Abstract
Nuclear reprogramming of differentiated cells can be induced by oocyte factors. Despite numerous attempts, these factors and mechanisms responsible for successful reprogramming remain elusive. Here, we identify one such factor, necessary for the development of nuclear transfer embryos, using porcine oocyte extracts in which some reprogramming events are recapitulated. After incubating somatic nuclei in oocyte extracts from the metaphase II stage, the oocyte proteins that were specifically and abundantly incorporated into the nuclei were identified by mass spectrometry. Among 25 identified proteins, we especially focused on a multifunctional protein, DJ-1. DJ-1 is present at a high concentration in oocytes from the germinal vesicle stage until embryos at the four-cell stage. Inhibition of DJ-1 function compromises the development of nuclear transfer embryos but not that of fertilized embryos. Microarray analysis of nuclear transfer embryos in which DJ-1 function is inhibited shows perturbed expression of P53 pathway components. In addition, embryonic arrest of nuclear transfer embryos injected with anti–DJ-1 antibody is rescued by P53 inhibition. We conclude that DJ-1 is an oocyte factor that is required for development of nuclear transfer embryos. This study presents a means for identifying natural reprogramming factors in mammalian oocytes and a unique insight into the mechanisms underlying reprogramming by nuclear transfer.
Keywords: oocyte extract and proteomics, reprogramming in mammalian oocytes
Embryonic cells differentiate into specific types of cells as development progresses. Once differentiated, the reversion of a differentiated cell state to an original undifferentiated state is strictly inhibited in normal development. However, it has been experimentally shown that differentiated nuclei can be returned to an undifferentiated embryonic state after nuclear transfer (NT) to enucleated eggs or oocytes (1, 2). Such experiments provide an opportunity to reprogram somatic cells as a means to prepare undifferentiated cells, which may be differentiated into any kinds of cells for cell-replacement therapy. Recently, nuclear reprogramming technology has been expanded by the production of induced pluripotent stem (iPS) cells (3). iPS cells can be obtained by overexpressing specific sets of transcription factors such as Oct4, Sox2, Klf4, and c-myc in cultured cells. The processes leading to establishment of iPS cell lines are being carefully examined and we are begining to understand how somatic cells acquire pluripotency by this method (4–6). The mechanisms leading to pluripotency may be different between iPS cells and NT embryos because somatic nuclei transferred into unfertilized metaphase II (MII) oocytes must undergo early embryonic development before the inner cell mass (ICM) can give rise to pluripotent embryonic stem (ES) cells. In addition, the molecules and mechanisms that induce somatic cell reprogramming are expected to be different between iPS cells and NT embryos (7, 8). A recent study has shown that nuclear transfer-mediated reprogramming resets differentiated cell states more efficiently than the iPS method (9). However, almost nothing is known about factors and mechanisms involved in the reprogramming of somatic cells by NT to mammalian oocytes.
Some proteins have been identified as reprogramming factors in Xenopus eggs and oocytes. These include ISWI (10), FRGY2a/b (11), BRG1 (12), Nucleoplasmin (13), and Histone B4 (14). Although the roles of these factors on nuclear remodeling and transcriptional reprogramming have been shown, it has not been demonstrated that these factors are crucial for nuclear reprogramming in NT embryos during embryonic development.
As shown above, reprogramming factors have been mainly identified in Xenopus. This is because Xenopus eggs and oocytes are extremely large and easily accessible, compared with mammalian oocytes. These properties allowed development of cell-free systems using Xenopus egg or oocyte extracts. In the extracts, at least some reprogramming events are induced on large numbers of cells (105 cells in 100 μL of extracts) (10, 15), allowing the study of reprogramming by biochemical means. In contrast to Xenopus, a system to study the molecular mechanisms underlying reprogramming has been lacking in mammalian oocytes. Recently, our group developed a cell-free system from porcine oocytes (16). We showed that a porcine oocyte extract from the MII oocytes is able to displace transcription factors from donor nuclei and deacetylate the histone tails of somatic nuclei. These reprogramming events related to the erasure of gene-expression memories are also observed in somatic nuclei transferred to MII oocytes, suggesting that a part of reprogramming events can be reproduced in the oocyte extracts. Besides the character of the MII extracts, somatic nuclei can be fully reprogrammed to totipotency when transplanted in MII oocytes. Reprogramming factors related to totipotency are also present in oocytes at the germinal vesicle (GV) stage (17), and GV oocyte extracts induce reprogramming events, which are different from reprogramming in MII extracts, such as activation of pluripotent genes (16). However, the ability of these GV factors to reprogram transferred nuclei toward totipotency has never been directly shown because cloned offspring have never been obtained by direct injection of nuclei to GV oocytes. Taken together, MII oocyte extracts are ideal to explore reprogramming factors important for development of NT embryos as these factors might be accumulated and active.
In this study, we use porcine oocyte extracts to identify a unique oocyte factor involved in nuclear reprogramming toward totipotency. We focused on protein exchange in extract-treated nuclei because the exchange of nuclear proteins between a donor nucleus and an oocyte cytoplasm is a prominent phenomenon during reprogramming (18–20). Additionally, some proteins, such as ISWI and nucleoplasmin, that are incorporated from Xenopus egg extracts into donor nuclei have been shown to have a role in reprogramming (10, 13). After incubation of somatic nuclei in oocyte extracts, proteins that become associated with somatic nuclei from the extracts were identified by mass spectrometry. The function of the identified proteins was then examined in NT embryos and cultured cells. These experiments revealed that PARK7 (DJ-1) is required for successful reprogramming in NT embryos.
Results
Identification of Oocyte Proteins That Associate with Somatic Nuclei.
