Abstract
The search for novel therapeutic interventions for viral disease is a challenging pursuit, hallmarked by the paucity of antiviral agents currently prescribed. Targeting of viral proteins has the inextricable challenge of rise of resistance. Safe and effective vaccines are not possible for many viral pathogens. New approaches are required to address the unmet medical need in this area. We undertook a cell-based high-throughput screen to identify leads for development of drugs to treat respiratory syncytial virus (RSV), a serious pediatric pathogen. We identified compounds that are potent (nanomolar) inhibitors of RSV in vitro in HEp-2 cells and in primary human bronchial epithelial cells and were shown to act postentry. Interestingly, two scaffolds exhibited broad-spectrum activity among multiple RNA viruses. Using the chemical matter as a probe, we identified the targets and identified a common cellular pathway: the de novo pyrimidine biosynthesis pathway. Both targets were validated in vitro and showed no significant cell cytotoxicity except for activity against proliferative B- and T-type lymphoid cells. Corollary to this finding was to understand the consequences of inhibition of the target to the host. An in vivo assessment for antiviral efficacy failed to demonstrate reduced viral load, but revealed microscopic changes and a trend toward reduced pyrimidine pools and findings in histopathology. We present here a discovery program that includes screen, target identification, validation, and druggability that can be broadly applied to identify and interrogate other host factors for antiviral effect starting from chemical matter of unknown target/mechanism of action.
Respiratory syncytial virus (RSV) (1) is an increasingly important pediatric pathogen (2). RSV affects all age groups (3), with symptoms ranging from mild rhinorrhea, cough and fever (4) to more serious bronchiolitis or pneumonia (5), and it is the single most common cause of childhood hospitalization (6). With the exception of the prophylactic antibody Synagis® (palivizumab) (7), for high-risk populations, there is no specific treatment for RSV, and the prospects for a safe and effective vaccine remain remote at this time (8). Overall, the effort to develop effective therapies for pathogenic viral infections is one of the most challenging in public health (9). The large-scale screening for natural products able to kill bacteria in vitro, which was the basis for the boom of antibiotics in the 1950s, was not successful for antivirals (10). Because of an increased understanding of the molecular mechanisms of viral life cycles, many viral proteins have emerged as targets for therapeutic intervention. However, antiviral drug discovery can be targeted at either viral proteins or cellular proteins as in the case of Selzentry® (maraviroc, targeting CCR5) for human immunodeficiency virus (HIV) (11). Although the former is likely to yield compounds with a narrow spectrum of antiviral activity, few effective and safe agents have emerged, and these face the inextricable challenge of high mutation rates that have confounded many conventional antiviral products. Targeting a cellular protein might afford antiviral compounds with a broader spectrum of activity and have the attractive property to lessen the chance of resistance development. However, targeting the host could result in toxicity, especially if the protein or pathway used is crucial for cell survival. This risk is mitigated if the virus uses the host target in a substantially different manner from the obligate cell process or the biological process is not essential. Therefore, an up-front understanding of antiviral mechanism of action (MoA) is required to mitigate such risks. Given the inherent challenges, we took an unbiased approach and sought chemical starting points that exert an antiviral phenotype in whole cells that could potentially reveal inhibitors of virtually any facet of the viral life cycle. By not restricting the screen to a biochemical assay for virally encoded protein, we did not preclude identifying inhibitors of such. Rather, we opened the additional possibility to identify host factors critical to propagation of the viral life cycle. Cognizant of the potential toxicity caveats associated with inhibition of host factors, we prioritized target identification to facilitate an assessment of druggability (12–14) of any such targets should they arise from our screen. We present here a small-molecule–driven paradigm to identify, and subsequently interrogate, the druggability of specific host factors for antiviral efficacy.
Results
Screening.
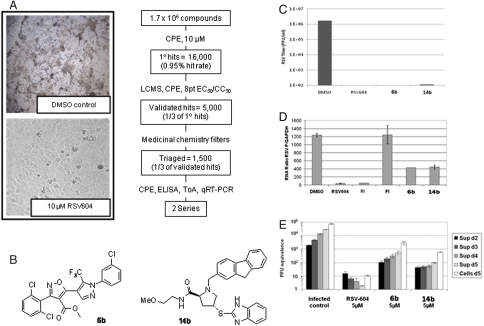
A whole cell screen in HEp-2 cells infected with RSV-A (Long) was performed using approximately 1.7 M compounds that identified > 16,000 primary hits (approximately 0.95% hit rate) with > 50% cell protection (denoted as CPE assay for cell cytopathic effect) at a single concentration (10 μM) (Fig. 1A). Hit confirmation (CPE, 10 μM, triplicate) delivered approximately 7,000 validated candidates (48% confirmation rate) that were further characterized to determine their purity/integrity (liquid chromatography–UV-MS), antiviral efficacy (EC50), and cell cytotoxicity (CC50) (eight-point dose response curve, half-log serial dilution) to deliver a working set of approximately 5,000 compounds (one-third of validated candidate list) from which two distinct chemical series were selected, pyrazole-isoxazole (6b) and proline (14b) (Fig. 1B).
Fig. 1.
