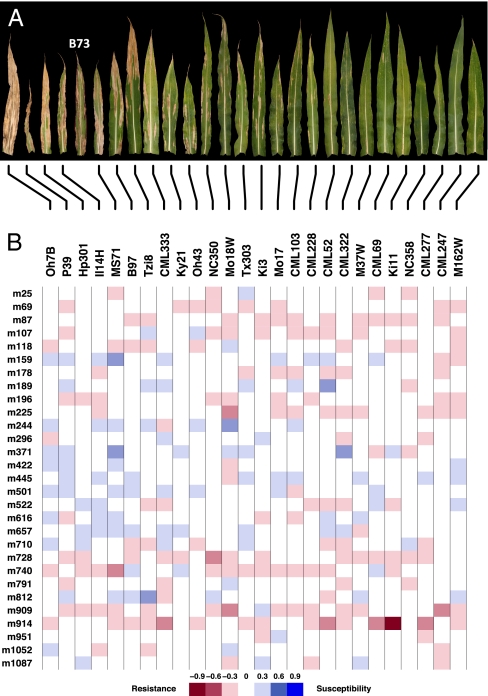
Fig. 1.
Allelic component and observed phenotype for founder lines of the NAM population. (A) Representative ear leaves from each founder inbred line and B73, the NAM reference parent. S. turcica, the causal agent of NLB, produces large cigar-shaped lesions that may coalesce, leading to complete blighting of the leaf in more susceptible inbred lines. (B) Heat map showing estimated allele effects for NLB resistance at 29 QTL identified in the NAM population for each founder inbred line. The large variation in the resistance phenotype results from the accumulation of alleles for resistance or susceptibility at multiple QTL in each founder line. Alleles for resistance are colored in red and alleles for susceptibility are in blue. The scale is shown on a square root transformation of percentage diseased leaf area.
Although NLB resistance is a complex trait, phenotypic observations were very consistent under our field conditions and we were able to produce very repeatable phenotypes. Broad-sense heritability estimates on a line mean basis within individual families averaged 0.63, 0.71, and 0.68 for the three ratings, respectively, and 0.77 for the NLB index (SI Appendix, Table S2). The NLB index averaged all three ratings, leading to more accurate phenotypic values and increased repeatability. As NAM captures variation across diverse founder lines as well as progeny segregation within families, the heritability for the population as a whole (0.87) was higher than for any of the individual families.
There were large differences observed among families (SI Appendix, Table S1 and Fig. S1), with family means for rating 3 ranging from 15.7 to 48.9% DLA. The average family performance was highly correlated (R2 = 0.80) with the phenotype of that family's respective founder (SI Appendix, Fig. S2), supporting the observation that NLB resistance is primarily conditioned by additive genetic variance (7, 27). We observed minimal genotype-by-environment interaction (GxE) for NLB resistance in NAM. GxE was on average 20%, 13%, and 14% of the genotype variance for the three ratings, respectively. There was very little GxE for family means, as the family-by-environment variance was less than 5% of the family genetic variance for all ratings. It should be noted that this study was conducted at a single location, and it is possible that GxE would be higher if the analysis had included more diverse locations. Because the population was inoculated with a single isolate of S. turcica race 1, inference cannot be made regarding possible genotype-by-pathogen interaction or the race specificity of the QTL identified.
We observed a strong negative correlation between flowering time (days to anthesis; DTA) and NLB resistance in the founder lines (r = −0.59), which is consistent with previous reports (7, 16, 28). Across NAM, this correlation was weaker (r = −0.32), although the family means for DTA and NLB resistance were still strongly correlated (r = −0.63). The correlation between DTA and NLB resistance within families was much lower. Of the 26 NAM families, only 8 had a significant correlation (P < 0.05) between DTA and NLB resistance. The strongest correlation (r = −0.26) was found in the CML322 x B73 family. This indicates that, although NLB resistance and relative maturity are confounded in the NAM founder lines, much of this correlation was lost due to genome shuffling during RIL development, and the modest negative correlation that remains is largely due to differences between families rather than within-family covariance.
We used a fixed-effects model with a main effect for family and marker effects nested within families in a joint linkage mapping strategy to identify QTL for the NLB index across NAM (22). Nesting markers within families restricted the analysis to estimating unique allele effects at each locus for each family, giving an estimated effect for each founder line at each QTL (26 founders × 29 QTL = 754 modeled allele effects). Although most founder lines did not have an allele effect significantly different from that of B73 at most QTL, this nested model allowed multiple allelic effects to be modeled for each QTL. As relatively maturity was correlated with NLB resistance, DTA was included as a covariate to reduce the confounding impact of flowering time on estimating allele effects for the NLB resistance index. Stepwise model selection identified 29 QTL that accounted for 77% of the variance in the BLUPs (SI Appendix, Table S3). After fitting the full QTL model, however, DTA was not a significant covariate. The reduced marginal effect of DTA in the full QTL model indicates that the markers identified for NLB resistance also explain variation in DTA (possible linkage or pleiotropy). This issue is examined in further detail below.
Although a few QTL with relatively large allele effects were detected, most of the QTL had small estimated effects. Only 24 of the 754 modeled QTL alleles were estimated to give an increase or decrease of more than 3% DLA, and only 3 QTL alleles had an estimated effect larger than ±5% (SI Appendix, Fig. S3). Most of the individual QTL allele effects were estimated as too small to be visually distinguished (a trained scorer can see a difference of about 4% DLA) without statistical inference. This is consistent with results obtained for flowering time in maize, for which most alleles had effect sizes that were smaller than the observation unit of 1 d (22).
