Abstract
Defining bacterial species remains a challenging problem even for the model bacterium Escherichia coli and has major practical consequences for reliable diagnosis of infectious disease agents and regulations for transport and possession of organisms of economic importance. E. coli traditionally is thought to live within the gastrointestinal tract of humans and other warm-blooded animals and not to survive for extended periods outside its host; this understanding is the basis for its widespread use as a fecal contamination indicator. Here, we report the genome sequences of nine environmentally adapted strains that are phenotypically and taxonomically indistinguishable from typical E. coli (commensal or pathogenic). We find, however, that the commensal genomes encode for more functions that are important for fitness in the human gut, do not exchange genetic material with their environmental counterparts, and hence do not evolve according to the recently proposed fragmented speciation model. These findings are consistent with a more stringent and ecologic definition for bacterial species than the current definition and provide means to start replacing traditional approaches of defining distinctive phenotypes for new species with omics-based procedures. They also have important implications for reliable diagnosis and regulation of pathogenic E. coli and for the coliform cell-counting test.
Keywords: evolution, genomics, species concept
The current definition of bacterial species (1), although pragmatic and universally applicable within the bacterial world (2), remains controversial: Technological limitations in identifying diagnostic traits make the definition difficult to implement and frequently result in the designation of species that are not adequately predictive of phenotype (3, 4). Further, and perhaps more importantly, it remains unclear whether the processes driving diversification and adaptation of bacteria produce sufficiently discrete groups of individuals (species) as opposed to a genetic continuum (4, 5) [referred to as “fuzzy” species (6)]. An improved understanding of the definition of bacterial species is important for reliable diagnosis of infectious disease agents, intellectual property rights, international and national regulations for transport and possession of pathogens, oversight and reporting of bioterrorism agents, and quarantine. Because the scientific, medical, regulatory, and legal communities, as well as the public, expect species to reflect the phenotype and ecology of an organism reasonably, efforts toward a more refined definition of a bacterial species are needed.
The case of Escherichia coli captures many of the problematic aspects of the bacterial species issue and has additional important ramifications for diagnostic microbiology and for assessing fecal pollution of natural ecosystems. Microbiological dogma is that E. coli strains live within the gastrointestinal tract of humans and other warm-blooded animals, are transmitted to a susceptible host via the fecal–oral route, and do not survive for extended periods outside the host. E. coli is phylogenetically distinct (monophyletic), as are the other known Escherichia species, E. fergusonii and E. albertii (7). Despite their phylogenetic cohesiveness, however, E. coli strains are ecologically and phenotypically heterogeneous (3, 7), and, in fact, a few strains have been assigned to a different genus (e.g., Shigella flexneri), based primarily on their distinct clinical presentation and importance as human pathogens (8). Currently, whether pathogens such as Shigella and other delineable groups of strains are assigned their own taxonomic classifications is based on subjective observations rather than on empirical ecologic or phylogenetic data, in part because of the lack of data concerning truly innocuous (nonpathogenic) strains that are more relevant for comparisons to the life-threatening, pathogenic strains (e.g., negative controls). Furthermore, recent environmental surveys repeatedly have recovered substantial E. coli populations from soils and freshwater habitats (9, 10), indicating that “naturalized” (innocuous) strains (9) may be widespread in nature. These findings also suggest that the current view of E. coli biodiversity and ecology might have been biased by the isolation procedures and/or the traditional focus on clinical samples. To what extend the latter populations represent truly autochthonous members of the natural communities sampled and how they differ genetically from host-associated E. coli remain elusive, however. Addressing these questions will have additional global consequences for the current practice of assessing fecal contamination based on E. coli cell counts (11).
Results and Discussion
Environmentally Adapted E. coli Lineages.