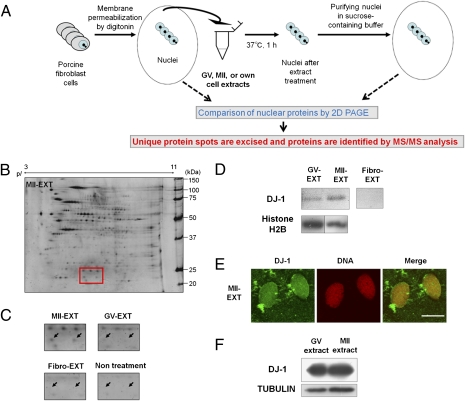
The strategy to identify a reprogramming factor in oocytes is diagramed in Fig. 1A. Briefly, porcine fibroblast cell nuclei were incubated in extracts from oocytes at the MII stage or the GV stage or extracts from fibroblast cells (own cell extracts). After the extract treatment, nuclei were collected and proteins were separated by 2D polyacrylamide gel electrophoresis (2D PAGE) (Fig. 1B and Fig. S1). As shown in Fig. S1, nuclear proteins were greatly changed after oocyte extract treatment, in agreement with a previous in vivo study in which nuclei were transferred into eggs (18). In particular, we focused on the proteins that were specifically and abundantly accumulated in nuclei after treatment with MII oocyte extracts. More than 60 unique protein spots were detected in MII extract-treated nuclei and these spots were excised and subjected to mass spectrometry. MALDI-TOF/TOF analysis and database searches revealed identity of 31 unique protein spots, and 25 proteins were finally identified (Fig. S2 and Table S1). Proteins were classified according to their function (Fig. S3A). Proteins with chaperone function were most commonly found, reflecting a marked change of nuclear proteins during reprogramming. Proteins related to DNA replication initiation were also found. This proteomic result agrees with a previous report (21) showing that reprogramming of somatic type replication forks to embryonic ones starts from the metaphase stage in oocyte extracts. Some examples of unique protein spots are shown in Fig. S3B.
Fig. 1.
Identification of protein DJ-1 as a candidate reprogramming factor present in the MII oocytes. (A) Schematic diagram of the strategy to identify reprogramming factors in oocytes. (B) Total protein of nuclei treated with MII oocyte extracts (MII-EXT) separated by 2D PAGE. The gel marked by the red square box (20–25 kDa) contains protein DJ-1 spots. pI, isoelectric point. (C) DJ-1 spots (arrows) in different nuclear samples from MII-EXT, GV oocyte extract-treated cells (GV-EXT), fibroblast extract-treated cells (Fibro-EXT), and permeabilized cells without extract treatment (nontreatment). (D) Western blotting on fibroblast nuclei after extract treatment. Incorporation of DJ-1 proteins into nuclei was detected using a DJ-1–specific antibody. Histone H2B was used as a loading control. (E) Immunofluorescence analysis of DJ-1 in nuclei after treatment with MII oocyte extracts. DNA was stained with propidium iodide. (Scale bar: 10 μm.) (F) Large amounts of DJ-1 are present in both GV and MII extracts, examined by Western blotting. Tubulin was used as a loading control.
DJ-1 Is Incorporated in Oocyte Extracts-Treated Nuclei.
Two unique protein spots (20–25 kDa and pI 5–7) were detected in the MII extract-treated nuclei [Fig. 1 B (red box) and C (arrows)]. MALDI-TOF/TOF analysis and database searches revealed that these two spots were both matched to PARK7, also known as DJ-1. This result was repeated in three independent experiments using different oocyte extracts. Two spots of different pI were identified as DJ-1, because oxidized DJ-1 has a lower pI (22). Incorporation of DJ-1 protein into fibroblast cell nuclei after treatment with MII oocyte extracts was also observed by Western blot (Fig. 1D) and immunofluorescence analysis (Fig. 1E). DJ-1 was also detected in nuclei treated with GV oocyte extracts (Fig. 1D), but the total amounts of incorporated DJ-1 proteins were higher in those treated with MII oocyte extracts than in those treated with GV oocyte extracts, agreeing with the 2D PAGE result (Fig. 1C). Considering that GV and MII oocyte extracts had similarly large amounts of DJ-1 (Fig. 1F) and both extracts have differential reprogramming abilities (16), greater incorporation of DJ-1 into cells treated with MII extracts may result from an active incorporation mechanism, implying that DJ-1 might have a specific role in MII oocyte extracts.
Expression Pattern of DJ-1.
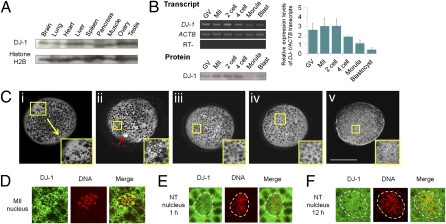
In addition to the unique incorporation pattern of oocyte DJ-1 proteins to somatic nuclei, DJ-1 has been reported to interact with a known reprogramming factor, c-myc (23, 24). Large amounts of DJ-1 are stored in oocytes (Fig. 1F), although its function in early embryonic development has never been examined. For these interesting characteristics of DJ-1, we asked whether DJ-1 plays a role in nuclear transfer embryos. We first investigated its expression pattern. DJ-1 was abundantly expressed in all porcine organs examined (Fig. 2A). DJ-1 expression was examined in porcine oocytes and NT embryos. These transcripts and proteins were highly expressed throughout oocyte maturation and persisted until the four-cell stage in NT embryos (Fig. 2B). Expression was down-regulated until the blastocyst stage (Fig. 2B). Generally, large amounts of DJ-1 proteins were detected throughout the oocyte cytoplasm, excluding lipid droplets (Fig. 2C). In oocytes at the GV stage, DJ-1 was present within the germinal vesicles as well as in cytoplasm [Fig. 2C, i, a germinal vesicle in the enlarged yellow box (Inset)]. It was dispersed in the MII oocyte cytoplasm (Fig. 2C, ii) and on condensed MII chromosomes (Fig. 2D). Incorporation of DJ-1 into transferred donor nuclei was observed from 1 h after nuclear transfer although the signal is weaker than that in cytoplasm (Fig. 2 C, iii and E). Three hours after nuclear transfer, nuclear DJ-1 is comparable to cytoplasmic DJ-1 (Fig. 2C, iv) and it remained in nuclei 12 h after transfer (Fig. 2 C, v and F), suggesting that DJ-1 has been already incorporated into nuclei before entering the first S phase (25). Together, maternally accumulated DJ-1 was incorporated into transferred somatic nuclei in NT embryos.
