Abstract
Microorganisms and their hosts communicate with each other through an array of signals. The plant hormone auxin (indole-3-acetic acid; IAA) is central in many aspects of plant development. Cyclodipeptides and their derivative diketopiperazines (DKPs) constitute a large class of small molecules synthesized by microorganisms with diverse and noteworthy activities. Here, we present genetic, chemical, and plant-growth data showing that in Pseudomonas aeruginosa, the LasI quorum-sensing (QS) system controls the production of three DKPs—namely, cyclo(l-Pro-l-Val), cyclo(l-Pro-l-Phe), and cyclo(l-Pro-l-Tyr)—that are involved in plant growth promotion by this bacterium. Analysis of all three bacterial DKPs in Arabidopsis thaliana seedlings provided detailed information indicative of an auxin-like activity, based on their efficacy at modulating root architecture, activation of auxin-regulated gene expression, and response of auxin-signaling mutants tir1, tir1 afb2 afb3, arf7, arf19, and arf7arf19. The observation that QS-regulated bacterial production of DKPs modulates auxin signaling and plant growth promotion establishes an important function for DKPs mediating prokaryote/eukaryote transkingdom signaling.
Keywords: root development, lateral roots, root hairs, phytostimulation, plant–bacteria interactions
The communication between bacteria and their hosts through interkingdom signaling is a recent field of research. This field evolved from the initial observation that bacteria can communicate through hormone-like signals—a process known as quorum sensing (QS) (1). The field expanded with the realization that these bacterial signals can modulate mammalian (2) and plant (3) cell-signal transduction, and that host hormones can cross-signal with QS molecules to modulate bacterial gene expression (4, 5). A predominant type of small-molecule autoinducer, N-acyl-l-homoserine lactone (AHL), is used by Gram-negative bacteria (6, 7). AHLs are synthesized from S-adenosyl methionine (SAM) and particular fatty acid carrier proteins by AHL synthases (1). AHLs all share the core homoserine lactone moiety, but distinct fatty acid side chains are incorporated into the signal molecules by their respective AHL enzymes. Small-to-medium–chained AHLs cross membranes freely and bind in the cytoplasm to transcription factors, which upon ligand binding, regulate the transcription of QS-controlled genes (1, 4).
Both pathogenic and symbiotic plant-associated bacteria require QS to successfully interact with their hosts (8, 9). However, plants have evolved multiple mechanisms to interpret these QS signals. Small concentrations of AHLs caused substantial changes in gene expression in Medicago truncatula and Arabidopsis thaliana, affecting primary metabolism, plant-hormone responses, and root system architecture (3, 10, 11). Bacteria that inhabit the rhizosphere may also influence plant growth by producing phytohormones, such as auxins (12). Application of indole-3-acetic acid (IAA) or IAA-related metabolites stimulates lateral root (LR) and root hair formation, which may increase water and nutrient acquisition, leading to increased biomass production (12). Consistent with this, several Arabidopsis mutants with defective auxin transport, perception, or signaling, including aux1, axr2, tir1, and tir3/doc1/big have been identified that show reduced root hair and LR formation or decreased plant size (13).
Plant/bacteria communication can be achieved by means of different metabolites, some of which can mimic the activity of endogenous phytohormones. Cyclodipeptides and their derivative diketopiperazines (DKPs) constitute a class of small molecules synthesized by a wide range of microorganisms that exhibit diverse and useful biological activities. For example, cyclo(l-Phe-l-Pro) and cyclo(l-Phe-trans-4-OH-l-Pro) act as antifungal compounds (14), and epipolythiodioxopiperazines show antitumor, antibacterial, antiviral, and immunosuppressive properties (15, 16). These compounds are synthesized by a family of tRNA-dependent peptide bond-forming enzymes termed cyclodipeptide synthases (17). Although DKPs are noteworthy bioactive molecules, there is limited information concerning the regulation of DKP biosynthesis in bacteria and its role in plant signaling.
Results
QS-Modulated Plant Growth Promotion by Pseudomonas aeruginosa.