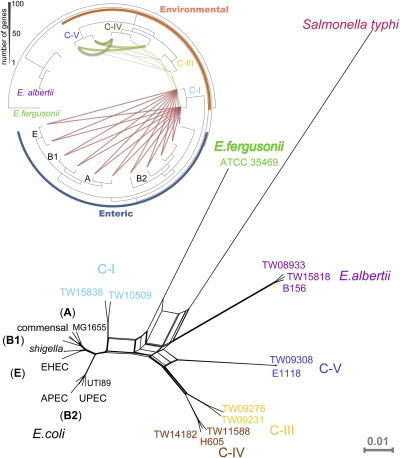
We recently described five Escherichia clades (C-I to C-V) that were recovered primarily from environmental sources and are indistinguishable from typical E. coli based on traditional phenotypic tests included in either the API20E Identification System (bioMerieux, Inc.) or the BBL Crystal Identification System (Becton, Dickinson and Company) (9). To provide genomic insights into the phylogenetic diversity and metabolic potential of these clades, we sequenced the genome of nine representatives from clades C-I, -III, -IV, and -V (Table 1, Table S1, and Fig. S1). Whole-genome phylogenetic analysis confirmed our earlier observations based on multilocus sequence typing that the clades span the phylogenetic tree between E. coli and E. albertii, forming a genetic continuum within the Escherichia genus. In particular, C-I appears to be a sister clade of typical E. coli, being only slightly more divergent than the B2 phylogenetic lineage that includes uropathogenic E. coli (UPEC). The remaining clades are more divergent from typical E. coli (Fig. 1). In agreement with previous phenotypic testing, the genomes of the strains of the four clades encode all genes shared by the available E. coli genomes (i.e., the E. coli core gene set) (Fig. 2A and Fig. S2). Thus, the clades appear to be phenotypically and genetically (e.g., in gene content) indistinguishable from typical E. coli. Based on this information and the current genomic standards for species demarcation (12), these clades would be justifiably classified as E. coli.
Table 1.
Genomes used in this study
| Strain | Lineage | Ecotype* | Pathotype† | Genome‡ | Origin | Sample | Type |
| MG1555 | E. coli | GIT | Commensal | NCBI | California | Human | Feces |
| HS | E. coli | GIT | Commensal | NCBI | Massachusetts | Human | Feces |
| SE11 | E. coli | GIT | Commensal | NCBI | Japan | Human | Feces |
| IAI1 | E. coli | GIT | Commensal | NCBI | France | Human | Feces |
| ED1a | E. coli | GIT | Commensal | NCBI | France | Human | Feces |
| Sakai | E. coli | GIT | EHEC | NCBI | Sakai, Japan | Human | Feces |
| EDL933 | E. coli | GIT | EHEC | NCBI | Michigan | Food | Ground beef |
| UTI89 | E. coli | GIT/UT | UPEC | NCBI | Unknown | Human | Unknown |
| 536 | E. coli | GIT/UT | UPEC | NCBI | Unknown | Human | Unknown |
| CFT073 | E. coli | GIT/UT | UPEC | NCBI | Massachusetts | Human | Blood |
| O1 | E. coli | GIT/Other | APEC | NCBI | United States | Chicken | Lung |
| ATCC35469 | E. fergusonii | Multiple | Multiple | NCBI | United States | Human | Feces |
| TW08933 | E. albertii | GIT | Serotype 7 | This study | Bangladesh | Human | Feces |
| TW15818 | E. albertii | GIT/Other | Diarrheic | This study | Australia | Poultry | Feces |
| B156 | E. albertii | GIT/Other | Avirulent | Broad Institute | Australia | Magpie | Feces |
| TW10509 | Escherichia clade I | GIT | ETEC | This study | India | Human | Feces |
| TW15838 | Escherichia clade I | GIT | Avirulent | This study | Australia | Environment | Freshwater sediment |
| TW09231 | Escherichia clade III | ENV | Avirulent | This study | Michigan | Environment | Freshwater beach |
| TW09276 | Escherichia clade III | ENV | Avirulent | This study | Michigan | Environment | Freshwater beach |
| H605 | Escherichia clade IV | ENV | Avirulent | Broad Institute | Australia | Human | Feces |
| TW14182 | Escherichia clade IV | ENV | Avirulent | This study | Michigan | Environment | Freshwater beach |
| TW11588 | Escherichia clade IV | ENV | Avirulent | This study | Puerto Rico | Environment | Soil |
| E1118 | Escherichia clade V | ENV | Avirulent | Broad Institute | Australia | Environment | Freshwater |
| TW09308 | Escherichia clade V | ENV | Avirulent | This study | Michigan | Environment | Freshwater beach |
| CT18 | Salmonella typhi | GIT | Typhoid | NCBI | Vietnam | Human | Unknown |
*Ecotype designation is based on the frequency of isolation from various hosts [gastrointestinal tract (GIT) or urinary tract (UT)] and the environment (ENV).