Identification of 2 series with anti-RSV antiviral effect in a whole-cell high-throughput screening (HTS). (A, Right) Flow chart of the HTS for RSV in HEp-2 cells using cell protection as efficacy determination. (A, Left) Images of healthy HEp-2 cells protected by RSV604 or killed by RSV infection. (B) The isoxazole-pyrazole (6b) and the proline (14b) series identified. (C) Effect of compounds (10 μM) in HEp-2 cells at 96 h postinoculation at an MOI 0.1 measured by plaque assay. (D) Time of addition experiments. Compounds (10 μM) were added at time of addition and 4 h postinoculation to HEp-2 cells at MOI 1. Virus was quantified by qRT-PCR from total cell RNA isolated from infected cells at 12 h postinoculation. (E) Efficacy in HBEC polarized. Virus was measured by qRT-PCR from the apical surface supernatant at days 2, 3, 4, and 5 postinoculation after 5 μM compound addition.
In Vitro Antiviral Activity.
Addition of 6b or 14b to HEp-2 cells 1 h prior to inoculation with RSV-A-Long at multiplicity of infection (MOI) 0.1 resulted in a reduction of 4 logs of virus production compared to DMSO control as measured by plaque assay at 96 h postinoculation. This strong reduction was similar to the effect by the replication inhibitor RSV604 (Fig. 1C). Time of addition studies were performed to determine at which step of the viral cycle the compounds were acting (15). The compound was added 4 h postinoculation at MOI 1 (Fig. 1D). Both 6b and 14b significantly reduced the viral copies measured by quantitative RT-PCR (qRT-PCR) from the whole cell RNA compared to DMSO control, similar to inhibitions observed with replication inhibitors RSV604 and RI. RSV604 targets the replication complex, whereas RI targets the L-polymerase. In contrast, FI, a fusion inhibitor that targets the F protein, could not suppress viral replication when added 6 h postinoculation. These data suggest that both 6b and 14b act at a postentry level and probably target virus replication.
Efficacy in Primary Cells.
RSV infection targets the lungs of patients. Therefore, we tested the effect of 6b and 14b in primary human bronchial epithelial cells (HBECs) grown at an air-liquid interface (16). These cells form a stratified polarized epithelium that develop ciliated and goblet cells that produce beating cilia and mucus that mimics the in vivo conditions. Addition of 5 μM of 6b or 14b to the basolateral chamber added at time of inoculation with RSV-A-Long at MOI 0.1 reduced 1.5 logs of viral titer as determined by qRT-PCR analysis of the supernatants collected at 2, 3, 4, and 5 d postinoculation (Fig.1E),confirming activity in primary polarized cells.
Broad-Spectrum Activity.
To test the specificity of the compounds against RSV, both 6b and 14b were screened against four different RNA virus families (Paramyxoviridae, Orthomyxoviridae, Flaviviridae, and Retroviridae). Potent EC50 values in the range 0.002 to 0.3 μM are observed for RSV, influenza virus, hepatitis C virus (HCV), dengue virus, yellow fever virus, and HIV (Table 1). The broad-spectrum activity was not only observed among the different viral families but within a family as well. The compounds are active against strains of RSV groups A and B, influenza virus A/PR/8/34, A/Udorn/72 and B/Lee, HCV infectious virus group 2a and replicon 1a, dengue virus 2 and replicon, yellow fever virus 17D, and the multidrug-resistant strain MDR769. In all assays the CC50 was > 10 μM. These unexpected results suggested that the target or targets are a cellular factor used by all RNA viruses. It would be highly unlikely that a compound would be active against viral proteins from such a large number of divergent virus families.
Table 1.
Broad-spectrum activity
| Family | Virus | Strain | EC506b, μM | EC5014b, μM | Cell type |
| Paramyxoviridae | RSV | A-Long | 0.007 | 0.086 | A549 |
| B-Washington | 0.009 | 0.098 | HEp-2 | ||
| Orthomyxoviridae | influenza H1N1 | A/PR/8/34 | 0.032 | 0.009 | MDCK |
| influenza H3N2 | A/Udorn/72 | 0.217 | 0.019 | MDCK | |
| influenza B | B/Lee | 0.117 | 0.342 | MDCK | |
| Flaviviridae | HCV | 1a replicon | 0.007 | 0.025 | Huh-7 |
| 2a | 0.0126 | 0.013 | Huh-7 | ||
| dengue | 2 | 0.005 | 0.06 | A549 | |
| replicon | 0.0026 | 0.044 | A549 | ||
| yellow fever | 17D | 0.0082 | 0.1 | A549 | |
| Retroviridae | HIV | MDR769 | 0.00097 | 0.023 | PBMC |
Affinity Purification/Analysis.
To identify the cellular efficacy target(s) through which 6b and 14b inhibit viral replication, we used a three-channel iTRAQ quantitative chemical proteomics approach as described previously (17, 18). In two separate experiments, this strategy is based on the immobilization of the bioactive analogues 6a and 14a, respectively, to affinity-capture cellular proteins from lysates of RSV-infected HEp-2 cells spiked with an excess amount (20 μM) of 6b (or 14b), the inactive analogues 6c (or 14c), or DMSO (Fig. 2A). Specific binding to the immobilized compound should be competed with 6b (or 14b), but not with 6c (or 14c). The protein with highest affinity to 6b was dihydroorotate dehydrogenase (DHODH) (Fig. 2B) (19). The protein with highest affinity to 14b was the trifunctional complex comprising carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase (collectively known as CAD) (Fig. 2C) (20). Interestingly, both enzymes belong to a common pathway, the de novo pyrimidine biosynthesis pathway (21).