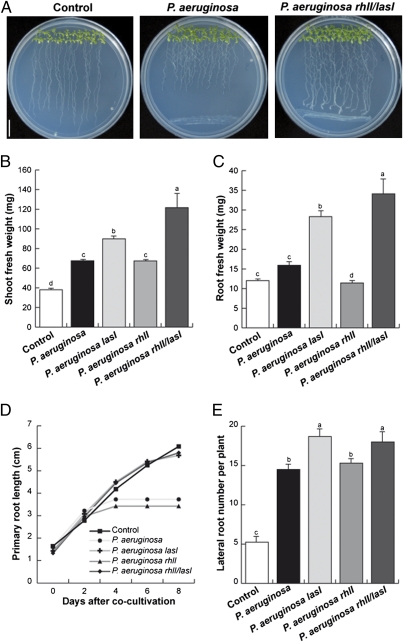
Pseudomonas aeruginosa has been used as a bacterial model to understand QS regulated by AHLs. P. aeruginosa has two mainly AHL-dependent QS systems—the las and rhl systems. In the las system, the LasI AHL synthase directs the synthesis of 3-oxo-C12-AHL, which interacts with the transcription factor LasR to target gene promoters. In the rhl system, the RhlI synthase directs the synthesis of C4-AHL, which interacts with the cognate regulator RhlR and controls transcription of target genes (6, 7, 18). We tested the in vivo effect of P. aeruginosa on plant growth by cocultivating 4-d-old Arabidopsis thaliana seedlings grown on agar plates containing 0.2× Murashige and Skoog (MS) medium with ∼2.8 × 108 cfu of P. aeruginosa PAO1 WT and the P. aeruginosa AHL synthase-deficient mutants lasI, rhlI, and rhlI/lasI double mutant by streaking the bacteria on the surface of the medium at a 5-cm distance from the primary root tip (Fig. 1A). After 8 d of growth in the presence of P. aeruginosa WT, a significant increase in shoot and root biomass production was observed (Fig. 1 B and C), which correlated with altered Arabidopsis root system architecture (RSA; Fig. 1 D and E). With comparable growth for all bacterial WT and mutant strains on the plant-bacteria interacting medium, the lasI single and rhlI/lasI double mutant exhibited lower primary root growth inhibition but greater formation of lateral roots and root hairs compared with the WT strain or the rhlI single P. aeruginosa mutant (Fig. 1 D and E and Fig. S1). Interestingly, altered RSA correlated with significantly increased shoot and root biomass production in plants cocultivated with P. aeruginosa lasI single mutants and with a nearly threefold-enhanced growth promotion by the rhlI/lasI double mutant (Fig. 1 B and C). This plant growth-promoting (PGP) effect could also be observed in plants cocultivated with bacteria at very close (1 cm) distance from the root tip, in which P. aeruginosa lasI and rhlI/lasI strains could directly contact the root system, and increased by fivefold shoot and root fresh weight (Fig. S2). These findings suggest that AHL signals produced by the AHL synthases LasI and RhlI modulate the production of compounds directly involved in biomass production, and cell division and differentiation processes in the root.
Fig. 1.
Effect of cocultivation with P. aeruginosa WT and QS mutant strains on root development and plant growth promotion. Four-day-old A. thaliana seedlings were cocultivated with WT P. aeruginosa or mutants defective on the AHL synthases LasI, RhlI, or LasI/RhlI at a distance of 5 cm from the primary root tip, and grown for an additional 8-d period. (A) Representative photographs of axenically grown Arabidopsis seedlings or seedlings cocultivated with WT P. aeruginosa and P. aeruginosa rhlI/lasI double mutant. (Scale bar = 1 cm.) (B) Effect of bacterial cocultivation on shoot biomass production or (C) root biomass production. Data from B and C show the means ± SD from three groups of 30 seedlings. (D) Effect of bacterial cocultivation on Arabidopsis primary root growth. Day 0 indicates the length reached by the primary root at the moment of bacterial application. Mean ± SD values were plotted at the indicated days in the kinetic experiment (n = 30). (E) Effect of bacterial cocultivation on lateral root formation. Date points represent mean ± SD (n = 30). These analyses were repeated three times with similar results. Different letters indicate means statistically different at P < 0.05.