†Pathotype refers to the interaction between a particular strain and its host. Commensal strains do not cause disease and commonly are found in the gastrointestinal tract of healthy humans; enterohemorrhagic E. coli (EHEC) strains cause bloody diarrhea in humans; urinary pathogenic E. coli (UPEC) cause urinary tract infections in humans and animals; avian pathogenic E. coli (APEC) cause a range of diseases in birds; enterotoxigenic E. coli (ETEC) cause watery diarrhea in humans; avirulent strains have not been associated with a particular disease or a commensal lifestyle.
‡Publically available genomes were downloaded from the National Center for Biotechnology Information (NCBI) or the Broad Institute.
Fig. 1.
Whole-genome phylogeny of the Escherichia genomes used in the study. The phylogenetic network shown was constructed with the SplitsTree software (27), using as input the concatenated alignment of 1,910 single-copy core genes. (Inset) The graph represents the amount of recent horizontal transfer of core genes between the genomes of the clades. The thickness of the line is proportional to the number of genes transferred (scale at upper left in figure).
Fig. 2.
Gene-content signatures of Escherichia clades. Heatmap of gene presence (yellow) and absence (blue) in 20 selected genomes, using all nonredundant genes that were found in at least two of the genomes as reference. (A) Genomes were clustered based on the presence/absence of genes; values in red represent bootstrap support from Jackknifing resampling with 100 replicates. (B) Genes and pathways distinguishing enteric and environmental genomes were expanded (underlying data are provided in Table S2). 1, acetylglucosamine transporter; 2, fructose transporter; 3, diol utilization operon; 4, lysozyme production. (C) Occurrence of the genes composing the Escherichia pangenome in the 20 genomes ranges from one (a genome-specific gene) to 20 (a core gene).
The orders-of-magnitude higher abundances of these clades in environmental samples relative to those in human feces and the clinic (9) indicate that they represent truly environmentally adapted organisms (meaning that they are not associated primarily with mammal hosts). Consistent with this interpretation, a recent study found that strains of clades C-III, -IV, and -V form biofilms more readily, outcompete typical E. coli strains at low temperatures (which characterize the environment compared with the gastrointestinal tract of warm-blooded hosts), and are nonpathogenic in a mouse model of septicemia (13). Furthermore, screening of 2,701 strains from humans, animals, and the environment identified an additional 57 environmental clade strains, and these strains were found more often in environmental and bird samples than in human samples (9). These studies consistently support the hypothesis that the environmental clades substantially expand the known ecological niche of E. coli.
Functions Important in the Gut.
Comparisons between the environmental genomes and their commensal or pathogenic (enteric) counterparts provided insights into the functional differentiation of E. coli strains. Consistent with the core gene results described above, we found almost no genes specific to enterics when queried against all genomes of environmental clades (Fig. S2). However, when the C-I clade was included in the enteric group (strains of C-I have been isolated from humans, and this clade does not appear to be overrepresented in environmental samples) and the stringency of the comparisons was relaxed to allow one or two genomes in each group not to encode the gene in question, we identified 84 and 120 genes as being specific to or highly enriched in the environmental and enteric groups, respectively (Fig. 2B and Table S2). The environment-specific gene set included several genes of unknown function as well as the complete pathway for diol utilization (energy substrate) and the gene for lysozyme production (hydrolysis of bacterial cell walls). These functions apparently are important for resource acquisition and survival in the environment. In contrast, the enteric-specific functions included genes involved in the transport and use of several nutrients that are thought to be abundant in the gut, such as N-acetylglucosamine, gluconate, and 5-C and 6-C sugars such as fucose (14). The latter genes were significantly enriched in the recently determined human microbiome (15), further corroborating their importance for colonization of the gut. Therefore, these genes characterize enteric E. coli strains relative to their environmental counterparts and may represent robust biomarkers for the development of molecular assays to count commensal E. coli cells in environmental samples more accurately than done by current methods. The enteric gene set also includes several prophage genes, consistent with recent findings from metagenomic studies indicating that the human virome is highly specialized to its host and differs from viromes of environmental ecosystems (16).