Fig. 2.
Target identification using affinity purification. (A) Chemoproteomics tool compounds for series 1 (6a/6c) and series 2 (14a/14c). (B) The scatter plots highlighting results for 6a/6b (B) and 14b/14c (C) relative to DMSO.
Target Validation.
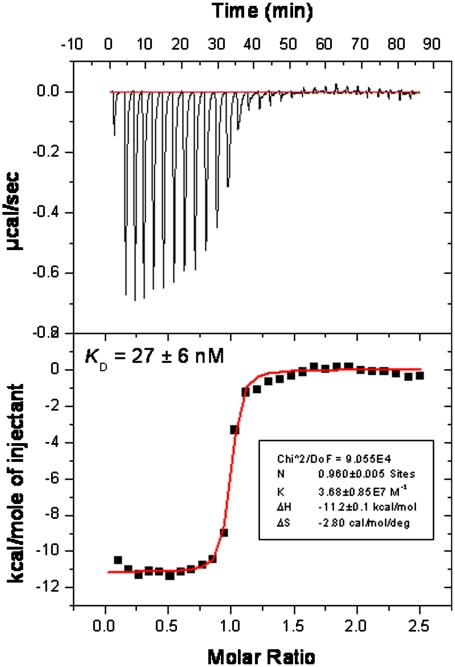
Because both DHODH and CAD are part of the same biochemical pathway, and the endpoint is the synthesis of pyrimidines (T/G for DNA, U/G for RNA) we tested whether addition of pyrimidine or purines could rescue the antiviral effect. Dose response curves were performed with RSV-A-Long with addition of 5 or 25 μM uridine (because RSV replication requires RNA and not DNA amplification) and as control the same concentrations of adenosine, which are not the endpoint of the pyrimidine de novo biosynthesis pathway. Both 6b and 14b lost antiviral activity in a dose-dependent manner when uridine was added but not when adenosine was supplemented (Table 2). When 25 μM of uridine was used, the antiviral phenotype was completely rescued. The same pattern was exhibited with brequinar, which targets DHODH (Table 2) (22). These data show that addition of pyrimidines can rescue the antiviral effect imparted by the depletion of pyrimidines by both 6b and 14b. In the case of 6b, we subsequently confirmed the biochemical inhibition of huDHODH via an isothermal titration calorimetry (ITC) study that measured the Kd = 27 ± 6 nM (Fig. 3).
Table 2.
Antiviral effect is rescued by uridine
| Series 1 |
Series 2 |
Brequinar |
Cells only |
||
| RSV* (EC50 of compound, μM) | CC50, μM | ||||
| Uridine †, μM (product of pyrimidine biosynthesis) | 0 | 0.004 | 0.101 | 0.053 | > 10 |
| 5 | 0.009 | 0.158 | 0.106 | > 10 | |
| 25 | > 10 | > 10 | > 10 | > 10 | |
| Adenosine, μM (product of purine biosynthesis) | 0 | 0.009 | 0.101 | 0.046 | > 10 |
| 5 | 0.008 | 0.158 | 0.053 | > 10 | |
| 25 | 0.010 | 0.142 | 0.051 | > 10 | |
*For clarity, only data for RSV shown. Data is consistent for HCV and influenza. Live virus used in all cases.
†Circulating levels in man are approximately 5–10 μM.
Fig. 3.
ITC data for 6b and huDHODH, with measured Kd = 27 ± 6 nM.
In Vitro Cytotoxicity Assessment.
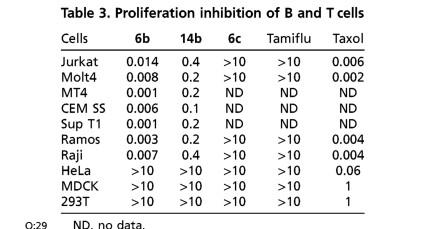
Because the pyrimidine de novo biosynthesis pathway is essential for the survival of highly replicating cells, we further investigated in vitro cell cytotoxicity. Dose response curves were obtained using a viability assay quantifying the levels of ATP, and no cytotoxicity was observed up to 10 μM in most continuous cell lines from different organs and species such as HeLa, HEp-2, A549, Huh-7, MDCK, and Vero, (Table 1). However, both 6b and 14b show activity against highly dividing T and B lymphoid-derived cells such as Jurkat, Molt4, MT4, CEMSS, SupT1, Ramos, and Raji (Table 3). These results suggest that highly dividing cells that rely on a substantial pool of pyrimidines for continued replication are susceptible to both inhibitors. These results raised the question of whether there could be in vivo cytotoxicity due to arrest of highly proliferating cells. Therefore, the assessment of safety as well as efficacy to establish a therapeutic index needed to be assessed in vivo.
Table 3.
Proliferation inhibition of B and T cells
ND, no data.
Animal Experiments.