Fig. 2.
DJ-1 expression in porcine tissues, oocytes, and nuclear transfer embryos. (A) DJ-1 proteins exist in various types of tissues examined by Western blotting. (B) Expression of DJ-1 transcripts and proteins was examined in oocytes (GV and MII stages) and NT embryos during preimplantation development. The graph shows the relative expression levels of DJ-1 transcripts examined by semiquantitative RT-PCR (normalized by ACTB transcripts). The numbers of oocytes and embryos used for Western blotting are as follows: GV, eight; MII, eight; two-cell, eight; four-cell, seven; Morula, two; Blast, three. Error bars, mean ± SEM. (C) Localization of DJ-1 protein in oocytes and NT embryos, observed by immunofluorescence analysis. i–v, a whole oocyte or a NT embryo, with an enlargement of the yellow boxed area (Inset) shown at the Lower Right. The enlarged areas (Insets) contain nuclei of oocytes or NT embryos. White areas show staining with anti–DJ1 antibody. i, GV oocyte; ii, MII oocyte; iii, NT embryo 1 h after nuclear transfer; iv, NT embryo 3 h after transfer; v, NT embryo 12 h after transfer. A red arrow in ii represents a first polar body in a MII oocyte. (Scale bar: 100 μm.) (D) Distribution of DJ-1 in a MII nucleus at high magnification. Yellow color on merged photos (Merge) represents nuclear localization of DJ-1. (E) DJ-1 distribution in transferred fibroblast nuclei 1 h after nuclear transfer into a MII oocyte. The border between nucleus and oocyte cytoplasm is marked with dashed white lines. (F) DJ-1 in transferred fibroblast nuclei 12 h after nuclear transfer.
Inhibition of DJ-1 Function in NT Embryos.
To test the role of DJ-1 on nuclear reprogramming, its function was inhibited in porcine NT embryos by several methods. Because DJ-1 is present in transferred nuclei 1 h after NT, its function should be inhibited before introduction of donor nuclei into the oocyte. However, neither siRNA injection nor antisense oligonucleotide injection during oocyte maturation caused significant degradation of maternal DJ-1 proteins, although maternal mRNA was decreased (Fig. S4 A and B), indicating that it is difficult to reduce DJ-1 proteins during oocyte maturation.
We then tried anti–DJ-1 antibody injection into MII oocytes before nuclear transfer, parthenogenetic activation (PA), and intracytoplasmic sperm injection (ICSI). The polyclonal antibody used here is raised against a peptide mapping near the N terminus of DJ-1 of human origin whose sequence matches perfectly to that of pig and is highly specific for recognizing the porcine DJ-1 among whole oocyte proteins (Fig. S4C). The antibody was injected into cytoplasm of MII oocytes >1 h before fusion with donor cells to allow binding to maternal DJ-1. The proportion of embryos that cleaved normally was less for NT embryos injected with anti–DJ-1 antibody (αDJ1-NT embryos) than for control NT embryos injected with IgG (IgG-NT embryos), at 24, 27, and 30 h after NT (Fig. S5), but the final proportion of normally cleaved embryos, judged 48 h after NT, was not different (Table 1). This result suggests that the first cleavage in αDJ1-NT embryos is delayed. Interestingly, the proportion of αDJ1-NT embryos that developed to the blastocyst stage was significantly lower than that of control IgG-NT and noninjected NT embryos (P < 0.004: 1.67 ± 1.67%, 13.40 ± 2.39% and 14.22 ± 2.92%, respectively) (Table 1). Similarly, significantly fewer embryos developed to the morula stage in αDJ1-NT embryos compared with controls (P < 0.002) (Table 1). Many αDJ1-NT embryos arrested at the two- or the four-cell stage. In contrast to that in NT embryos, anti–DJ-1 antibody injection to ICSI and PA embryos did not affect development to the morula and blastocyst stages (Table 1).
Table 1.