Ecologic Barriers to Gene Flow Within Escherichia.
The availability of several genome sequences that span the Escherichia tree provided the opportunity to evaluate the importance of interclade genetic flow for E. coli evolution with greater phylogenetic coverage than previously achieved (7, 17). To this end, we devised a strategy to assess recent genetic exchange events based on embedded quartet decomposition analysis (EDCA) (details are given in Materials and Methods and Figs. S3 and S4). We focused on recent events only because historic genetic exchange of core genes (mediated by homologous recombination) frequently was impossible to detect robustly because of multiple (old) recombination events on the same segment of the genome and the process of amelioration of the newly introduced DNA sequence into the recipient cell (18).
We observed detectable genetic exchange of core genes within the environmental clades, within enterics, and between C-I and enterics but not between enterics and the remaining environmental clades or E. albertii (Fig. 1 Inset and Fig. S5). The core genes exchanged were distributed randomly in the genome and did not show any strong biases in terms of function when compared with the rest of the genome (Fig. S6 and Table S3). These findings are consistent with a generalized mechanism for the transfer of genetic material (e.g., transformation or conjugation) and incorporation into the recipient genome via homologous recombination. They also confirm the closer affiliation of C-I with typical E. coli relative to the other clades and reveal reduced genetic flow between environmental and enteric genomes, presumably because of ecological barriers.
Nonetheless, the number of core genes exchanged within the evolutionary time that corresponded to 0.02 synonymous substitutions per site (the divergence time typically separating the genomes of the same clade) accounted for only a small portion of the total core genes in the genome (0.06–2.33%). We also observed that noncore (auxiliary) genes were exchanged among the clades less frequently than core genes (Fig. S5 and Table S3). Given also that more than 50% of the total unique genes of the E. coli pangenome are genome or clade specific (Fig. 2C), our observations suggest that asexual divergence coupled with clade-specific gene acquisition or deletion dominates interclade recombination in driving Escherichia evolution.
Test of the Fragmented Speciation Model.
It has been proposed recently that organisms of the Escherichia genus evolve according to a fragmented speciation model (19) and that the model may be applicable to additional bacterial groups (20). If the model were true, one would expect that genomic islands that differentiate two ecologically distinct populations to be flanked by regions of increased nucleotide divergence, because such population-specific islands are free from the homogenizing effects of recombination. In other words, because interpopulation homologous recombination is halted around the genomic island (the sequence is not conserved in the population that does not carry the island), the genetic variation of the flanking DNA would be increased between the two populations compared to within either of the individual populations (Fig. 3 gives a graphical representation of the expected signature of the model).
Fig. 3.
Lack of evidence in support of the fragmented speciation model. A representative example of the nucleotide divergence patterns, measured as the number of SNPs (y axis) observed in the flanking regions of a genomic island (x axis) that differentiates environmental from enteric genomes. Note the difference between the SNP level expected for the environmental genomes according to the fragmented speciation model (ecologically distinct population) relative to the observed level (for the enteric group, the genome average SNP level represents the expected SNP level according to the model). The island shown encodes the genes for utilization of fucose, a sugar commonly found in the glycan structures of the cell wall of animals.
Our results strongly indicate that the environmentally adapted genomes are ecologically differentiated as compared with their enteric counterparts and thus are more appropriate for testing the model directly than are the divergent Salmonella genomes used previously (19). Although several candidate (ecologically relevant) genomic islands were identified (such as the islands encoding the fucose and gluconate utilization operons), and these islands were flanked by DNA sequences that were conserved and syntenic in the environmental strains, no island showed the predicted signature of the fragmented speciation model. Instead, the level of nucleotide divergence in the flanking regions of the islands covaried between the environmental and enteric genome (Fig. 3). Similar patterns were observed when the analysis was restricted to commensal vs. pathogenic E. coli for the genomic islands that encode the known pathogenicity factors of the latter genomes (Fig. S7). Thus, the predicted signature of the model was not observed even in comparisons of genomes that show both higher genetic relatedness and genetic flow than observed between environmental and enteric strains. In a few of the genomic islands examined, the flanking genes did show increased nucleotide divergence between ecologically distinct genomes. However, this pattern typically was associated with genes that were interrupted by the insertion of mobile elements; because of relaxed functional constraints, the truncation of gene(s), rather than the action of recombination, presumably caused an increased accumulation of mutations. Such truncated genes or their remnants may underlie some of the incongruent phylogenetic signal observed previously (20).