P. aeruginosa Produces DKPs Capable of Stimulating LR Development in Arabidopsis.
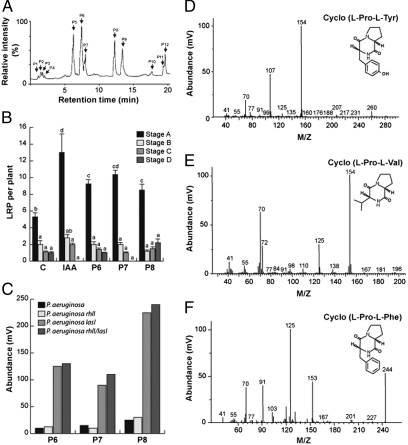
Diverse bacterial species possess the ability to produce the auxin phytohormone IAA (12). To search for IAA or IAA-related substances, EtOAc extracts of WT P. aeruginosa cell-free culture supernatants were assayed for their ability to stimulate LR initiation by counting lateral root primordia (LRP; Fig. 2 A and B). Three active fractions were identified (P6, P7, and P8) with peak retention times of 7, 7.5, and 12 min, respectively. The supply of purified fractions strongly increased stage A LRP production (Fig. 2B). The corresponding active peaks dramatically accumulated in P. aeruginosa lasI and rhlI/lasI mutant extracts (Fig. 2C). The molecular identity of purified peaks 6, 7, and 8 was resolved by GC/mass spectrometry and further confirmed by 1H NMR and 13C NMR spectra analysis as diketopiperazines (DKPs) cyclo(l-Pro-l-Tyr), cyclo(l-Pro-l-Val), and cyclo(l-Pro-l-Phe), respectively (Fig. 2 D–F and Fig. S3). 13C NMR (100 MHz) spectra analysis and δ-values data of each purified compound show that the carbon number is in agreement with DKP molecular structures obtained for MS and 1H NMR analysis (Fig. S3). Our finding that in the P. aeruginosa lasI and rhlI/lasI mutants all three DKPs increase in concentration suggests that DKP biosynthesis is regulated by the LasI/LasR QS system.
Fig. 2.
Identification and characterization of DKPs produced by P. aeruginosa. (A) Representative HPLC semipreparative chromatogram from culture supernatants of WT P. aeruginosa. Arrows indicate the collected peaks that were tested for activity on lateral root development in A. thaliana. (B) Arabidopsis Col-0 seedlings were germinated and grown for 6 d on the surface of agar plates containing 0.2× MS medium and transferred into 24-well cell culture plates (10 seedlings per well) and grown in 0.2× MS liquid medium containing 3 μM IAA or 30 μM of each DKP for 12 h, then cleared and LRP recorded according to Zhang et al. (39) for 10 independent roots. Data points represent mean ± SD. (C) Relative abundance of P6, P7, and P8 in ethyl acetate extracts from 1-L cultures of WT P. aeruginosa or mutants defective on the AHL synthases LasI, RhlI, or LasI/RhlI subjected to GC/mass spectrometry. Data points represent the mean relative abundance (×107 mV). (D–F) Mass spectra of P6, P7, and P8 fractions purified by HPLC and analyzed under GC/mass spectrometry. (D) P6, cyclo(l-Pro-l-Tyr; m/z = 260), (E) P7, cyclo(l-Pro-l-Val; m/z = 196), and (F) P8, cyclo(l-Pro-l-Phe; m/z = 244).
Chemical Complementation of P. aeruginosa lasI and rhlI/lasI Mutants.
Application of commercially available C4-AHL and 3-oxo-C12-AHL compounds to the growth medium of WT and P. aeruginosa mutant strains cocultivated with A. thaliana seedlings showed that only 3-oxo-C12-AHL normalized primary root growth inhibition and root hair development by lasI or rhlI/lasI strains, as observed in seedlings cocultivated with WT P. aeruginosa or rhlI single mutant (Fig. S4). Chemical complementation of single lasI or double rhlI/lasI mutants by 3-oxo-C12-AHL revealed that regulation of plant growth and development by P. aeruginosa is likely controlled by the LasI QS system.