In vitro development of anti–DJ-1 antibody-injected embryos to the blastocyst stage
| No. trials | Total no. embryos | First cleavage (%) | Morulae (%) | Blastocysts (%) | Embryos arrested at the two- and four-cell stages (%) | |
| NT embryos | ||||||
| a) Anti–DJ1-injected embryos | 6 | 103 | 70 (68.04 ± 5.04) | 3 (2.50 ± 2.50) [b, d, e, f, g] | 2 (1.67 ± 1.67) [d, f, g] | 59 (57.61 ± 4.27) [b, d, f] |
| b) Anti–DJ1-injected (+Pifithrin-α) | 3 | 35 | 26 (75.37 ± 6.94) | 8 (21.10 ± 5.09) [a] | 6 (12.98 ± 9.40) | 11 (34.78 ± 7.97) [a] |
| c) Anti–DJ1-injected (+DMSO) | 3 | 33 | 26 (75.56 ± 8.89) | 3 (7.22 ± 3.89) [d, f, g] | 2 (4.44 ± 4.44) | 20 (61.11 ± 5.56) [d, f] |
| d) IgG-injected embryos | 6 | 111 | 73 (64.76 ± 4.52) | 26 (25.41 ± 4.81) [a, c] | 15 (13.40 ± 2.39) [a] | 36 (30.31 ± 5.34) [a, c] |
| e) DJ1(L166P) mRNA-injected embryos | 4 | 74 | 53 (71.07 ± 5.23) | 10 (13.04 ± 1.96) [a, f, g] | 5 (6.25 ± 2.39) | 35 (46.43 ± 7.92) |
| f) EGFP mRNA-injected embryos | 4 | 83 | 57 (68.30 ± 5.48) | 19 (23.16 ± 1.90) [a, c, e] | 11 (13.08 ± 3.47) [a] | 30 (35.65 ± 4.81) [a, c] |
| g) Control (noninjected embryos) | 4 | 134 | 93 (68.94 ± 2.43) | 28 (21.07 ± 1.05) [a, c, e] | 19 (14.22 ± 2.92) [a] | ND |
| ICSI embryos | ||||||
| Anti–DJ1-injected embryos | 5 | 63 | 21 (33.06 ± 0.85) | 18 (28.55 ± 1.82) | 18 (28.55 ± 1.82) | 3 (4.51 ± 2.09) |
| IgG-injected embryos | 5 | 60 | 21 (34.58 ± 1.33) | 17 (27.86 ± 1.56) | 17 (27.86 ± 1.56) | 4 (6.72 ± 1.88) |
| Control (Noninjected embryos) | 5 | 68 | 25 (36.15 ± 2.36) | 21 (30.82 ± 0.65) | 21 (30.82 ± 0.65) | 4 (5.33 ± 2.26) |
| PA embryos | ||||||
| Anti–DJ1-injected embryos | 3 | 63 | 49 (76.18 ± 3.21) | ND | 33 (51.99 ± 5.03) | ND |
| IgG-injected embryos | 3 | 72 | 58 (79.39 ± 2.56) | ND | 39 (51.49 ± 6.79) | ND |
Three experimental groups are set as nuclear transfer (NT) embryos, intracytoplasmic sperm injection (ICSI) embryos, and parthenogenetically activated (PA) embryos. DJ-1–injected embryos represent the embryos injected with antibody against DJ-1. Similarly, IgG-, DJ1(L166P) mRNA-, and EGFP mRNA-injected embryos are shown. a–g, samples in NT embryos: Samples that show significant differences within the same column in NT embryos are indicated in brackets (P < 0.05). For example, sample f shows significant differences from a, c, and e in development to the morula stage. ICSI and PA embryos do not show any significant differences. Percentages are expressed as mean± SEM. ND, not determined.
Finally, DJ-1 was inhibited by injecting mRNA of the DJ-1 dominant negative form [DJ1(L166P)] (26). Similar to the result from antibody injection, a lower proportion of NT embryos injected with DJ1(L166P) mRNA developed to the morula and blastocyst stages, compared with EGFP mRNA-injected or noninjected NT embryos, although the proportion of cleaved embryos did not differ (Table 1).
P53 Activation Is Involved in Developmental Arrest After Inhibition of DJ-1 Function.
To uncover the basis for developmental arrest in DJ-1 antibody-injected NT embryos, global changes in gene expression were examined by Affymetrix porcine microarrays. NT embryos were collected 28 h after nuclear transfer when many of the embryos had just reached the two-cell stage. We chose this stage because major zygotic genome activation occurs at the four-cell stage, which might mask altered expression of direct target genes, and because DJ-1 seems to be active before the two- or the four-cell stage.
Global transcripts were compared among αDJ1-NT, IgG-NT, and noninjected NT embryos and donor cells. Comparing αDJ1-NT embryos and donor cells, 6,411 probes (26.78%) were underexpressed and 4,256 probes (17.78%) were overexpressed among a total of 23,937 probes (≥2.0-fold), indicating a significant reprogramming in a large number of genes. To find genes affected specifically by DJ-1 inhibition, we examined differentially expressed probes comparing αDJ1-NT and IgG-NT embryos and 401 down-regulated probes (1.68%) and 349 up-regulated probes (1.46%) were found. These genes were involved in RNA metabolism, oxidative stress, signaling pathways, and cell metabolism. DJ-1 has been reported to be engaged in these cellular processes (27–31). In particular, previous reports identified a set of DJ-1–related genes using cultured cells, and the expression of these genes is largely disturbed in αDJ1-NT embryos [Ras oncogenes (RABL3, 2.3-fold down; RAP2C, 2.1-fold down; RAB26, 12.5-fold up) and serine/cysteine protease inhibitor (SERPINB1, 3.3-fold up)] (27, 32), suggesting that antibody injection specifically inhibited DJ-1 function in NT embryos.
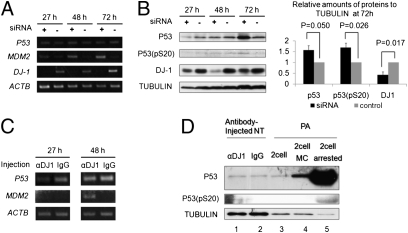
Microarray analysis also showed that the expression of genes involved in the P53 pathway was largely disturbed in αDJ1-NT embryos compared with IgG-NT as summarized in Fig. S6 (genes whose expression was up-regulated or down-regulated are marked with an asterisk). Moreover, it is reported that DJ-1 inhibition is correlated with P53 activation (33, 34). We therefore decided to examine P53 expression after DJ-1 inhibition. The correlation between P53 and DJ-1 expression was first examined in cultured cells. DJ-1 expression in porcine fibroblasts was efficiently and significantly inhibited at both the transcriptional and the translational levels by siRNA against pig DJ-1 (Fig. 3 A and B). Lipid droplets reminiscent of cellular stress appeared in the knockdown cells, but not control cells (Fig. S7, arrows). Up-regulation of MDM2, a main regulator of P53, was observed in DJ-1 knockdown cells (Fig. 3A). Concomitantly, P53 and phosphorylated P53 at Serine 20 [P53(pS20)], which is a stabilized form of P53 (35), were slightly up-regulated in DJ-1 knockdown cells (Fig. 3B). These results suggest that DJ-1 inhibition induces activation of P53 in cultured cells.