Conclusions and Perspectives.
Our results collectively suggest that asexual divergence coupled with clade-specific gene acquisition or deletion has a much stronger influence on the evolution of the Escherichia genus than homologous recombination (sexual reproduction). These results differ quantitatively from those reported previously (7, 17). The difference is caused, at least in part, by the different genomes and methods used in the analysis. For instance, the previous studies evaluated the intraclade level, whereas our analysis was focused on more divergent genomes, an approach that is advantageous for unequivocally detecting recent gene exchange and recombination events (21, 22). Although our results do not rule out the existence of high levels of recombination within a clade, they do reveal that genetic exchange between incipient ecologically distinct clades of E. coli may not be as pronounced or prolonged as would be expected by the fragmented speciation model (19), and this reduced level of exchange probably accounts for the lack of evidence in support of the model.
Data described here concerning the environmental Escherichia clades show that justifiable species, which are ecologically distinct, sexually isolated, and phylogenetically tractable, may be identifiable even in cases of apparent phenotypic identity or a genetic continuum (such as revealed within the Escherichia genus in Fig. 1). These findings, which also are consistent with recent metagenomic studies of natural populations (22), suggest that a more ecologic definition for species is more appropriate than the current definition that is heavily based on genetic distinctiveness alone. Comparative genomic analyses linked a substantial fraction of the clade-specific gene acquisitions (and deletions) to the unique ecology of the clade (e.g., Fig. 2B). These findings further corroborate the notion that it is time to start replacing traditional approaches of defining diagnostic phenotypes for new species with omics-based procedures.
What the preferred ecological niche or host (if any) of clades II–V is and whether the clades actually can persist in the external environment in the absence of fecal inputs (i.e., represent truly free-living bacteria) remain elusive, and additional data need to be collected before more robust conclusions can emerge. For instance, strains of clades II–V have been recovered occasionally from birds and ruminant mammals (9), but the extent to which these results are influenced by the processes of strain migration and extinction (as opposed to persistence within the host) is unclear. What our genomic data as well as data from physiological studies and environmental surveys performed previously (9, 13) suggest is that clades II–V are better at surviving in the external environment than is commensal E. coli and are poor competitors in the human gastrointestinal tract relative to successful clonal complexes such as those represented by CFT073 and MG1555 strains. Therefore, clades II–V are highly unlikely to represent a risk to public health.
Of practical significance, the cryptic clades represent microorganisms that show worldwide distribution (Table S1) and have been readily identified as typical E. coli by expert microbiologists in the laboratory and by managers of water quality who use this organism to assess fecal pollution of surface waters. However, these organisms probably should not be considered E. coli and are highly unlikely to represent an environmental hazard, according to our analyses. These findings underscore the need to reevaluate coliform testing and the microbiologic dogma that the niche of enteric microbes, such as E. coli, is the mammalian intestinal tract.
Materials and Methods
Information for each of the 25 Escherichia genomes used in this study is provided in Table 1. Twelve of the genomes (nine Escherichia spp. and two E. albertii) were sequenced as part of this study, using either the Illumina GA-II genome analyzer or the Roche 454 Sequencer available at the Genomic Facility at Michigan State University (Table S1). For sequencing, a pair-ended sequencing strategy (76-bp–long reads, 300-bp library insert size) was used that yielded ∼300× coverage for each genome (one genome per Illumina lane). The accession numbers of the genomes sequenced in this study are provided in Table S1.