Bacterial DKPs Modulate Auxin Responses in Arabidopsis.
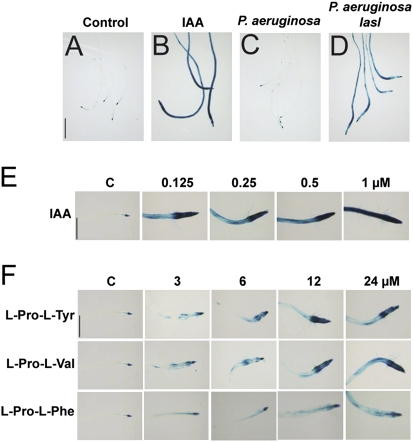
Lateral root growth and root hair formation are tightly regulated by auxin (13). The peculiar heterocyclic system of DKPs can be found in IAA and other compounds with auxin activity (19). The finding that cocultivation of Arabidopsis seedlings with lasI and rhlI/lasI mutants also leads to plants with enhanced lateral root and root hair formation prompted us to evaluate whether DKPs could act as auxin signal mimics. To determine if the enhanced production of DKPs by P. aeruginosa lasI mutants could affect auxin signaling in plants, Arabidopsis transgenic seedlings expressing the auxin-inducible DR5:uidA marker (20) were cocultivated with WT P. aeruginosa or lasI mutant. In aseptically grown seedlings, DR5:uidA is expressed primarily in the root tip region (Fig. 3A). DR5:uidA seedlings supplied with 3 μM IAA showed a strong GUS activity throughout the primary root (Fig. 3B), indicating activated auxin responses. The pattern of GUS expression in DR5:uidA seedlings cocultivated with WT P. aeruginosa remained similar to that observed in axenically grown plants (Fig. 3C). In contrast, in plants cocultivated with P. aeruginosa lasI mutant, there was a very clear increase in expression of this marker in the entire primary root (Fig. 3), indicating that the LasI QS system regulates the biosynthesis of compound(s) with auxin activity. Next, we tested the activity of all three DKPs on DR5:uidA expression in root tips by transferring 6-d-old seedlings grown on 0.2× MS solidified medium to 0.2× MS liquid medium supplied with IAA or DKPs, respectively. Fig. 3 E and F shows histochemical staining for transgenic DR5:uidA seedlings that were treated with IAA or the different DKPs. A dose-dependent GUS expression in plants treated with cyclo(l-Pro-l-Tyr) and cyclo(l-Pro-l-Val) was clearly observed, whereas cyclo(l-Pro-l-Phe) showed less activity (Fig. 3F). In a similar assay, a second auxin response marker—namely, BA3:uidA (21)—was activated by IAA and all three DKPs (Fig. S5). These results show that bacterial DKPs can activate auxin-inducible gene expression in Arabidopsis seedlings.
Fig. 3.
Effect of bacterial DKPs on auxin responses in A. thaliana. (A–F) Twelve hours of β-glucuronidase (GUS) staining of DR5:uidA primary roots supplied with the solvent (A), with 3 μM IAA (B), cocultivated with WT P. aeruginosa for 8 d (C) or with P. aeruginosa lasI mutant for 8 d (D). (E) Effect of IAA or (F) purified DKPs on DR5:uidA gene expression in transgenic seedlings grown on 0.2× MS agar medium for 6 d and then transferred into 24-well cell culture plates (10 seedlings per well) containing 2 mL 0.2× MS liquid medium supplied with the indicated concentrations of compounds and incubated for 10 h. Seedlings were stained for GUS activity and cleared for microscopy analysis. Photographs show representative individuals from at least 30 stained plants. (Scale bars = 500 μm.)
The biological activity of IAA and cyclo(l-Pro-l-Tyr) was also tested in relation to primary root growth. IAA inhibited primary root growth in nanomolar concentrations, whereas much greater concentrations of cyclo(l-Pro-l-Tyr) were required for growth-repressing effects (Fig. S5), indicating weak auxin activity for this DKP.
DKPs Enhance Aux/IAA Protein Degradation and Require a Canonical Auxin Signaling Pathway for Activity.