Fig. 3.
Activation of P53 after inhibition of DJ-1. (A) RT-PCR analysis of cultured porcine fibroblast cells after knockdown of DJ-1 by specific siRNA. DJ-1 expression was specifically inhibited by siRNA and, at the same time, activation of MDM2 was observed. Duration (h) after siRNA transfection is indicated. (B) (Left) Activation of P53 after knockdown of DJ-1 in porcine fibroblasts was examined by Western blotting analysis. (Right) Relative band intensities of P53, P53(pS20), and DJ-1 when standardized with Tubulin. P53 and phosphorylated P53 at serine 20 [P53(pS20)] were significantly up-regulated, whereas DJ-1 was successfully down-regulated by knockdown. Error bars, mean ± SEM (t test, n = 3). (C) Transcripts in NT embryos injected with anti–DJ-1 antibody (αDJ1) or IgG (IgG) were examined by RT-PCR analysis. MDM2 was detected only in embryos injected with DJ-1 antibody. (D) Activation of P53 was examined in NT and PA embryos by Western blotting analysis. Anti–DJ-1 antibody was injected into NT embryos (αDJ1, lane 1) and IgG was injected as a control (IgG, lane 2). NT embryos were collected 72 h after nuclear transfer. For Western blots of PA embryos, three types of two-cell stage PA embryos were collected: Nontreated embryos were collected 48 h after parthenogenetic activation (2 cell, lane 3); Mitomycin C was added to the culture medium from 24 to 48 h after activation, and sampling was performed at 48 h (2 cell MC, lane 4); and embryos arrested at the two-cell stage were collected at 120 h (2 cell arrested, lane 5).
We examined whether P53 is similarly activated in DJ-1–deficient NT embryos. RT-PCR analysis showed MDM2 transcripts were detected only in porcine αDJ1-NT embryos (Fig. 3C), agreeing with the result from DJ-1 knockdown in cultured cells. Faint bands of P53 and P53(pS20) proteins were detected in αDJ1-NT embryos although their expression did not show a clear difference between αDJ1-NT and control IgG-NT embryos (Fig. 3D, lanes 1 and 2). We further tested the correlation between P53 activation and early embryonic arrest; P53 protein expression was compared between normal and arrested PA embryos at the two-cell stage. Mitomycin C treatment (10 μM) with the two-cell embryos caused developmental arrest of embryos and up-regulated P53 proteins (Fig. 3D, lanes 3 and 4). Furthermore, when P53 expression was compared between the two-cell embryos (day 2) and the arrested two-cell embryos (day 5), large amounts of P53 and P53(pS20) were accumulated in the arrested embryos (Fig. 3D, lanes 3 and 5). These results suggest that P53 activation is correlated with early embryonic arrest. Finally, we asked whether P53 inhibition on αDJ1-NT embryos can rescue the early embryonic arrest. P53 was inhibited by a specific inhibitor, Pifithrin-α, at a concentration of 1 μM (36, 37). Remarkably, Pifithrin-α treatment successfully rescued the embryonic arrest of αDJ1-NT (Table 1). Pifithrin-α did not improve development of normal nuclear transfer embryos to the blastocyst stage (38), suggesting that a rescued phenotype observed by this inhibitor treatment should be specific to αDJ1-NT. These results indicate that embryonic arrest of αDJ1-NT can, at least to some extent, be attributed to P53 activation. Considering that DJ-1 inhibition did not change amounts of P53 transcripts (Fig. 3 A and C) and weakly changed total P53 protein amounts (Fig. 3 B and D), regulation of P53 activation by DJ-1 may be carried out at the posttranslational level. This idea agrees with previous reports in which DJ-1 represses P53 transcriptional activity through direct binding to P53 proteins rather than repressing P53 transcription (39, 40).
Discussion
Factors necessary for the successful reprogramming of somatic cells by mammalian oocytes have been explored since the first successful production of the cloned sheep (2). Although more than a decade has passed, they remain largely unknown, partly because many biochemical analyses have not been applied to oocytes and embryos whose quantities are limited. Here, we used functional oocyte extracts to overcome this issue and combined them with proteome analysis. We identified an oocyte factor, DJ-1, necessary for the successful development of porcine NT embryos.
DJ-1 was initially identified as an oncogene that transformed mouse NIH 3T3 cells weakly on its own and more strongly in combination with Ras (24). Further studies revealed that DJ-1 has multiple functions (Fig. S6). DJ-1 modulates the PTEN/Akt survival pathway (30, 41), suppresses Ask1-mediated apoptosis (42), has RNA binding activity (28) and chaperone activity (43), and protects cells from oxidative stress (44). DJ-1 is a dimeric protein and has some structural similarities with heat-shock proteins (45). Deletion and a point mutation (L166P) of DJ-1 that disrupts normal dimer formation are responsible for the onset of familiar Parkinson's disease (46). In germ cells, high amounts of expression in testis and sperm have been reported and disturbed testicular DJ-1 expression is related to male infertility (47). We find that large amounts of DJ-1 are accumulated in oocytes during oogenesis (Fig. 2 B and C). Nevertheless, nothing is known about the role of maternally stored DJ-1 in early embryonic development.