The 76-bp–long pair-ended reads first were clustered into two groups based on their quality score and length using the K-means algorithm, and the low-quality group was discarded. Sequences were trimmed further on both the 5′ and 3′ ends, based on a threshold of Q = 20, and were assembled using the Velvet algorithm (23). The K-mer parameter was varied to maximize the N50 of the resulting assembly for each genome (high stringency). Detailed statistics of each genome assembly are provided in Table S1. Comparisons of the assembly of genome TW10509 and the assembly performed at the Broad Institute based on independent, high-coverage 454 data revealed that our contigs had very low sequencing error (<0.01%) and contained no misassemblies or contaminating sequences (Fig. S1). Our in silico evaluation also suggested that our assemblies recovered at least 98% of the core and 95% of the total genes in the genome (Fig. S1). The few genes missing from our assemblies did not affect our conclusions because our analyses were based primarily on core genes recovered intact in all genome sequences. Genes on the assembled contigs were identified by the GeneMark pipeline (24) and annotated as previously described (25).
After all mobile elements (transposase, integrases, and so forth) and truncated gene sequences were removed, an all-versus-all BLAST search was carried out using all protein-coding genes annotated in all genomes. Alignments with coverage lower than 85% of the length of the query protein sequence were discarded. The analysis identified 1,910 genes that constituted reciprocal best matches in all pair-wise genome comparisons (core orthologs). These genes subsequently were aligned using ClustalW2 (26), and the resulting alignments were concatenated to provide the whole-genome alignment. The phylogeny of all genomes was reconstructed using the latter alignment and the Neighbor-Net algorithm (27) of the SplitsTree package and is shown in Fig. 1. It should be noted that the set of 1,910 genes represents a subset of the total core genes shared among the genomes analyzed (estimated to be around 2,200–2,500 genes, given that about 20–25 core genes were missed in each genome assembly and that we analyzed 12 draft genomes; Fig. S1); it does not include truncated genes or genes not recovered in our assemblies. Nonetheless, the missing genes are highly unlikely to have a significant impact on the derived whole-genome phylogeny (because of the large number of genes included in the underlying alignment) or on the results of the horizontal gene transfer (HGT) analysis (see below), because they represented a small number of the total core genes in the genome and were distributed randomly around the genome (Fig. S6).
To identify genes that recently were exchanged horizontally among the Escherichia clades, we used the approach outlined in Fig. S3. In brief, the protein sequences of core orthologs (1,910 genes) were aligned using ClustalW2 (26). The corresponding nucleotide sequences of the aligned protein sequences subsequently were aligned, codon by codon, using the pal2nal script, with “remove mismatched codons” enabled and the protein alignment as the guide (28). Synonymous substitutions per site (Ks) were calculated based on the method described by Goldman and Yang (29) using KaKs_Calculator (30). To capture only recent HGT events, a Ks-based filter was applied to qualify orthologous genes that (i) had Ks values ≤0.02 (recent HGT events); (ii) were not short (i.e., <300 bp) or truncated; and (iii) had a sequence that was not typically highly conserved within the Escherichia genus (i.e., the genes did not rank in the lower 15% of Ks values in all pair-wise genome comparisons). The cutoff Ks = 0.02 was used because it represented the average Ks among orthologs of genomes of the same lineage; hence, it was optimal for evaluating interlineage HGT events (we did not assess intralineage HGT). In addition, genes in the low Ks ranks that represented informational genes, such as the ribosomal genes and DNA/RNA polymerases, were removed manually from further analysis because it could not be established whether the identity patterns observed were caused by genetic exchange or high sequence conservation. Fewer than 100 genes were removed. Embedded quartet decomposition analysis (EQDA) (31) was used subsequently to infer interclade HGT events as follows. Embedded quartet analysis was applied to two clades at a time, using two genomes per clade (i.e., four genomes in total). The resulting phylogeny was bootstrapped and compared with the whole-genome tree topology. Only quartets incongruent with the genome topology and at least 95/100 bootstrap support were selected to represent HGT events. Noncore genes shared by at least two clades were assessed in the same way as core genes (Fig. S3).