Reflecting the continuum of the resistance phenotypes in NAM, the founder genomes were found to be a mosaic of loci for resistance and susceptibility. Most founders had alleles for both resistance and susceptibility (Fig. 1B), and individual families had between 3 and 14 segregating QTL, with an average of 8.7. Even the very susceptible inbred line Oh7b had three alleles for resistance, whereas the most resistant genotypes incorporated favorable alleles at only 5–8 of the QTL. At 21 of the 29 QTL (72%), estimated allele effects were higher, lower, and equivalent to that of the reference B73 allele, indicating that at least three alleles are present at these QTL in the founder lines (SI Appendix, Fig. S4). This supports the hypothesis that multiple alleles are present at many QTL (22). We tested epistasis as two-way marker interactions and did not detect epistatic effects between QTL markers with additive effects or between QTL markers with additive effects and all other marker loci. To evaluate the impact of environment on the identified QTL, we fit a similar fixed-effects model with main effects for family and environment, markers nested within families, and additional QTL-by-environment interaction terms for each QTL. There were no significant QTL-by-environment interactions detected by this model, again confirming that NLB resistance in NAM can be largely attributed to simple additive genetic variance.
We hypothesized that incubation period (IP), a resistance component for NLB measured as the number of days after inoculation when disease lesions first appear, could be used to identify resistance that is effective during the early stages of pathogenesis. We measured IP for NAM during the first year of this study and modeled these data with a Cox proportional hazards model, because IP is analogous to survival data from clinical trials. Using stepwise model selection, four QTL for IP were identified in NAM. The three most significant QTL for IP colocalized with large-effect QTL for DLA on chromosomes 1, 6, and 8. The remaining QTL for IP was identified on chr. 3 (m352; 61.8 cM; 132.3 Mb) at a location not associated with disease severity. This QTL could therefore be effective during initial pathogen infection but not during the later stages of pathogen development.
Presumably due to greater statistical power in NAM as well as the ability to survey a broad range of germplasm, additional QTL were identified (8.7 per family) compared with previous studies (6.4 per biparental population) (6). We identified a large-effect QTL on chr. 8 at 152.2 Mb segregating in multiple families. This is likely to be Ht2, as this position is consistent with the physical location identified through fine mapping (19). Ht2 is race-specific but does not condition complete immunity, and can be observed as a large-effect QTL. A bimodal phenotypic distribution was not observed even in the family segregating for the largest-effect allele at this QTL (Ki11 x B73), and there was no other phenotypic evidence of a “major” gene. The S. turcica isolate used in this study was virulent on Ht1, and no large QTL were observed at the Ht1 region of chr. 2.
The numerous QTL identified here, each with small allelic effects, explained a substantial proportion of the observed phenotypic variance in the founder lines. There was a strong and significant correlation between the empirical phenotypic observations and the predicted founder phenotypes calculated as the sum of all QTL effects for each founder (SI Appendix, Fig. S5). The coefficient of determination for the regression of the BLUP phenotype on the sum of significant allele effects (P < 0.05) was 0.60. Including all allele effects, both significant and nonsignificant, increased the correlation (R2 = 0.71), indicating that very small (nonsignificant) allele effects at QTL contributed to the resistance phenotype. The broad-sense heritability of the NLB index for the NAM founders was 0.74, and the full QTL model was able to explain 96% (0.71/0.74) of this variation.
We examined each QTL for correlated allele effects between DTA and NLB resistance but did not find evidence of pleiotropic effects of DTA on NLB resistance. Only one QTL was found to have correlated effects for NLB resistance and DTA (P < 0.05). This QTL (m25; 17.5 Mb on chr. 1) had a positive correlation between NLB allele effects and DTA allele effects, in contrast to the negative phenotypic correlation seen between the traits in NAM. Allele effects for the large-effect QTL identified on chr. 8 in the region of Vgt1, a flowering-time QTL in maize (29), did not show a correlation between DTA and NLB resistance, indicating that the resistance is not the result of a pleiotropic effect of Vgt1. These observations, along with the observation of reduced phenotypic correlations within families, support the conclusion that the strong phenotypic association between NLB resistance and relative maturity in the founder lines is predominantly due to confounding population structure for NLB resistance and DTA, and to a limited extent genetic linkage within families, rather than pleiotropic effects of flowering time on NLB resistance.
Most tropical maize lines have a higher level of NLB resistance (and disease resistance in general) than their temperate counterparts, which likely reflects the favorable conditions for disease development in the tropics and high incentive for breeders to select for resistance in these environments. We examined the relationship between allelic effects at each QTL and quantitative measures of allelic origin. There were five QTL that correlated with tropical/subtropical estimates of inbred-line origin. The tropical allele conditioned resistance for four of these QTL, whereas the alternate allele was resistant at one QTL (SI Appendix, Fig. S6). We observed that one QTL was correlated with nonstiff-stalk origin, and the nonstiff-stalk allele at this locus was associated with increased susceptibility. These correlations were not strong, and lend only limited support to the hypothesis of preferential selection for disease resistance alleles in tropical material.