Auxin is perceived by direct binding to the transport inhibitor response 1 (TIR1) protein, a member of a small family of F-box proteins (22, 23). This interaction accelerates the Skp1, Cdc53/Cullin1, F-box protein ubiquitin ligase-catalyzed degradation of Aux/IAA repressor proteins, allowing derepression of auxin-regulated genes by auxin response transcription factors (ARFs) (24). We next compared the effect of IAA and DKPs on auxin-mediated degradation of Aux/IAA proteins using the Arabidopsis HS::AXR3NT-GUS line (24). Seedlings expressing the HS::AXR3NT-GUS construct were heat shocked at 37 °C for 2 h and further treated with 3 μM IAA, or 30 μM of cyclo(l-Pro-l-Tyr), cyclo(l-Pro-l-Val), and cyclo(l-Pro-l-Phe) for 60 min. Treatment with DKPs showed enhanced degradation of the fusion protein in a similar way to IAA, but greater concentrations of the compounds were required to achieve the same effect on HS::AXR3NT-GUS degradation (Fig. S5). Our data indicate that DKPs likely act in the auxin-mediated signaling pathway, possibly by direct binding to an auxin receptor, which rapidly destabilizes the AXR3 protein.
We performed a computational molecular docking analysis of DKP affinity to the Arabidopsis TIR1 receptor using the published crystallized TIR1 structure with the Aux/IAA7 peptide. This analysis revealed only one conformation cluster for all three DKPs with the same orientation into TIR1, which mimics the binding of IAA or 2,4-D (Fig. S6), suggesting that DKPs can fit in the TIR1 binding pocket.
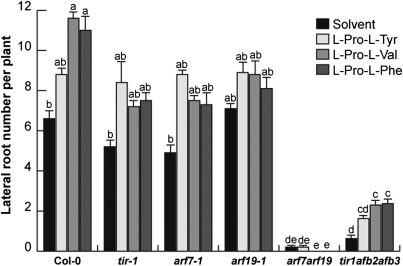
To determine whether the TIR1 family of auxin receptors and ARFs are involved in Arabidopsis responses to DKPs, we analyzed LR formation in response to cyclo(l-Pro-l-Tyr), cyclo(l-Pro-l-Val), and cyclo(l-Pro-l-Phe) in WT Col-0 Arabidopsis seedlings and in tir1-1 and tir1afb2afb3, single and triple mutants, respectively, and in arf7-1, arf19-1, and arf7arf19 mutants. In solvent-treated WT seedlings, cyclo(l-Pro-l-Val) and cyclo(l-Pro-l-Phe) increased total LR number per seedling (Fig. 4). tir1-1 mutants had a roughly 25% reduction in LR number, observed in WT seedlings in solvent media. Interestingly, the increase in LR formation observed in WT seedlings when treated with DKPs was clearly reduced in tir1-1 mutants (Fig. 4). When a triple tir1afb2afb3 mutant was analyzed, it was found that LR formation was not stimulated by DKP treatment (Fig. 4). The single arf7-1 mutant displayed a significant reduction in LR number compared with WT seedlings in solvent media; in addition, the stimulation of LR formation by DKPs was reduced in both arf7-1 and arf19-1 single mutants. The arf7arf19 double mutant was completely insensitive to DKPs in terms of increased lateral root formation (Fig. 4). A kinetic experiment monitoring primary root growth and lateral root formation revealed that WT P. aeruginosa similarly inhibited primary root growth in WT and all five auxin-related mutant lines tested, whereas arf7arf19 and tir1afb2afb3 lines were resistant to lateral root induction both by P. aeruginosa WT or lasI mutants (Fig. S7).
Fig. 4.
A. thaliana WT and tir1-1, arf7-1, arf19-1, arf7arf19, and tir1afb2afb3 mutant lines were germinated and grown for 6 d on 0.2× MS agar medium and transferred into 24-well cell culture plates (10 seedlings per well) and grown in 2 mL 0.2× MS liquid medium supplemented with 30 μM of each DKP for two additional days. Data points show the mean lateral root number per plant ± SD. Different letters indicate means that differ statistically at P < 0.05. The experiment was repeated twice with similar results.