Inhibition of maternal DJ-1 function indicated that DJ-1 is indispensable for early embryonic development in NT embryos (Table 1). Furthermore, microarray analysis revealed that the P53 pathway is disturbed in DJ1-NT embryos compared with control IgG-NT embryos (Fig. S6). P53 responds to diverse cellular stresses and induces cell-cycle arrest, apoptosis, cell senescence, or DNA repair. P53 expression is mainly regulated by MDM2, which binds to and targets p53 for ubiquitin-dependent proteolysis (48, 49). It has been generally considered that P53 and MDM2 exist as an autoregulatory feedback loop; MDM2 transcription is induced by P53 and in turn inactivates P53 by ubiquitination. Therefore, activation of MDM2 is often due to elevated P53 activity as was observed in this study (Fig. 3A). P53 activation critically affects embryonic viability (50, 51). Its activation also plays a crucial role in reprogramming somatic cells toward iPS cells (52–56). However, it has not been revealed whether P53 activation is involved in reprogramming of NT embryos. Our study shows that P53 activation is implicated in embryonic arrest in NT embryos. Interestingly, other identified oocyte proteins that are associated with somatic nuclei (Table S1), such as HSP27 (57), 14–3-3-zeta (58), and HSP90 (59), are reported to repress P53 activity. Several oocyte factors may synergistically inhibit P53 activation in NT embryos.
One of the interesting properties that DJ-1 possesses is that its function is necessary only for NT embryos, but not for ICSI and PA embryos (Table 1). NT embryos undergo higher rates of apoptosis (60) and are less tolerant to the in vitro culture environment than fertilized embryos (61). Lower levels of gene expression related to cell protection from stress and apoptosis have been reported in NT embryos, compared with fertilized embryos (62). Considering that NT embryos are experimentally produced whereas fertilized embryos are made by the natural process, it is reasonable to think that transferred somatic nuclei undergo higher cellular stress than sperms. This notion is supported by the fact that NT embryos exhibit higher rates of apoptosis and weaker stress-coping functions than fertilized embryos (60–62). Nevertheless, the proportion of NT embryos that develop to the blastocyst stage is similar to that of in vitro fertilized embryos in many reports. This comparable developmental ability to the blastocyst stage implies that oocytes intrinsically possess robust mechanisms to relieve cellular stresses and these mechanisms might work as a safeguard to ensure preimplantation development of NT embryos. These stress-coping mechanisms are probably relying on many different molecules because we found a lot of antistress proteins by mass spectrometry (Table S1). DJ-1 is one of the key players among these proteins and the lack of DJ-1 function has a detrimental effect on NT embryos, suggesting that the other redundant factors with stress-coping functions may not be sufficient to complement DJ-1 function in NT embryos. In contrast, the loss of DJ-1 in fertilized embryos might not be as harmful as in NT embryos because fertilized embryos have less cellular stress and/or stronger stress-coping functions than NT embryos as described above.
Thus far, many studies have focused on chromatin remodeling proteins and transcription factors as reprogramming-related factors in oocytes. DJ-1 acts as a positive regulator of cell survival by modulating signaling pathways rather than by directly modifying chromatin structures. Our results show that such a process is critical for nuclear reprogramming in embryos. This notion is further supported by a recent publication in which inhibition of HDAC6 that is involved in induction of apoptosis responding to cellular stress greatly improves cloning efficiency in mice (63). During reprogramming, original cell states are forced to change. This process must be accompanied by a lot of cellular stresses. DJ-1 may relieve those stresses and protect NT embryos. Exploring downstream networks of DJ-1 in embryos might provide further clues to the long-standing question of why cloned embryos are able to develop to term.
Materials and Methods
Oocyte Extract Treatment.
Porcine oocyte extracts were prepared as described previously (16). Briefly, 1,000–1,200 GV oocytes or 800 in vitro matured MII oocytes were collected. After denudation and zona pellucida removal, oocytes were suspended in 4–5 μL of extraction buffer (50 mM KCl, 5 mM MgCl2, 5 mM EGTA, 2 mM 2-mercaptoethanol, 0.1 mM PMSF, protease inhibitor mixture, and 50 mM Hepes, pH 7.6) containing an energy-regenerating system (2 mM ATP, 20 mM phosphocreatine, 20 units/mL creatine kinase, and 2 mM GTP). Oocytes were disrupted by ultracentrifugation and the oocyte components were extracted. These extract solutions were kept on ice before use. Porcine fibroblast cells were treated with 30 μg/mL digitonin (Calbiochem) for permeabilization (64). Cell nuclei (2 × 105) were incubated in the oocyte extracts or fibroblast cell extracts for 1 h at 37 °C. After incubation, nuclei were suspended in sucrose buffer (75 mM KCl, 4 mM MgCl2, 250 mM sucrose, 0.5 mM spermidine trihydrochloride, 0.15 mM spermine tetrahydrochloride, 1 mM DTT, 10 mM β-glycerophosphate, protease-inhibitor mixture, and 10 mM Hepes, pH 7.8) and collected by centriguation. Nuclear pellets were dissolved in 150 μL of the lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, bromophenol blue, protease inhibitor, and phosphatase inhibitor) and these samples were used for 2D PAGE. For the purpose of protein identification, five different samples were pooled, which equals ∼4,000 oocytes.
Statistical Analysis.
The differences in developmental abilities of embryos were analyzed by Student's t test. P < 0.05 was considered statistically significant. SI Materials and Methods provides complete information on materials and methods, including primer sequences (Table S2).