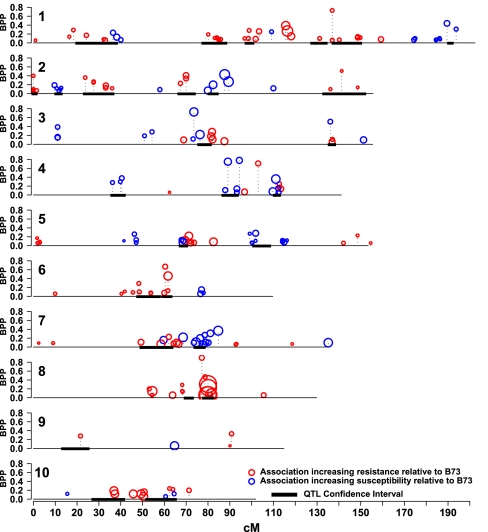
We used a nested association mapping approach, leveraging the linkage disequilibrium (LD) among founder lines for a genome-wide association study (GWAS) of NLB resistance (30). We imputed 1.6 million SNPs from the first-generation maize HapMap scored on the NAM founder lines to RIL progeny using pedigree and linkage marker information (24, 30, 31). Trait–marker associations were then evaluated by chromosome. On a per-chromosome basis, we used residual phenotypic values after accounting for all QTL on other chromosomes, and fit 100 stepwise linear models using bootstrap samples (32). The bootstrap posterior probability (BPP) was calculated as the number of bootstrap samples in which an SNP was selected out of the 100 total. Through bootstrap analysis, 208 SNPs were identified as significantly associated with the NLB index (BPP > 0.05; Fig. 2), and 56 of these had posterior probabilities of greater than 0.2. Thirty-nine percent (82 of 208) of the SNP associations were within QTL confidence intervals, and 28 of 29 QTL had one or more SNP associations. Exact overlap of linkage mapping and GWAS is not expected, as linkage mapping tests markers within a family whereas GWAS tests marker effects across families, leading to different strengths and weaknesses in each approach (30). Predicted genes containing or directly adjacent to SNP associations were evaluated as potential candidate genes for QDR (Table 1 and Dataset S1).
Fig. 2.
Using the NAM reference design, 1,606,526 SNPs were imputed from the NAM founders and tested for association with resistance to NLB. A bootstrap permutation of stepwise SNP selection for each chromosome was run for 100 iterations and the bootstrap posterior probability for each SNP was calculated. The BPP is shown on the y axis, with red and blue circles showing the position and BPP of SNP associations that increase and decrease resistance, respectively, compared with B73. The diameter of each bubble circle represents the size of the estimated effect of that SNP on NLB. Regions within confidence intervals of QTL are shown as solid lines under the SNP profile.
Table 1.
A subset of the 208 SNP loci found to be associated with resistance to NLB by nested association mapping
| SNP no. | Chromosome | Physical position (AGP v.1) | BPP | Average SNP effect | cM | Inside QTL CI | Candidate gene |
| 4 | 1 | 12136678 | 0.17 | −0.346 | 24.26 | Yes | Serine-threonine protein kinase (47) |
| 6 | 1 | 16387974 | 0.06 | −0.395 | 32.85 | Yes | Germin (11) |
| 8 | 1 | 21699987 | 0.13 | 0.490 | 38.21 | No | Antifreeze (41, 44–46) |
| 10 | 1 | 76772901 | 0.19 | −0.260 | 80.13 | Yes | Cytochrome P450 (48, 49) |
| 12 | 1 | 88927678 | 0.12 | −0.277 | 84.11 | Yes | Serine-threonine protein kinase (47) |
| 19 | 1 | 183754852 | 0.28 | −0.299 | 98.87 | Yes | Mov34/MPN/PAD-1 |
| 33 | 1 | 264713872 | 0.08 | −0.418 | 159.29 | No | Antifreeze (41, 44–46) |
| 35 | 1 | 280343673 | 0.10 | 0.300 | 174.88 | No | Basic leucine zipper transcription factor (52) |
| 40 | 1 | 289465566 | 0.08 | 0.239 | 186.39 | No | Antifreeze (41, 44–46) |
| 43 | 2 | 1210644 | 0.40 | −0.265 | −0.05 | No | Mlo (54, 55) |
| 44 | 2 | 1221350 | 0.06 | −0.233 | 0.00 | No | Mlo (54, 55) |
| 49 | 2 | 3852515 | 0.12 | 0.253 | 10.67 | Yes | Serine-threonine protein kinase (47) |
| 52 | 2 | 4500004 | 0.13 | 0.251 | 12.73 | No | Serine-threonine protein kinase (47) |
| 55 | 2 | 9394756 | 0.27 | −0.276 | 27.58 | Yes | RLK (35) |
| 64 | 2 | 160834095 | 0.20 | 0.607 | 82.36 | Yes | RLK (35) |
| 66 | 2 | 179024847 | 0.27 | 0.760 | 89.43 | No | BTB/POZ-like (53) |
| 71 | 3 | 3382179 | 0.15 | 0.389 | 11.08 | No | Antifreeze (41, 44–46) |
| 72 | 3 | 3382266 | 0.39 | 0.356 | 11.08 | No | Antifreeze (41, 44–46) |
| 93 | 4 | 177670891 | 0.75 | 0.572 | 89.12 | Yes | Antifreeze (41, 44–46)/PR transcriptional factor and ERF (50, 51) |
| 111 | 5 | 13659256 | 0.11 | 0.174 | 41.49 | No | Antifreeze (41, 44–46) |
| 125 | 5 | 190880589 | 0.07 | 0.251 | 100.08 | No | PR transcriptional factor and ERF (50, 51) |
| 126 | 5 | 191109424 | 0.06 | 0.267 | 100.26 | No | Serine-threonine protein kinase (47) |
| 129 | 5 | 203735206 | 0.12 | 0.305 | 114.14 | No | RLK (35) |
| 130 | 5 | 203735229 | 0.09 | 0.307 | 114.14 | No | RLK (35) |
| 132 | 5 | 204103356 | 0.08 | 0.442 | 114.92 | No | Antifreeze (41, 44–46) |
| 139 | 6 | 116055358 | 0.11 | −0.222 | 41.88 | Yes | RLK (35) |
| 143 | 6 | 138675782 | 0.07 | −0.274 | 53.79 | Yes | Antifreeze (41, 44–46) |