To determine the druggability of targeting the de novo pyrimidine pathway for antiviral efficacy, the RSV cotton rat model was chosen (23). Prior to assessment, in vivo PK was established for 6b via subcutaneous and oral routes at 10 mg/kg to enable study design for efficacy studies. Pharmacokinetics in cotton rats confirmed that although subcutaneous administration of 6b showed slightly higher dose-normalized area under the curve and maximum concentration values, plasma and lung exposure following oral administration could be achieved. Cotton rats were dosed orally with 6b or brequinar at 25 or 50 mg/kg twice a day for 7 d, and lungs were harvested on either day 4 or 7 postinoculation. Plasma samples were collected at the termination of the study to confirm anticipated exposure based upon the pharmacokinetic studies. No viral load reduction compared to vehicle dosing was observed with either compound (Table 4). At day 4, animals dosed with 6b had similar titers as the control group, and those dosed brequinar actually showed higher titers (Table 4). By day 7, both 6b- and brequinar-treated animals had up to 2.5-logs higher titers compared to vehicle control. This suggests that both compounds are inhibiting the clearance of virus because at day 7 the infection in the vehicle-treated animals is almost resolved and titers are undetectable or just at the detection limit of 102 pfu/mL. Ribavirin was used as an internal control and gave the expected 1-log reduction at day 4.
Table 4.
Cotton rat efficacy model for RSV; viral load reduction in lungs (log)
| Dose | 6b | BRQ | ||
| Day 4 | Day 7 | Day 4 | Day 7 | |
| 50 mg/kg | −0.37 | −1.22 | −0.48 | > -1.41 |
| 100 mg/kg | −0.35 | −1.26 | −0.87 | > -2.52 |
Histopathology.
Microscopic changes for both compounds were similar, albeit brequinar’s effects were somewhat more pronounced, especially in the highest dose. At this highest dose, animals had single-cell necrosis/apoptosis in the intestine, lymphoid depletion/increased apoptosis in thymus, and decreased cellularity/single-cell necrosis in bone marrow. Bone marrow changes were reflected in the peripheral blood as moderate to marked decreases in absolute reticulocyte counts. Three out of eight animals in the high-dose 6b group died prior to scheduled necropsy. One of these animals was necropsied and the cause of death was determined to be mucosal necrosis of the duodenum and cecum with secondary peritonitis. At the termination of the study, mucosal changes were also present in the duodenum and cecum of another animal (Fig. 4). Overall, the pathology observed in the animals administered either brequinar or 6b was consistent with on-target toxicity of highly proliferating cells by inhibition of the de novo pyrimidine biosynthesis pathway.
Fig. 4.
Apoptosis in jejunum after administration of brequinar 25 mg/kg per d for 7 d
Discussion
Natural products are a unique source of structural complexity and stereodiversity, and they have been exploited by many in the pursuit of new medicine (24). However, the large-scale screening for natural products able to kill bacteria in vitro, which was the basis for the boom of antibiotics in the 1950s, was not successful for antivirals (10). One reason for this might be that we have simply yet to isolate, purify, characterize, and screen the next wave of natural-product candidates; a pursuit that, although underway, will take considerable time. Although organic synthesis has been used to enrich the pool of molecular starting points with natural product–like complexity and diversity (25), the screening of compound libraries for novel chemical scaffolds as leads for drugs remains limited by the overall diversity, not number, of candidates that are screened (26). To address the urgent need for novel treatments for RSV, we took a different approach and sought chemical starting points that exert an antiviral phenotype in whole cells that could potentially reveal inhibitors of virtually any facet of the viral life cycle through a phenotypic screening paradigm using a library of both natural and nonnatural products. Inhibitors of RSV were identified from two distinct chemical scaffolds. Extensive in vitro profiling revealed 6b and 14b to be inhibitors of postentry events of viral replication, active against both RSV serogroups (A and B) with no evidence of cell cytotoxicity. Importantly, the two series exhibit antiviral effect in primary HBECs grown in ALI, a differentiated, stratified, ciliated epithelium model that recapitulates the lining of the human lung (16). We were surprised to find the antiviral activity of both series was not specific to RSV but that the compounds were also active against influenza virus and HCV. Upon testing of a broader panel, we consistently found nanomolar antiviral activities against multiple subtypes of multiple RNA viruses in multiple different cell types, including the multidrug-resistant MDR769 strain of HIV. The broad-spectrum activity of these compounds highly suggested that a cellular target was involved in the MoA because they were active among positive- and negative-sense RNA, and retroviruses. In addition, resistant viruses were not able to be generated in the presence of both series, pointing again to a possible cellular target.
We were intrigued with identifying two proteins that share a common pathway, de novo pyrimidine biosynthesis (27). The protein identified by 6b was dihydroorotate dehydrogenase (DHODH) (19), and that of 14b was the multifunctional polypeptide termed CAD (20) [comprising the catalytic triad carbamoyl phosphatase (CSPase), aspartate transcarbamylase (ATCase) and dihydroorotase (DHOase)]. CAD initiates and regulates de novo pyrimidine biosynthesis, delivering dihydroorotate to DHODH, the fourth and rate-limiting enzyme in the de novo pathway (21). Together, both lead to the production of uridine that serves as an activated precursor of RNA and DNA, CDP-diacylglycerol phosphoglyceride, and UDP sugars for protein glycosylation/glycogen synthesis (28). There is precedence of a hereditary orotic aciduria disease that is triggered by a congenital disfunction in the pyrimidine metabolism transmitted as an autosomal recessive trait. Hematological remission of the condition was observed in patients after the dosing of uridine as supplement (29). We hypothesized that if DHODH and CAD are the protein targets of series 1 and 2, addition of exogenous uridine to the assay media should result in rescue of the antiviral phenotype. Conversely, addition of adenosine, the precursor for de novo purine biosynthesis, should have no effect. This was found to be the case, thus validating the chemoproteomic identification of DHODH and CAD as the molecular targets of 6b and 14b, respectively. We were subsequently able to demonstrate binding of 6b to DHODH using biochemical and biophysical methods as further confirmation of the chemoproteomics results.