Discussion
LasI QS-Controlled DKP Production Enhances Plant Growth Promotion Capability of P. aeruginosa
In this work, we explored genetically whether AHL QS is involved in growth and development of Arabidopsis evaluating the effects of cocultivation with P. aeruginosa lasI, rhlI and rhlI/lasI single and double mutants on plant biomass production and root architectural changes. We found that both shoot and root biomass production increased in Arabidopsis seedlings cocultivated with WT P. aeruginosa. Interestingly, growth promotion capability was further potentiated in lasI and rhlI/lasI mutants (Fig. 1 and Fig. S2), which correlated with decreased primary root growth inhibitory effect with the mutants compared with WT P. aeruginosa (Fig. 1). This primary root growth normalizing effect is apparently independent of the second bacterial AHL QS system RhlI, which produces C4-AHL and can be reverted by the inclusion of 3-oxo-C12-AHL in the agar medium (Fig. S4). It was noticeable that lasI and rhlI cocultivation strongly promoted LR formation and root hair development in Arabidopsis WT seedlings in a way that suggests that the effects of the bacteria are mediated by auxin (Fig. 1 and Figs. S1 and S2).
Our data are apparently contradictory to previous reports showing that P. aeruginosa is pathogenic to Arabidopsis (25–27). However, in their initial screen, Rahme et al. (25) evaluated a collection of 75 P. aeruginosa strains (30 human, 20 soil, and 25 plant isolates) for their ability to cause disease on leaves of four different A. thaliana ecotypes. Most strains elicited no symptoms, and only two strains, UCBPP-PA14, a human isolate, and UCBPP-PA29, a plant isolate, caused severe soft-rot symptoms in leaves of some, but not all, of the ecotypes tested (25). It is also important to note that P. aeruginosa pathogenicity tests have focused mainly on leaves, infiltrating thousands of bacteria into plant tissues, which are able to secrete a variety of potent degradative enzymes and virulence factors (25–27). In contrast, accumulating information shows the potential of P. aeruginosa as a phytostimulant (28). It is tempting to speculate that the issue of whether P. aeruginosa is a pathogen or a plant growth promoting bacterium or both would depend upon the specific assays and even with the initial concentrations of the inoculums.
Our detailed study is unique in that it characterizes the responses of Arabidopsis roots to P. aeruginosa. Our data shows that cocultivating P. aeruginosa in the vicinity of the primary root did not cause cell death or cell damage (Fig. S8), as revealed by CycB1:uidA (29) and AtPRZ:uidA (30) marker gene expression in the primary root meristem and expression of the cell nuclei marker AtHistH2B:YFP (31) by confocal laser scanning microscopy in seedlings stained with propidium iodide (PI). This finding suggests that root architecture remodeling under these conditions is unlikely due to a toxic effect, but instead by induction of cell differentiation processes at the root meristem region. Secretion of 3-oxo-C12-AHL by WT P. aeruginosa likely contributes to primary root inhibition by decreasing proliferative cell activity in the meristem, as application of the purified compound arrested cell division in the root meristem (Fig. S9). In consonance with these results, no symptoms of chlorosis or necrosis were detected in leaves of plants cocultivated with WT P. aeruginosa or QS-related mutants (Fig. S2).
In our experiments, we were unable to detect IAA from bacterial extracts, but instead found that three DKPs are produced by WT P. aeruginosa and are negatively regulated by the LasI AHL QS-controlled pathway. Each compound was purified to homogeneity by semipreparative HPLC, and its structure confirmed by MS and NMR spectroscopy as cyclo(l-Pro-l-Tyr), cyclo(l-Pro-l-Val), and cyclo(l-Pro-l-Phe; Fig. 2 and Fig. S3). Certain DKPs from P. aeruginosa and other Gram-negative bacteria, including cyclo(l-Ala-l-Val), cyclo(l-Pro-l-Tyr), and cyclo(l-Pro-l-Phe), have been described as QS factors affecting bioluminescence and swarming motility (32). Whether these molecules modulate AHL-mediated QS in the producer organism affecting pathogenic or symbiotic relationships with plants, or in other organisms occupying a similar ecological niche, remains to be established.