| 144 | 6 | 138675953 | 0.08 | −0.289 | 53.79 | Yes | Antifreeze (41, 44–46) |
| 145 | 6 | 148066145 | 0.08 | −0.395 | 59.98 | Yes | RLK (35) |
| 159 | 7 | 125153323 | 0.10 | −0.554 | 65.62 | No | Peptidase/serine-threonine protein kinase (47) |
| 162 | 7 | 135130480 | 0.10 | 0.412 | 73.54 | No | Basic leucine zipper transcription factor (52) |
| 173 | 7 | 153450268 | 0.07 | −0.252 | 93.05 | No | Antifreeze (41, 44–46) |
| 187 | 8 | 151397247 | 0.06 | −1.365 | 80.16 | Yes | ABC transporter (9) |
| 193 | 9 | 12502229 | 0.28 | −0.324 | 21.56 | Yes | RLK (35), CERK1 (36, 37), EFR (38) |
| 205 | 10 | 136256827 | 0.24 | −0.291 | 62.36 | Yes | Multiantimicrobial extrusion protein (56) |
The gene associations listed are those with plausible roles in plant defense based on the published literature. A complete list of resistance-associated SNPs with further details and corresponding genes is given in Dataset S1. cM, centimorgan imputed on NAM map; QTL CI, quantitative trait locus confidence interval; BPP, bootstrap posterior probability.
There were five SNP associations in or adjacent to LRR receptor-like kinase (RLK) genes and gene clusters and one additional association with a sixth LRR-related gene. LRR domains have long been implicated in plant disease resistance and are considered a hallmark of canonical (NB-LRR) “R-genes,” which also contain NB domains missing in RLKs (2). Other notable examples of RLKs include the “atypical” resistance genes Xa21 and Xa3/Xa6 (33, 34), as well as pattern-recognition receptors involved in basal resistance such as FLS2 (35), CERK1 (36, 37), and EFR (38). It has been hypothesized that quantitative resistance could share mechanisms with basal defense in the form of RLKs (2, 3), which is supported here and by recent GWAS for southern leaf blight resistance (39). Multiple associations (n = 11) identified candidate genes with antifreeze domains. This type of gene has high similarity to pathogenesis-related (PR) proteins and has been shown to enhance disease resistance (40–42). PR proteins both with (41, 43, 44) and without (45) antifreeze activity have been identified, and include β-1,3-glucanases, chitinases, and thaumatin-like and polygalacturonase inhibitor proteins (41, 44–46). The candidate genes identified here support the overlap in both form and function between PR and antifreeze proteins (40).
Several other candidate genes identified by GWAS included genes with recognizable roles in plant defense (Table 1). Several serine-threonine protein kinase genes were implicated; this family of genes is known to be involved in plant defense responses (47). A germin-like protein was associated with a QTL on chr. 6; germin-like proteins have been shown to be involved in QDR (11). A phytochrome P450 was implicated; P450 genes are known to be involved in phytoalexin production and other defense responses (48, 49). Several transcription factors were found to be associated with NLB resistance, including two PR transcription factors of the ERF (ethylene response factor) family. ERFs are activated at the convergence of the ethylene and jasmonate pathways and are involved in defense signaling for necrotrophic pathogens (50, 51). We identified two classes of genes known to interact with NPR1, including two basic leucine zipper transcription factors (52) and two BTB/POZ-like genes (53). An association on chr. 2 (1.2 Mb) contained an Mlo-like gene and a second gene with multiple transmembrane domains. The recessive mlo mutation in barley confers broad-spectrum resistance to biotrophic Erysiphe graminis f. sp. hordei, the causal pathogen of powdery mildew disease (54, 55). There was also an association with a multiantimicrobial extrusion protein (56).
On chr. 8, at the QTL that colocalizes with Ht2 (19), an association was found with posterior probability of 0.90. This resistance-associated SNP fell outside the QTL confidence interval, in a 160-kb region that has no predicted genes, but ∼73 kb upstream of a predicted MAP-kinase gene (GRMZM2G007848), a type of gene involved in defense signaling (57, 58). Causal polymorphisms may be distant from the functional gene, as in the case of vgt1 in maize, a QTL for flowering time located in a noncoding sequence 70 kb upstream of the functional transcription factor (29). Within the confidence interval of this QTL, however, there were associations adjacent to an ABC transporter/ATPase domain-containing gene and an RLK gene (GRMZM2G316907), both of which are credible candidate genes (9, 35–38).