In addition to the de novo biosynthesis, cells can acquire pyrimidine nucleotides via salvage pathways; most cells have multiple passive and active transporters that facilitate uptake of circulating pyrimidine pools (30, 31), with the relative contribution of the de novo and salvage pathways depending on cell type and developmental stage. In general, resting or fully differentiated cell types have little need for de novo source, because the demand for pyrimidines is largely satisfied by salvage pathways (32). Evidence to suggest that allosteric regulation of CAD governs pyrimidine biosynthesis in vivo comes, in part, from studies showing the enzyme is subject to feedback inhibition by the end product UTP (27). When intracellular UTP levels drop, catalytic activity of CAD increases to achieve homeostasis (33).
We speculate that the viral replication machinery utilizes intracellular pyrimidine pools to suffice vRNA replication (34), during which the demand for pyrimidines likely exceeds supply through salvage by approximately 15–30-fold (35). This likely triggers the activation of the de novo synthesis pathway through unbinding of UTP from CAD, although the exact mechanism remains unknown. There is always the possibility that one of the viral encoded proteins up-regulates the pathway as well. In any scenario, inhibition of the de novo pathway would result in an antiviral phenotype by restricting the virus of pyrimidines necessary for replication. There are multiple in vitro data implicating de novo pyrimidine biosynthesis pathway as an antiviral candidate for adenovirus, vaccinia virus, influenza B virus, paramyxovirus, picornavirus, reovirus, herpes viruses, and BK virus (36–39). There are also reports of efficacy studies in animal models with banzi virus, influenza B virus, and vaccinia virus (36) that showed either a lack of or very limited efficacy. Clinical trials in humans were also reported for HIV, CMV, and BK virus (38, 40–43), with no conclusive results. It is reported that mean levels of circulating uridine in man are approximately 6 μM (44), spanning a range between 2.6 μM (45) and 21 μM (46). In our hands, in vitro uridine rescue was observed within this range. We grew concerned of the possibility that circulating levels of uridine would suffice reversal of the antiviral phenotype, and, in turn, would negate the possibility to achieve antiviral efficacy in vivo with 6b or 14b.
Of greater concern was knowing that proliferating cells typically require a 3–8-fold increase in pyrimidine levels to undergo cell division (32), that is largely satisfied by use of the de novo pathway (47). Indeed, inhibitors of DHODH such as Arava® (leflunomide) are used clinically as antiproliferative agents for the treatment of rheumatoid arthritis (48). In placebo-controlled trials, adverse events include GI symptoms, skin rash and reversible alopecia that are consistent with an antiproliferative MoA (49). We had observed a lack of cell cytotoxicity in multiple nonproliferating cell types, even under nonconfluent conditions. However, we did observe profound cytotoxicity against highly dividing T and B cell lymphoid-derived cells, confirming the antiproliferative phenotype for 6b and 14b in vitro. We therefore questioned whether administration of 6b or 14b in vivo would lead to a restricted safety margin.
To experimentally determine the druggability of pyrimidine biosynthesis for in vivo antiviral efficacy, we sought evidence for toxicity in the context of an efficacy study through incorporation of uridine and histopathology as biomarkers for safety. It seemed that the MoA of the de novo pyrimidine as a drug target would suit more for a rapidly replicating RNA virus. Indeed, for RSV there were two reports that used the mouse model of RSV and showed that the active metabolite of leflunomide did not have an antiviral effect but did block the immuno- and lung pathology associated with the disease (50, 51). However, none of these animal experiments addressed the pharmacokinetics of the drug and, most importantly, the possible toxicity associated with on-target effects. There are a number of animal models available for studying RSV, including primates, cows, cotton rats, and mice, each with its own set of advantages and disadvantages (52). In terms of rodents, we chose to use the cotton rat, because they have been shown to be at least 50-fold, and as much as 1,000-fold, more permissive than mouse strains for RSV (23), with the virus replicating in the nasal epithelium and bronchiolar epithelium of the animal (53). Because the in vitro antiviral effect of CAD phenocopies that of DHODH (the rate-limiting step), we chose to profile DHODH inhibition in the cotton rat model.