Bacterial DKPs Show Auxin-Like Activity in Arabidopsis.
The structure/activity relationship of auxin has been extensively investigated. Among more than 200 auxinic compounds identified, only two common features can be recognized as critical for auxin activity: a planar aromatic ring structure and a carboxyl group-containing side chain (19). The ring structure and its attached atoms on known auxinic compounds can vary significantly, suggesting a large degree of promiscuity. However, the two common features alone do not necessarily give rise to an auxin-like molecule. The DKPs identified in this work possess a heterocyclic system also found in IAA and other compounds with auxin activity. The effects of IAA, cyclo(l-Pro-l-Val), cyclo(l-Pro-l-Tyr), and cyclo(l-Pro-l-Phe) on auxin-regulated gene expression suggests that all three DKPs show a weak auxin activity. Cocultivation of transgenic Arabidopsis seedlings expressing the auxin-inducible reporter constructs DR5:uidA or BA3:uidA with P. aeruginosa lasI mutant, or treatment with DKPs, clearly activated GUS expression in the root system. However, greater concentrations than IAA were required for DKPs to activate auxin-inducible gene expression (Fig. 3 and Fig. S5). Four additional lines of evidence indicate that DKPs may act as auxin signal mimics: (i) the effect of cyclo(l-Pro-l-Tyr) inhibiting primary root growth (Fig. S5); (ii) the effects of cyclo(l-Pro-l-Val), cyclo(l-Pro-l-Tyr), and cyclo(l-Pro-l-Phe) on Aux/IAA stability using the Arabidopsis HS::AXR3NT-GUS line (Fig. S5); (iii) the promotion of LR formation by DKP application to Arabidopsis WT seedlings; and (iv) the finding that DKPs no longer stimulate lateral root formation in auxin receptor mutants tir1afb2afb3 and in arf7arf19 (Fig. 4 and Fig. S7). Molecular docking analysis further predicted that DKPs might interact with the TIR1 auxin receptor (Fig. S6). These data suggest that the planar structure of DKPs is likely responsible of their activity as auxin signal mimics. These compounds might bind to the promiscuous auxin binding pocket of TIR1 with different affinities, as shown for synthetic auxins with varied structure such as naphthalene acetic acid (1-NAA), 2,4-dichlorophenoxyacetic acid (2,4-D), and 4-amino-3,5,6-trichoropicolinic acid (picloram) (19, 33). The interaction between the F-box–containing protein (SCF) and the Aux/IAA was previously demonstrated in a pull-down assay in which TIR1-myc was recovered from plant extracts using recombinant Aux/IAA proteins in the absence and presence of auxin (34). These experiments showed that the interaction between TIR1 and the Aux/IAA proteins is dramatically enhanced by auxin. Whether DKPs elicit auxin responses by direct binding to TIR1 or AFB auxin receptors remains to be determined.
Role of DKPs in Plant/Bacteria Interactions.
Plant hormones control plant growth by affecting the spatial and temporal expression of genes involved in cell division, elongation, and differentiation. The potential of single bacterial strains to interfere with plant hormone levels remains one of the major challenges toward better understanding, predicting, and possibly controlling plant hormone responses in complex plant-associated bacterial communities. We propose a model to explain the effects of P. aeruginosa in Arabidopsis (Fig. S10). WT P. aeruginosa alters RSA by at least two mechanisms: one that likely involves 3-oxo-C12-AHL and is independent of auxin signaling leads to primary root growth inhibition, and one where it promotes root branching interacting with auxin as it requires normal functioning of auxin receptors TIR1, AFB2, and AFB3 and transcriptional regulators ARF7 and ARF19 acting downstream (Fig. S7). In our model, the primary root growth inhibition is no longer observed in plants cocultivated with lasI or rhlI/lasI, but instead an increase in root hair and lateral root formation occurs that correlates with plant growth promotion and production of DKPs. Root branching induced by lasI or rhlI/lasI mutants indeed required normal auxin signaling and could be a particular response to all three DKPs secreted by the mutants (Figs. S7 and S10). Our results showing the involvement of DKPs in RSA modulation add to the plethora of potential functions of these intriguing molecules. Based on their auxin-like activity, DKPs can be regarded as broad-spectrum molecules used to modulate the activity of both prokaryotic and eukaryotic cells, and thus represent a novel class of signals enabling interkingdom communication. An interesting study by Degrassi et al. (35) showed that plant growth-promoting Pseudomonas putida WCS358 produces and secretes four DKPs, which as shown in the present work may be involved in plant growth promotion. Manipulating AHL-dependent QS signaling and DKP biosynthesis may be a promising strategy for development of bacterial inoculants to enhance crop yields by means of auxin signaling and root architecture modulation.