GWAS using NAM has recently been demonstrated as an effective tool for gene identification in maize (30). Although many compelling candidate gene associations with defense-related genes were identified, it should be recognized that there are limitations for using NAM in gene finding. Given the very low LD in maize and the relatively limited number of SNPs used for this study, we are likely to miss associations. An order-of-magnitude higher SNP coverage (>10 M) will be needed to sufficiently capture LD throughout the maize genome (30, 31), improving power and reducing false positives. Polymorphisms for copy-number variation and presence–absence variation were also not tested here but are ubiquitous in maize (59, 60), and likely contribute to functional variation in QDR.
Limited knowledge of QDR restricts the power of candidate approaches. It is expected that the genes conditioning QDR will cover a broad range of mechanisms, including classes of genes previously unassociated with disease resistance (3). As little is known about the molecular basis of QDR, it is difficult to assess the plausibility of many of the gene associations identified for NLB resistance. Furthermore, experience has shown that the causal polymorphism for a QTL can be distant from the functional gene (29). While recognizing these limitations, it is nonetheless notable that several previously known pathogen defense-related genes, particularly genes involved in basal resistance, were associated with QDR using genome-wide nested association mapping (39, this study). This raises questions concerning the pathogen specificity of QDR. Indeed, several lines of evidence point to multiple disease resistance of disease QTL (9, 11, 15, 61, 62). Nonetheless, many or most QTL have pathogen-specific resistance. Thus, a complementary hypothesis can be presented that pathogen features that are conserved within a species correspond to QTL that are race-nonspecific but effective against only a single disease. Detailed characterization of QTL in near-isogenic backgrounds can allow these and other hypotheses to be addressed (39, 63).
Despite the importance of QDR in crop production and ecology, the genetic basis of QDR remains largely unknown. With the genetic resources available in maize, we see this important crop species emerging as a model system for the study of QDR. Coupled with a history of robust, large-scale field trials and community expertise in quantitative genetics, maize pathosystems can serve as an excellent tool for dissection of the genetics underlying QDR while providing valuable information for resistance breeding in this staple crop. Uncovering the molecular mechanisms of complex disease resistance in plants will assist in the development of durably resistant crop cultivars, increasing food and economic security.
Materials and Methods
Plant Materials.
The maize nested association mapping population is described in detail by Yu et al. (24), Buckler et al. (22), and McMullen et al. (23).
Phenotypic Evaluation for Resistance to NLB.
Field trials were conducted during 2007, 2008, and 2009 at Cornell University's Robert B. Musgrave Research Farm in Aurora, NY. Trials were planted on May 15, 2007; May 14, 2008; and May 18, 2009. Lines were planted as single-row plots 2.1 m in length with 0.76 m between rows. Plots were overplanted and thinned to 10 plants per row. Trials were laid out in an augmented incomplete block design with one replication in each year. For each trial, lines were grouped by family with augmented incomplete blocks within each family. Each incomplete block consisted of 20 RILs and two checks: B73 and the second parent for the respective family. All trials were artificially inoculated with E. turcicum race 1 as described by Chung et al. (19) for field experiments. Individual plants were inoculated at the six- to eight-leaf stage, which corresponded to July 2, 2007; June 27, 2008; and July 16, 2009.
In 2007, two disease phenotypes were evaluated: incubation period (IP) and disease severity (DS). In 2008 and 2009, only DS was evaluated due to the extreme time requirements for evaluation of IP. For the evaluation of IP, all plots were evaluated daily. IP was measured as the number of days after inoculation that the first water-soaked lesion was observed on 50% of the plants in a plot. For DS, all plots were evaluated at three time points during the season at 10-d intervals, with the first rating corresponding to shortly after anthesis for B73. DS ratings were conducted by visually evaluating each plot and rating the percentage of total DLA using a 0–100% rating scale with 1% increments. DTA was measured as the number of days from planting when 50% of the plants in a row were shedding pollen.
The trait distribution for DLA was skewed toward resistance, so a square root transformation was used to normalize the trait distribution before further analysis was conducted. To account for year and field effects, a multivariate mixed model was run in ASReml (VSN International) with a unique unstructured covariance for each family. The model solution gave BLUPs for each NAM RIL at each rating. For each RIL, the BLUPs were then averaged to give an NLB index. Broad-sense heritabilities for the line BLUPs were estimated for NAM and for each individual family. With the multivariate response, heritability was calculated on a line mean basis for each rating and for the NLB index (64).
Joint General Linear Model.
A joint general linear model (GLM) was selected using stepwise model selection with Proc GLMselect in SAS v9.1.3 software as described by Buckler et al. (22). Stepwise selection was conducted with effect selection/removal set at P = 1 × 10−4. To determine this threshold, permutation analysis was conducted by randomizing the NLB index values within each population and then identifying the most significant marker effect using stepwise selection as described above. This was repeated for 1,000 iterations to determine an experimental α of 0.05, which corresponded to a selection threshold of P = 10−4. DTA was included as a covariate in model selection to reduce the confounding effect of relative maturity on disease resistance (22). A fixed-effects model was used with a main effect for family and marker effects nested within families. This nested model estimates a unique allele effect for each family. This approach gives increased power by allowing modeling of multiple alleles at each QTL across NAM. Although it is unlikely that there is a unique QTL allele for each family at every QTL, this nested model provides a statistical framework for modeling multiple alleles at any given QTL. Therefore, based on this model, multiple allelic effects, as opposed to only two, are reported for each QTL. Selected marker effects were then fit into a GLM and dropped individually to confirm significance. A final GLM was fit using R statistical software (65).