We were unable to demonstrate antiviral efficacy in this model using either 6b or the known inhibitor brequinar in the assay. Instead, we observed viral titers marginally increase over days 4–7 of the study. We speculate this is due to the immunosuppressive phenotype of DHODH inhibition impacting the host ability to clear the virus (22). It is intriguing that there seems to be a marked difference in the strong phenotype observed in vitro with the lack of efficacy observed in vivo. One possible explanation is that given the ability of uridine to rescue the antiviral phenotype in vitro, the lack of efficacy could be attributed to the high levels of circulating uridine observed in cotton rats (approximately 20 μM). The fact that Davis et al. were not able to demonstrate efficacy in mice provides further evidence that inhibition of this pathway to result in an antiviral effect is not attainable in vivo. Although plasma concentrations of uridine were not changed by the administration of 6b, there was a trend toward decreasing levels of uridine following brequinar dosing over 4 and 7 d. This decrease suggests that despite not impacting the viral titer, brequinar concentrations were adequate to inhibit the DHODH pathway and thereby impact pyrimidine biosynthesis. Unfortunately, we did not observe efficacy at high doses of the drug, but we observed cytotoxicity negating a therapeutic index. In hematology studies, brequinar administration appeared to result in a decrease in blood neutrophil and monocyte counts, suggesting a decrease in the inflammatory response to RSV infection. In the study with 6b, single-cell necrosis of crypt epithelium was not a prominent feature of changes in contrast to the study with brequinar that produced single-cell necrosis in most animals. However, mucosal necrosis of duodenum and/or cecum was present at doses ≥50 mg/kg per d. Together, these experiments provide a thorough druggability assessment of efficacy and on-target toxicity for the de novo pyrimidine biosynthesis as an antiviral target.
In summary, through phenotypic screening for inhibitors of a single RNA virus (RSV), we have identified compounds that exhibit broad-spectrum antiviral activity. Chemoproteomic identification of the protein targets as inhibitors of de novo pyrimidine biosynthesis led us to interrogate whether this host pathway is druggable for antiviral therapy. Monitoring biomarkers for antiproliferation during an antiviral efficacy study in vivo leads us to conclude this is not the case. A retrospective analysis of the in vitro profiling reveals dividing T- and B-type lymphoid-derived cells (e.g., Jurkat) as a possible indicator of the outcome in vivo. In order to identify a promising clinical candidate, the relative therapeutic index of any host-targeted compound will need to be an integral part of the early discovery process. We speculate that greater success might be seen by targeting host processes that directly interact with virally encoded gene products, rather than essential host enzymes that play a critical role in normally proliferating cells.
Materials and Methods
RSV strains A-Long (VR-26) and B-Washington (RSV-9320 VR-955) and influenza virus strains A/PuertoRico/8/34 (VR-1469) and B/Lee/40 (VR-101) were obtained from ATCC. Influenza A/Udorn/72 and HIV-1 strains MDR52-52, br/92/020, SF162, JR-CSF, MDR807, and MDR769 were supplied by Southern Research Institute. HCV 2a carrying the renilla-luciferase gene was obtained from Chiron. Dengue virus and yellow fever strains were supplied by Novartis Institutes for Tropical Diseases. A detailed description of the protocols and procedures to accompany the Results section is available in SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. Qing Yin Wang, Novartis Institutes for Tropical Diseases, for kindly providing the dengue and yellow fever data for compounds 6b and 14b.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017142108/-/DCSupplemental.
References
- 1.Chanock R, Roizman B, Myers R. Recovery from infants with respiratory illness of a virus related to chimpanzee coryza agent (CCA): Isolation, properties and characterization. Am. J. Epidemiol. 1957;66:281–290. doi: 10.1093/oxfordjournals.aje.a119901. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Respiratory syncytial virus activity—United States, 1999–2000 season. JAMA J Am Med Assoc. 2001;285:287–288. [PubMed] [Google Scholar]
- 3.Falsey AR, et al. Respiratory syncytial virus infection in elderly and high-risk adults. New Engl J Med. 2009;352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 4.Shay DK, et al. Bronchiolitis-associated hospitalizations among US children, 1980–1996. JAMA J Am Med Assoc. 1999;282:1440–1446. doi: 10.1001/jama.282.15.1440. [DOI] [PubMed] [Google Scholar]