Supplementary Material
Acknowledgments
This research was supported by a grant from the Japan Society for the Promotion of Science (Research Fellowship Program) (to K. Miyamoto) and a grant from the Ministry of Agriculture, Forestry, and Fisheries (Research Project for Utilizing Advanced Technologies in Agriculture, Forestry, and Fisheries) and Grant-in-Aid for Scientific Research (B) (to H. Imai).
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in the GenBank database (accession no. GSE18104).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013634108/-/DCSupplemental.
References
- 1.Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64–65. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- 2.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brambrink T, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sridharan R, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gurdon J, Murdoch A. Nuclear transfer and iPS may work best together. Cell Stem Cell. 2008;2:135–138. doi: 10.1016/j.stem.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Kim K, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kikyo N, Wade PA, Guschin D, Ge H, Wolffe AP. Active remodeling of somatic nuclei in egg cytoplasm by the nucleosomal ATPase ISWI. Science. 2000;289:2360–2362. doi: 10.1126/science.289.5488.2360. [DOI] [PubMed] [Google Scholar]
- 11.Gonda K, et al. Reversible disassembly of somatic nucleoli by the germ cell proteins FRGY2a and FRGY2b. Nat Cell Biol. 2003;5:205–210. doi: 10.1038/ncb939. [DOI] [PubMed] [Google Scholar]
- 12.Hansis C, Barreto G, Maltry N, Niehrs C. Nuclear reprogramming of human somatic cells by xenopus egg extract requires BRG1. Curr Biol. 2004;14:1475–1480. doi: 10.1016/j.cub.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 13.Tamada H, et al. Chromatin decondensation and nuclear reprogramming by nucleoplasmin. Mol Cell Biol. 2006;26:1259–1271. doi: 10.1128/MCB.26.4.1259-1271.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jullien J, Astrand C, Halley-Stott RP, Garrett N, Gurdon JB. Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation. Proc Natl Acad Sci USA. 2010;107:5483–5488. doi: 10.1073/pnas.1000599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyamoto K, et al. Reprogramming events of mammalian somatic cells induced by Xenopus laevis egg extracts. Mol Reprod Dev. 2007;74:1268–1277. doi: 10.1002/mrd.20691. [DOI] [PubMed] [Google Scholar]
- 16.Miyamoto K, et al. Cell-free extracts from mammalian oocytes partially induce nuclear reprogramming in somatic cells. Biol Reprod. 2009;80:935–943. doi: 10.1095/biolreprod.108.073676. [DOI] [PubMed] [Google Scholar]
- 17.Bui HT, et al. The cytoplasm of mouse germinal vesicle stage oocytes can enhance somatic cell nuclear reprogramming. Development. 2008;135:3935–3945. doi: 10.1242/dev.023747. [DOI] [PubMed] [Google Scholar]
- 18.Gurdon JB, Laskey RA, De Robertis EM, Partington GA. Reprogramming of transplanted nuclei in amphibia. Int Rev Cytol Suppl. 1979:161–178. doi: 10.1016/s0074-7696(08)60902-x. [DOI] [PubMed] [Google Scholar]
- 19.Gao S, et al. Rapid H1 linker histone transitions following fertilization or somatic cell nuclear transfer: Evidence for a uniform developmental program in mice. Dev Biol. 2004;266:62–75. doi: 10.1016/j.ydbio.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Teranishi T, et al. Rapid replacement of somatic linker histones with the oocyte-specific linker histone H1foo in nuclear transfer. Dev Biol. 2004;266:76–86. doi: 10.1016/j.ydbio.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Lemaitre JM, Danis E, Pasero P, Vassetzky Y, Méchali M. Mitotic remodeling of the replicon and chromosome structure. Cell. 2005;123:787–801. doi: 10.1016/j.cell.2005.08.045. [DOI] [PubMed] [Google Scholar]
- 22.Mitsumoto A, et al. Oxidized forms of peroxiredoxins and DJ-1 on two-dimensional gels increased in response to sublethal levels of paraquat. Free Radic Res. 2001;35:301–310. doi: 10.1080/10715760100300831. [DOI] [PubMed] [Google Scholar]
- 23.Kim YC, Kitaura H, Iguchi-Ariga SM, Ariga H. DJ-1, an oncogene and causative gene for familial Parkinson's disease, is essential for SV40 transformation in mouse fibroblasts through up-regulation of c-Myc. FEBS Lett. 2010;584:3891–3895. doi: 10.1016/j.febslet.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 24.Nagakubo D, et al. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem Biophys Res Commun. 1997;231:509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- 25.Miyamoto K, Hoshino Y, Minami N, Yamada M, Imai H. Effects of synchronization of donor cell cycle on embryonic development and DNA synthesis in porcine nuclear transfer embryos. J Reprod Dev. 2007;53:237–246. doi: 10.1262/jrd.18085. [DOI] [PubMed] [Google Scholar]
- 26.Moore DJ, Zhang L, Dawson TM, Dawson VL. A missense mutation (L166P) in DJ-1, linked to familial Parkinson's disease, confers reduced protein stability and impairs homo-oligomerization. J Neurochem. 2003;87:1558–1567. doi: 10.1111/j.1471-4159.2003.02265.x. [DOI] [PubMed] [Google Scholar]
- 27.Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP. DJ-1, a cancer- and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci USA. 2006;103:15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Brug MP, et al. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc Natl Acad Sci USA. 2008;105:10244–10249. doi: 10.1073/pnas.0708518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hod Y, Pentyala SN, Whyard TC, El-Maghrabi MR. Identification and characterization of a novel protein that regulates RNA-protein interaction. J Cell Biochem. 1999;72:435–444. [PubMed] [Google Scholar]