To examine the impact of QTL-by-environment interaction, a fixed-effects model incorporating environment-by-population-by-marker effects was modeled in R. For this model, actual phenotypic observations from each environment were used, as the previous models were fit using line BLUPs that averaged phenotypes across environments. The model was as before with a main effect for family, a covariate for DTA, and marker effects nested within family. Additional terms for environment, family by environment, and QTL by family by environment for each QTL were included. Marginal F tests were used to determine the significance of each term.
A Cox proportional hazards model was fit in R statistical software (65) for IP. As with disease severity, DTA was included as a covariate to reduce the effect of maturity. Markers were again nested within population. Stepwise model selection was conducted with the selection of marker effects determined by the P value from a Wilk's F test. The threshold for effect selection was P = 10−4.
For each QTL position identified, confidence intervals were constructed by sequentially examining flanking markers. This procedure was conducted in both directions from the QTL marker to determine the full confidence interval. Starting with the first flanking marker in one direction, the QTL marker and the flanking marker were both fit into the full linear model. The P value from the marginal F test for the QTL marker was then determined. If the QTL marker did not have a significant contribution to the model (P < 0.05), the flanking marker was considered equivalent to the QTL marker and within the 95% confidence interval. The flanking marker was then moved outward and the test repeated until the QTL marker significantly contributed to the model. This procedure was then repeated on the other side of the QTL marker. To compensate for regions of low marker density, pseudomarkers were imputed where consecutive markers were farther apart than 1 cM.
To examine the possible pleiotropic effects of flowering time on disease resistance, the QTL model using markers identified for NLB resistance was fit to DTA (after removing DTA as a covariate in the QTL model). The estimated DTA allele effects for each founder line were then determined and compared with the estimated allele effects for NLB from corresponding founders at each of the QTL markers.
Genome-Wide SNP Association.
Single-nucleotide polymorphisms from the first-generation maize HapMap (http://www.panzea.org) for the 25 founder lines, Mo17 and B73, were tested for association with NLB resistance (31). Missing SNP genotypes were imputed using fastPHASE software (30, 66). SNP positions were referenced to the B73 AGP v.1 physical map, and the complete SNP dataset was then imputed to the NAM RILs using pedigree information and the 1,106 reference SNPs on the genetic map. Trait–marker association was conducted by chromosome after accounting for QTL on other chromosomes. One hundred nonparametric bootstrap samples, each sampling 80% of each family, were analyzed by stepwise regression. SNPs detected in more than 5% of the samples (BPP > 0.05) were examined as polymorphisms in LD with potential candidate genes from the B73 filtered gene set (MaizeSequence release 4a.53; http://www.maizesequence.org).
Supplementary Material
Acknowledgments
We had valuable assistance from past and present members of the R.J.N. laboratory with field trials for NAM: Oliver Ott, Ellie Walsh, Chia-Lin Chung, Lin-Si Hsieh, Judith Kolkman, Joy Longfellow, Tiffany Jamann, Santiago Mideros, Kristen Kennedy, Kerri Lyons, Cassilyn Schweighofer, Ariel Fialko, and Sara Heins. Oliver Ott and Ellie Walsh greatly contributed to phenotyping incubation period in 2007. Nick Lepak assisted with seed sources and Dallas Kroon assisted with data management. Help from Jim Holland on ASReml mixed models was greatly appreciated. The valuable input of three anonymous reviewers led to many improvements in this manuscript. This work was supported by the Generation Challenge Program, The McKnight Foundation, and Cornell University. Development of NAM was supported by US National Science Foundation grants (DBI-0820619, 0321467, 0703908, and 0638566) and US Department of Agriculture-Agricultural Research Service.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010894108/-/DCSupplemental.