- 5.Smyth RL, Openshaw PJM. Bronchiolitis. Lancet. 2006;368:312–322. doi: 10.1016/S0140-6736(06)69077-6. [DOI] [PubMed] [Google Scholar]
- 6.Stensballe LG, Devasundaram JK, Simoes EA. Respiratory syncytial virus epidemics: The ups and downs of a seasonal virus. Pediatr Infect Dis J. 2003;22:S21–S32. doi: 10.1097/01.inf.0000053882.70365.c9. [DOI] [PubMed] [Google Scholar]
- 7.Moler FW, Brown RW, Faix RG, Gilsdorf JR. Comments on palivizumab (Synagis) Pediatrics. 1999;103:495–496. doi: 10.1542/peds.103.2.495. [DOI] [PubMed] [Google Scholar]
- 8.Schickli J, Dubovsky F, Tang RS. Challenges in developing a pediatric RSV vaccine. Hum Vaccines. 2010;5:582–591. doi: 10.4161/hv.9131. [DOI] [PubMed] [Google Scholar]
- 9.King DA, et al. Infectious diseases: Preparing for the future. Science. 2006;313:1392–1393. doi: 10.1126/science.1129134. [DOI] [PubMed] [Google Scholar]
- 10.Rappuoli R. From Pasteur to genomics: Progress and challenges in infectious diseases. Nat Med. 2004;10:1177–1185. doi: 10.1038/nm1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Clercq E. Strategies in the design of antiviral drugs. Nat Rev Drug Discov. 2002;1:13–25. doi: 10.1038/nrd703. [DOI] [PubMed] [Google Scholar]
- 12.Schmidtke P, Barril X. Understanding and predicting druggability. A high-throughput method for detection of drug binding sites. J Med Chem. 2010;53:5858–5867. doi: 10.1021/jm100574m. [DOI] [PubMed] [Google Scholar]
- 13.Brown D, Superti-Furga G. Rediscovering the sweet spot in drug discovery. Drug Discov Today. 2003;8:1067–1077. doi: 10.1016/s1359-6446(03)02902-7. [DOI] [PubMed] [Google Scholar]
- 14.Owens J. Determining druggability. Nat Rev Drug Discov. 2007;6:187–187. [Google Scholar]
- 15.Van Acker KLA, et al. Time-of-addition assay for identifying anti-viral compounds. WO/2005/028669. World Intellectual Property Organization; 2005
- 16.Tristram DA, Hicks W, Jr, Hard R. Respiratory syncytial virus and human bronchial epithelium. Arch Otolaryngol Head Neck Surg. 1998;124:777–783. doi: 10.1001/archotol.124.7.777. [DOI] [PubMed] [Google Scholar]
- 17.Bantscheff M, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 18.Bantscheff M, et al. Robust and sensitive ITRAQ quantification on an LTQ orbitrap mass spectrometer. Mol Cell Proteomics. 2008;7:1702–1713. doi: 10.1074/mcp.M800029-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu S, et al. Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents. Structure. 2000;8:25–33. doi: 10.1016/s0969-2126(00)00077-0. [DOI] [PubMed] [Google Scholar]
- 20.Coleman PF, Suttle DP, Stark GR. Purification from hamster cells of the multifunctional protein that initiates de novo synthesis of pyrimidine nucleotides. J Biol Chem. 1977;252:6379–6385. [PubMed] [Google Scholar]
- 21.Baumgartner R, et al. Dual binding mode of a novel series of DHODH inhibitors. J Med Chem. 2006;49:1239–1247. doi: 10.1021/jm0506975. [DOI] [PubMed] [Google Scholar]
- 22.Makowka L, Sher LS, Cramer DV. The development of brequinar as an immunosuppressive drug for transplantation. Immunol Rev. 1993;136:51–70. doi: 10.1111/j.1600-065x.1993.tb00654.x. [DOI] [PubMed] [Google Scholar]
- 23.Niewiesk S, Prince G. Diversifying animal models: The use of hispid cotton rats (Sigmodon Hispidus) in infectious diseases. Lab Anim. 2002;36:357–372. doi: 10.1258/002367702320389026. [DOI] [PubMed] [Google Scholar]
- 24.Newman DJ, Cragg GM. Natural product scaffolds as leads to drugs. Future Med Chem. 2009;1:1415–1427. doi: 10.4155/fmc.09.113. [DOI] [PubMed] [Google Scholar]
- 25.Galloway WRJD, Isidro-Llobet A, Spring DR. Diversity-oriented synthesis as a tool for the discovery of novel biologically active small molecules. Nat Commun. 2010;1 doi: 10.1038/ncomms1081. 80. [DOI] [PubMed] [Google Scholar]
- 26.Hajduk PJ, Galloway WRJD, Spring DR. Drug discovery: A question of library design. Nature. 2011;470:42–43. doi: 10.1038/470042a. [DOI] [PubMed] [Google Scholar]
- 27.Jones ME. Pyrimidine nucleotide biosynthesis in animals: Genes, enzymes, and regulation of UMP biosynthesis. Annu Rev Biochem. 1980;49:253–279. doi: 10.1146/annurev.bi.49.070180.001345. [DOI] [PubMed] [Google Scholar]
- 28.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 29.Becroft DM, Phillips LI. Hereditary orotic aciduria and megaloblastic anaemia: A second case, with response to uridine. Br Med J. 1965;1:547–552. doi: 10.1136/bmj.1.5434.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cass CE, Young JD, Baldwin SA. Recent advances in the molecular biology of nucleoside transporters of mammalian cells. Biochem Cell Biol. 1998;76:761–770. doi: 10.1139/bcb-76-5-761. [DOI] [PubMed] [Google Scholar]
- 31.Mascia L, Turchi G, Bemi V, Ipata PL. Uracil salvage pathway in PC12 cells. Biochim Biophys Acta. 2000;1524:45–50. doi: 10.1016/s0304-4165(00)00139-2. [DOI] [PubMed] [Google Scholar]