- 30.Kim RH, et al. DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell. 2005;7:263–273. doi: 10.1016/j.ccr.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Vasseur S, et al. DJ-1/PARK7 is an important mediator of hypoxia-induced cellular responses. Proc Natl Acad Sci USA. 2009;106:1111–1116. doi: 10.1073/pnas.0812745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishinaga H, et al. Expression profiles of genes in DJ-1-knockdown and L 166 P DJ-1 mutant cells. Neurosci Lett. 2005;390:54–59. doi: 10.1016/j.neulet.2005.07.053. [DOI] [PubMed] [Google Scholar]
- 33.Giaime E, et al. Loss of function of DJ-1 triggered by Parkinson's disease-associated mutation is due to proteolytic resistance to caspase-6. Cell Death Differ. 2010;17:158–169. doi: 10.1038/cdd.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bretaud S, Allen C, Ingham PW, Bandmann O. p53-dependent neuronal cell death in a DJ-1-deficient zebrafish model of Parkinson's disease. J Neurochem. 2007;100:1626–1635. doi: 10.1111/j.1471-4159.2006.04291.x. [DOI] [PubMed] [Google Scholar]
- 35.Chehab NH, Malikzay A, Stavridi ES, Halazonetis TD. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci USA. 1999;96:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komarov PG, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 37.Murphy PJ, et al. Pifithrin-alpha inhibits p53 signaling after interaction of the tumor suppressor protein with hsp90 and its nuclear translocation. J Biol Chem. 2004;279:30195–30201. doi: 10.1074/jbc.M403539200. [DOI] [PubMed] [Google Scholar]
- 38.Esteves TC, et al. Somatic cell nuclear reprogramming of mouse oocytes endures beyond reproductive decline. Aging Cell. 2011;10:80–95. doi: 10.1111/j.1474-9726.2010.00644.x. [DOI] [PubMed] [Google Scholar]
- 39.Fan J, et al. Sumoylation is critical for DJ-1 to repress p53 transcriptional activity. FEBS Lett. 2008;582:1151–1156. doi: 10.1016/j.febslet.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Fan J, et al. DJ-1 decreases Bax expression through repressing p53 transcriptional activity. J Biol Chem. 2008;283:4022–4030. doi: 10.1074/jbc.M707176200. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y, et al. Inactivation of Drosophila DJ-1 leads to impairments of oxidative stress response and phosphatidylinositol 3-kinase/Akt signaling. Proc Natl Acad Sci USA. 2005;102:13670–13675. doi: 10.1073/pnas.0504610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Junn E, et al. Interaction of DJ-1 with Daxx inhibits apoptosis signal-regulating kinase 1 activity and cell death. Proc Natl Acad Sci USA. 2005;102:9691–9696. doi: 10.1073/pnas.0409635102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shendelman S, Jonason A, Martinat C, Leete T, Abeliovich A. DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol. 2004;2:e362. doi: 10.1371/journal.pbio.0020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taira T, et al. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004;5:213–218. doi: 10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li HM, Niki T, Taira T, Iguchi-Ariga SM, Ariga H. Association of DJ-1 with chaperones and enhanced association and colocalization with mitochondrial Hsp70 by oxidative stress. Free Radic Res. 2005;39:1091–1099. doi: 10.1080/10715760500260348. [DOI] [PubMed] [Google Scholar]
- 46.Bonifati V, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 47.Wagenfeld A, Yeung CH, Strupat K, Cooper TG. Shedding of a rat epididymal sperm protein associated with infertility induced by ornidazole and alpha-chlorohydrin. Biol Reprod. 1998;58:1257–1265. doi: 10.1095/biolreprod58.5.1257. [DOI] [PubMed] [Google Scholar]
- 48.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 49.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 50.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 51.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 52.Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Utikal J, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marión RM, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O'Callaghan-Sunol C, Gabai VL, Sherman MY. Hsp27 modulates p53 signaling and suppresses cellular senescence. Cancer Res. 2007;67:11779–11788. doi: 10.1158/0008-5472.CAN-07-2441. [DOI] [PubMed] [Google Scholar]
- 58.Danes CG, et al. 14-3-3 zeta down-regulates p53 in mammary epithelial cells and confers luminal filling. Cancer Res. 2008;68:1760–1767. doi: 10.1158/0008-5472.CAN-07-3177. [DOI] [PubMed] [Google Scholar]
- 59.Regan PL, et al. Hsp90 inhibition increases p53 expression and destabilizes MYCN and MYC in neuroblastoma. Int J Oncol. 2011;38:105–112. [PMC free article] [PubMed] [Google Scholar]
- 60.Hao Y, et al. Apoptosis and in vitro development of preimplantation porcine embryos derived in vitro or by nuclear transfer. Biol Reprod. 2003;69:501–507. doi: 10.1095/biolreprod.103.016170. [DOI] [PubMed] [Google Scholar]
- 61.Boiani M, et al. Variable reprogramming of the pluripotent stem cell marker Oct4 in mouse clones: Distinct developmental potentials in different culture environments. Stem Cells. 2005;23:1089–1104. doi: 10.1634/stemcells.2004-0352. [DOI] [PubMed] [Google Scholar]
- 62.Rodriguez-Osorio N, et al. Transcriptional reprogramming of gene expression in bovine somatic cell chromatin transfer embryos. BMC Genomics. 2009;10:190. doi: 10.1186/1471-2164-10-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ono T, et al. Inhibition of class IIb histone deacetylase significantly improves cloning efficiency in mice. Biol Reprod. 2010;83:929–937. doi: 10.1095/biolreprod.110.085282. [DOI] [PubMed] [Google Scholar]
- 64.Miyamoto K, et al. Reversible membrane permeabilization of mammalian cells treated with digitonin and its use for inducing nuclear reprogramming by Xenopus egg extracts. Cloning Stem Cells. 2008;10:535–542. doi: 10.1089/clo.2008.0020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.