References
- 1.Ayliffe M, Singh R, Lagudah E. Durable resistance to wheat stem rust needed. Curr Opin Plant Biol. 2008;11:187–192. doi: 10.1016/j.pbi.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Sacco MA, Moffett P. Disease resistance genes: Form and function. In: Bouarab K, Brisson N, Daayf F, editors. Molecular Plant-Microbe Interactions. Wallingford, UK: CABI; 2009. pp. 94–141. [Google Scholar]
- 3.Poland JA, Balint-Kurti PJ, Wisser RJ, Pratt RC, Nelson RJ. Shades of gray: The world of quantitative disease resistance. Trends Plant Sci. 2009;14:21–29. doi: 10.1016/j.tplants.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Singh RP, Huerta-Espino J, Manilal HM. Genetics and breeding for durable resistance to leaf and stripe rusts in wheat. Turk J Agric For. 2005;29:121–127. [Google Scholar]
- 5.Rosewarne GM, Singh RP, Huerta-Espino J, Rebetzke GJ. Quantitative trait loci for slow-rusting resistance in wheat to leaf rust and stripe rust identified with multi-environment analysis. Theor Appl Genet. 2008;116:1027–1034. doi: 10.1007/s00122-008-0736-0. [DOI] [PubMed] [Google Scholar]
- 6.Wisser RJ, Balint-Kurti PJ, Nelson RJ. The genetic architecture of disease resistance in maize: A synthesis of published studies. Phytopathology. 2006;96:120–129. doi: 10.1094/PHYTO-96-0120. [DOI] [PubMed] [Google Scholar]
- 7.Welz HG, Geiger HH. Genes for resistance to northern corn leaf blight in diverse maize populations. Plant Breed. 2000;119:1–14. [Google Scholar]
- 8.Fu D, et al. A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science. 2009;323:1357–1360. doi: 10.1126/science.1166289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krattinger SG, et al. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science. 2009;323:1360–1363. doi: 10.1126/science.1166453. [DOI] [PubMed] [Google Scholar]
- 10.Fukuoka S, et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science. 2009;325:998–1001. doi: 10.1126/science.1175550. [DOI] [PubMed] [Google Scholar]
- 11.Manosalva PM, et al. A germin-like protein gene family functions as a complex quantitative trait locus conferring broad-spectrum disease resistance in rice. Plant Physiol. 2009;149:286–296. doi: 10.1104/pp.108.128348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perkins JM, Pedersen WL. Disease development and yield losses associated with northern leaf blight on corn. Plant Dis. 1987;71:940–943. [Google Scholar]
- 13.Raymundo AD, Hooker AL. Measuring the relationship between northern corn leaf blight and yield losses. Plant Dis. 1981;65:325–327. [Google Scholar]
- 14.Ullstrup AJ, Miles SR. The effects of some leaf blights of corn on grain yield. Phytopathology. 1957;47:331–336. [Google Scholar]
- 15.Zwonitzer JC, et al. Mapping resistance quantitative trait loci for three foliar diseases in a maize recombinant inbred line population—Evidence for multiple disease resistance? Phytopathology. 2010;100:72–79. doi: 10.1094/PHYTO-100-1-0072. [DOI] [PubMed] [Google Scholar]
- 16.Balint-Kurti PJ, Yang J, Van Esbroeck G, Jung J, Smith ME. Use of a maize advanced intercross line for mapping of QTL for northern leaf blight resistance and multiple disease resistance. Crop Sci. 2010;50:458–466. [Google Scholar]
- 17.Bentolila S, et al. Identification of an RFLP marker tightly linked to the Ht1 gene in maize. Theor Appl Genet. 1991;82:393–398. doi: 10.1007/BF00588588. [DOI] [PubMed] [Google Scholar]
- 18.Yin X, et al. Fine mapping of the Ht2 (Helminthosporium turcicum resistance 2) gene in maize. Chin Sci Bull. 2003;48:165–169. [Google Scholar]
- 19.Chung C-L, Jamann T, Longfellow J, Nelson R. Characterization and fine-mapping of a resistance locus for northern leaf blight in maize bin 8.06. Theor Appl Genet. 2010;121:205–227. doi: 10.1007/s00122-010-1303-z. [DOI] [PubMed] [Google Scholar]
- 20.Zaitlin D, Demars SJ, Gupta M. Linkage of a second gene for NCLB resistance to molecular markers in maize. Maize Genet Coop Newsl. 1992;66:69–70. [Google Scholar]
- 21.Simcox KD, Bennetzen JL. The use of molecular markers to study Setosphaeria turcica resistance in maize. Phytopathology. 1993;83:1326–1330. [Google Scholar]
- 22.Buckler ES, et al. The genetic architecture of maize flowering time. Science. 2009;325:714–718. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]
- 23.McMullen MD, et al. Genetic properties of the maize nested association mapping population. Science. 2009;325:737–740. doi: 10.1126/science.1174320. [DOI] [PubMed] [Google Scholar]
- 24.Yu J, Holland JB, McMullen MD, Buckler ES. Genetic design and statistical power of nested association mapping in maize. Genetics. 2008;178:539–551. doi: 10.1534/genetics.107.074245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flint-Garcia SA, et al. Maize association population: A high-resolution platform for quantitative trait locus dissection. Plant J. 2005;44:1054–1064. doi: 10.1111/j.1365-313X.2005.02591.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee M, et al. Expanding the genetic map of maize with the intermated B73 x Mo17 (IBM) population. Plant Mol Biol. 2002;48:453–461. doi: 10.1023/a:1014893521186. [DOI] [PubMed] [Google Scholar]
- 27.Hughes GR, Hooker AL. Gene action conditioning resistance to northern leaf blight in maize. Crop Sci. 1971;11:180–184. [Google Scholar]
- 28.Welz HG, Schechert AW, Geiger HH. Dynamic gene action at QTLs for resistance to Setosphaeria turcica in maize. Theor Appl Genet. 1999;98:1036–1045. doi: 10.1007/s001220051280. [DOI] [PubMed] [Google Scholar]
- 29.Salvi S, et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci USA. 2007;104:11376–11381. doi: 10.1073/pnas.0704145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian F, et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet. 2011;43:159–162. doi: 10.1038/ng.746. [DOI] [PubMed] [Google Scholar]