- 32.Fairbanks LD, Bofill M, Ruckemann K, Simmonds HA. Importance of ribonucleotide availability to proliferating T-lymphocytes from healthy humans. Disproportionate expansion of pyrimidine pools and contrasting effects of de novo synthesis inhibitors. J Biol Chem. 1995;270:29682–29689. [PubMed] [Google Scholar]
- 33.Simmons AJ, Rawls JM, Piskur J, Davidson JN. A mutation that uncouples allosteric regulation of carbamyl phosphate synthetase in Drosophila. J Mol Biol. 1999;287:277–285. doi: 10.1006/jmbi.1999.2618. [DOI] [PubMed] [Google Scholar]
- 34.Evers DL, et al. Inhibition of human cytomegalovirus signaling and replication by the immunosuppressant FK778. Antiviral Res. 2005;65:1–12. doi: 10.1016/j.antiviral.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Biron KK, et al. Metabolic activation of the nucleoside analog 9-{[2-hydroxy-1-(hydroxymethyl)ethoxy]methyl}guanine in human diploid fibroblasts infected with human cytomegalovirus. Proc Natl Acad Sci USA. 1985;82:2473–2477. doi: 10.1073/pnas.82.8.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smee DF, et al. Novel pyrazolo[3,4-d]pyrimidine nucleoside analog with broad-spectrum antiviral activity. Antimicrob Agents Chemother. 1987;31:1535–1541. doi: 10.1128/aac.31.10.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farasati NA, Shapiro R, Vats A, Randhawa P. Effect of leflunomide and cidofovir on replication of BK virus in an in vitro culture system. Transplantation. 2005;79:116–118. doi: 10.1097/01.tp.0000149338.97084.5f. [DOI] [PubMed] [Google Scholar]
- 38.Knight DA, et al. Inhibition of herpes simplex virus type 1 by the experimental immunosuppressive agent leflunomide. Transplantation. 2001;71:170–174. doi: 10.1097/00007890-200101150-00031. [DOI] [PubMed] [Google Scholar]
- 39.Wachsman M, Hamzeh FM, Assadi NB, Lietman PS. Antiviral activity of inhibitors of pyrimidine de-novo biosynthesis. Antiviral Chem Chemother. 1996;7:7–13. [Google Scholar]
- 40.Josephson MA, et al. Treatment of renal allograft polyoma BK virus infection with leflunomide. Transplantation. 2006;81:704–710. doi: 10.1097/01.tp.0000181149.76113.50. [DOI] [PubMed] [Google Scholar]
- 41.Avery RK, et al. Use of leflunomide in an allogeneic bone marrow transplant recipient with refractory cytomegalovirus infection. Bone Marrow Transpl. 2004;34:1071–1075. doi: 10.1038/sj.bmt.1704694. [DOI] [PubMed] [Google Scholar]
- 42.Battiwalla M, et al. Leflunomide failure to control recurrent cytomegalovirus infection in the setting of renal failure after allogeneic stem cell transplantation. Transpl Infect Dis. 2007;9:28–32. doi: 10.1111/j.1399-3062.2006.00170.x. [DOI] [PubMed] [Google Scholar]
- 43.Kelly LM, Lisziewicz J, Lori F. “Virostatics” as a potential new class of HIV drugs. Curr Pharm Des. 2004;10:4103–4120. doi: 10.2174/1381612043382495. [DOI] [PubMed] [Google Scholar]
- 44.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 45.Karle JM, Anderson LW, Erlichman C, Cysyk RL. Serum uridine levels in patients receiving N-(phosphonacetyl)-L-aspartate. Cancer Res. 1980;40:2938–2940. [PubMed] [Google Scholar]
- 46.Dudman NPB, Deveski WB, Tattersall MHN. Radioimmunoassays of plasma thymidine, uridine, deoxyuridine, and cytidine/deoxycytidine. Anal Biochem. 1981;115:428–437. doi: 10.1016/0003-2697(81)90029-4. [DOI] [PubMed] [Google Scholar]
- 47.Evans David R, Guy Hedeel I. Mammalian pyrimidine biosynthesis: Fresh insights into an ancient pathway. J Biol Chem. 2004;279:33035–33038. doi: 10.1074/jbc.R400007200. [DOI] [PubMed] [Google Scholar]
- 48.Schorlemmer HU, Kurrle R, Schleyerbach R. A77-1726, leflunomide's active metabolite, inhibits in vivo lymphoproliferation in the popliteal lymph node assay. Int J Immunother. 1998;14:205–211. doi: 10.1007/s000110050542. [DOI] [PubMed] [Google Scholar]
- 49.Schiff MH, Strand V, Oed C, Loew-Friedrich I. Leflunomide: Efficacy and safety in clinical trials for the treatment of rheumatoid arthritis. Drugs Today. 2000;36:383–394. doi: 10.1358/dot.2000.36.6.584259. [DOI] [PubMed] [Google Scholar]
- 50.Davis IC, et al. Nucleotide-mediated inhibition of alveolar fluid clearance in BALB/c mice after respiratory syncytial virus infection. Am J Physiol Lung C. 2004;286:L112–L120. doi: 10.1152/ajplung.00218.2003. [DOI] [PubMed] [Google Scholar]
- 51.Davis IC, et al. Post-infection A77-1726 blocks pathophysiologic sequelae of respiratory syncytial virus infection. Am J Respir Cell Mol Biol. 2007;37:379–386. doi: 10.1165/rcmb.2007-0142OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Byrd LG, Prince GA. Animal models of respiratory syncytial virus infection. Clin Infect Dis. 1997;25:1363–1368. doi: 10.1086/516152. [DOI] [PubMed] [Google Scholar]
- 53.Cianci C, et al. Oral efficacy of a respiratory syncytial virus inhibitor in rodent models of infection. Antimicrob Agents Chemother. 2004;48:2448–2454. doi: 10.1128/AAC.48.7.2448-2454.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.