Although it is possible that our approach did not filter out a few informational genes that show high sequence conservation, this possibility should have no effect on our conclusions about the relative importance of HGT between commensal and environmental genomes, because HGT was assessed based on the same core genes for all genomes and genome quartets that showed comparable intergenome evolutionary relatedness. We also evaluated the extent to which EQDA analysis might be affected by the sequences used in the analysis; for instance, whether orthologous sets with high sequence similarity showed more false positives than more divergent orthologs because of the weak phylogenetic signal resulting from highly identical sequences. Our results, which are summarized in Fig. S4, suggested that our EQDA is impervious to such artifacts and that our approach did not underestimate the number of recently exchanged genes.
Supplementary Material
Acknowledgments
We thank the personnel of the Genomics Facility at Michigan State University for their help with sequencing the Escherichia genomes. This project was supported in part by National Science Foundation Award DEB 0516252 and in part by Contract HHSN2722009000018C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database. For a list of accession numbers, see Table S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015622108/-/DCSupplemental.
References
- 1.Stackebrandt E, et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52:1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- 2.Rosselló-Mora R, Amann R. The species concept for prokaryotes. FEMS Microbiol Rev. 2001;25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 3.Konstantinidis KT, Ramette A, Tiedje JM. The bacterial species definition in the genomic era. Philos Trans R Soc Lond B Biol Sci. 2006;361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gevers D, et al. Opinion: Re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 5.Fraser C, Hanage WP, Spratt BG. Recombination and the nature of bacterial speciation. Science. 2007;315:476–480. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanage WP, Fraser C, Spratt BG. Fuzzy species among recombinogenic bacteria. BMC Biol. 2005;3:6. doi: 10.1186/1741-7007-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Touchon M, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan R, Reeves PR. When does a clone deserve a name? A perspective on bacterial species based on population genetics. Trends Microbiol. 2001;9:419–424. doi: 10.1016/s0966-842x(01)02133-3. [DOI] [PubMed] [Google Scholar]
- 9.Walk ST, et al. Cryptic lineages of the genus Escherichia. Appl Environ Microbiol. 2009;75:6534–6544. doi: 10.1128/AEM.01262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishii S, Ksoll WB, Hicks RE, Sadowsky MJ. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl Environ Microbiol. 2006;72:612–621. doi: 10.1128/AEM.72.1.612-621.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.American Public Health Associaton . Standard Methods for the Examination of Water and Wastewater. 18th Ed. Washington, DC: American Public Health Association; 1992. [Google Scholar]
- 12.Goris J, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 13.Ingle DJ, et al. Biofilm formation, thermal niche and virulence characteristics of Escherichia spp. Appl Environ Microbiol. 2011 doi: 10.1128/AEM.02401-10. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang DE, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci USA. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin J, et al. MetaHIT Consortium. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wirth T, et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence JG, Ochman H. Amelioration of bacterial genomes: Rates of change and exchange. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 19.Retchless AC, Lawrence JG. Temporal fragmentation of speciation in bacteria. Science. 2007;317:1093–1096. doi: 10.1126/science.1144876. [DOI] [PubMed] [Google Scholar]
- 20.Retchless AC, Lawrence JG. Phylogenetic incongruence arising from fragmented speciation in enteric bacteria. Proc Natl Acad Sci USA. 2010;107:11453–11458. doi: 10.1073/pnas.1001291107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eppley JM, Tyson GW, Getz WM, Banfield JF. Genetic exchange across a species boundary in the archaeal genus Ferroplasma. Genetics. 2007;177:407–416. doi: 10.1534/genetics.107.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konstantinidis KT, DeLong EF. Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. ISME J. 2008;2:1052–1065. doi: 10.1038/ismej.2008.62. [DOI] [PubMed] [Google Scholar]
- 23.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konstantinidis KT, Braff J, Karl DM, DeLong EF. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific subtropical gyre. Appl Environ Microbiol. 2009;75:5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 27.Bryant D, Moulton V. Neighbor-net: An agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- 28.Suyama M, Torrents D, Bork P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34(Web Server issue):W609–612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z, et al. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhaxybayeva O, Gogarten JP, Charlebois RL, Doolittle WF, Papke RT. Phylogenetic analyses of cyanobacterial genomes: Quantification of horizontal gene transfer events. Genome Res. 2006;16:1099–1108. doi: 10.1101/gr.5322306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.