- 31.Gore MA, et al. A first-generation haplotype map of maize. Science. 2009;326:1115–1117. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- 32.Valdar W, et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet. 2006;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- 33.Andaya CB, Ronald PC. A catalytically impaired mutant of the rice Xa21 receptor kinase confers partial resistance to Xanthomonas oryzae pv. oryzae. Physiol Mol Plant Pathol. 2003;62:203–208. [Google Scholar]
- 34.Sun X, et al. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 2004;37:517–527. doi: 10.1046/j.1365-313x.2003.01976.x. [DOI] [PubMed] [Google Scholar]
- 35.Gómez-Gómez L, Boller T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 36.Miya A, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan J, et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 39.Kump KL, et al. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat Genet. 2011;43:163–168. doi: 10.1038/ng.747. [DOI] [PubMed] [Google Scholar]
- 40.Griffith M, Yaish MWF. Antifreeze proteins in overwintering plants: A tale of two activities. Trends Plant Sci. 2004;9:399–405. doi: 10.1016/j.tplants.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 41.Hon WC, Griffith M, Mlynarz A, Kwok YC, Yang DS. Antifreeze proteins in winter rye are similar to pathogenesis-related proteins. Plant Physiol. 1995;109:879–889. doi: 10.1104/pp.109.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seo PJ, et al. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 2010;61:661–671. doi: 10.1111/j.1365-313X.2009.04091.x. [DOI] [PubMed] [Google Scholar]
- 43.Huang T, Duman JG. Cloning and characterization of a thermal hysteresis (antifreeze) protein with DNA-binding activity from winter bittersweet nightshade, Solanum dulcamara. Plant Mol Biol. 2002;48:339–350. doi: 10.1023/a:1014062714786. [DOI] [PubMed] [Google Scholar]
- 44.Meyer K, Keil M, Naldrett MJ. A leucine-rich repeat protein of carrot that exhibits antifreeze activity. FEBS Lett. 1999;447:171–178. doi: 10.1016/s0014-5793(99)00280-x. [DOI] [PubMed] [Google Scholar]
- 45.Hiilovaara-Teijo M, Hannukkala A, Griffith M, Yu X-M, Pihakaski-Maunsbach K. Snow-mold-induced apoplastic proteins in winter rye leaves lack antifreeze activity. Plant Physiol. 1999;121:665–674. doi: 10.1104/pp.121.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeh S, et al. Chitinase genes responsive to cold encode antifreeze proteins in winter cereals. Plant Physiol. 2000;124:1251–1264. doi: 10.1104/pp.124.3.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J, Loh Y-T, Bressan RA, Martin GB. The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell. 1995;83:925–935. doi: 10.1016/0092-8674(95)90208-2. [DOI] [PubMed] [Google Scholar]
- 48.Takemoto D, Hayashi M, Doke N, Nishimura M, Kawakita K. Molecular cloning of a defense-response-related cytochrome P450 gene from tobacco. Plant Cell Physiol. 1999;40:1232–1242. doi: 10.1093/oxfordjournals.pcp.a029511. [DOI] [PubMed] [Google Scholar]
- 49.Zhou N, Tootle TL, Glazebrook J. Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell. 1999;11:2419–2428. doi: 10.1105/tpc.11.12.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15:165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berrocal-Lobo M, Molina A, Solano R. Constitutive expression of ETHYLENE RESPONSE FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002;29:23–32. doi: 10.1046/j.1365-313x.2002.01191.x. [DOI] [PubMed] [Google Scholar]
- 52.Després C, DeLong C, Glaze S, Liu E, Fobert PR. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell. 2000;12:279–290. [PMC free article] [PubMed] [Google Scholar]
- 53.Boyle P, et al. The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell. 2009;21:3700–3713. doi: 10.1105/tpc.109.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Büschges R, et al. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell. 1997;88:695–705. doi: 10.1016/s0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 55.Humphry M, Consonni C, Panstruga R. mlo-based powdery mildew immunity: Silver bullet or simply non-host resistance? Mol Plant Pathol. 2006;7:605–610. doi: 10.1111/j.1364-3703.2006.00362.x. [DOI] [PubMed] [Google Scholar]
- 56.Nawrath C, Heck S, Parinthawong N, Métraux J-P. EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell. 2002;14:275–286. doi: 10.1105/tpc.010376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jonak C, Okrész L, Bögre L, Hirt H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol. 2002;5:415–424. doi: 10.1016/s1369-5266(02)00285-6. [DOI] [PubMed] [Google Scholar]
- 58.Nakagami H, Pitzschke A, Hirt H. Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 2005;10:339–346. doi: 10.1016/j.tplants.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 59.Lai J, et al. Genome-wide patterns of genetic variation among elite maize inbred lines. Nat Genet. 2010;42:1027–1030. doi: 10.1038/ng.684. [DOI] [PubMed] [Google Scholar]
- 60.Swanson-Wagner RA, et al. Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 2010;20:1689–1699. doi: 10.1101/gr.109165.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wisser RJ, Sun Q, Hulbert SH, Kresovich S, Nelson RJ. Identification and characterization of regions of the rice genome associated with broad-spectrum, quantitative disease resistance. Genetics. 2005;169:2277–2293. doi: 10.1534/genetics.104.036327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murali Mohan S, et al. Identification of quantitative trait loci associated with resistance to foliar diseases in sorghum. Euphytica. 2010;176:199–211. [Google Scholar]
- 63.Chung C-L, et al. Resistance loci affecting distinct stages of fungal pathogenesis: Use of introgression lines for QTL mapping and characterization in the maize–Setosphaeria turcica pathosystem. BMC Plant Biol. 2010;10:103. doi: 10.1186/1471-2229-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin CY, Allaire FR. Heritability of a linear combination of traits. Theor Appl Genet. 1977;51:1–3. doi: 10.1007/BF00306054. [DOI] [PubMed] [Google Scholar]
- 65.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 66.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.