Materials and Methods
Plant Material and Growth Conditions.
Arabidopsis thaliana (Col-0); the transgenic lines DR5::uidA (20), BA3::uidA (21), HS::AXR3NT-GUS (22), CycB1:uidA (29), AtPRZ:uidA (30), and AtHistH2B:YFP (31); and the mutant lines tir1-1, tir1afb2afb3 (36), arf7-1, arf19-1, and arf7arf19 (37) were used for all experiments. Seeds were surface sterilized with 95% (vol/vol) ethanol for 5 min and 20% (vol/vol) bleach for 7 min. After five washes with sterile distilled water, seeds were germinated and grown on agar plates containing 0.2× Murashige and Skoog medium (Murashige and Skoog basal salts mixture, M5524; Sigma). The suggested formulation is 4.3 g·L−1 of salts for 1× medium; we used 0.9 g·L−1, which we consider and refer to as 0.2× MS. This medium lacks amino acids and vitamins. Phytagar (micropropagation grade) was purchased from Phytotechnology. Plants were placed in a plant growth chamber (Percival Scientific AR-95L) with a photoperiod of 16 h of light, 8 h of darkness, a light intensity of 100 μmol·m2·s−1, and a temperature of 22 °C.
In Vitro Plant/Bacteria Cocultivation Assay.
Bacterial strains used in this work were P. aeruginosa PAO1 (WT), P. aeruginosa lasI, rhlI, and rhlI/lasI single and double mutants, respectively (38). The bacterial strains were evaluated in vitro for their plant growth-promotion ability, using the Arabidopsis Col-0 ecotype. Bacterial densities of 2.5 × 108 cfu were cocultivated by streaking on agar plates containing 0.2× MS medium. Six-day-old germinated Arabidopsis seedlings (20 seedlings per plate) were grown to one side of the plate, opposite to the bacterial streak site at a 5-cm distance from the root tip. The seedlings were grown for a further 8-d period by placing the plates in the growth chamber in a completely randomized design. All experiments were replicated at least three times.
Hormone Treatments.
For all experiments, 0.2× MS medium was supplemented with IAA or DKPs. Ethanol-dissolved compounds were added to cooled (50 °C) molten medium and poured into plates. Control plates were supplied with the greatest concentration of ethanol used in the AHL treatments. IAA was purchased from Sigma. DKPs were directly purified from WT P. aeruginosa and lasI mutant cultures.
Aux/IAA Protein Degradation Assay.
Six-day-old HS::AXR3NT-GUS Arabidopsis transgenic seedlings were incubated on liquid 0.2× MS medium for 2 h at 37 °C, followed by transfer of the seedlings into liquid 0.2× MS medium supplied with the different DKP or IAA compounds for 60 min at 22 °C. The seedlings were washed with fresh 0.2× MS medium and for 12–14 h histochemically stained for GUS activity.
Analysis of Growth, Statistics, and Histochemical and Microscopy Analysis.
The detailed analysis of growth, purification, and chemical characterization of cyclodipeptides, statistics, histochemical, and microscopy analysis are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Peter Doerner, Christian Luschnig, Athanasios Theologis, Tom Guilfoyle, and Mark A. Estelle for kindly providing seeds of transgenic and mutant lines. Dr. Barbara Iglewski is acknowledged for her kind donation of WT P. aeruginosa and mutant strains. This work was supported by Consejo Nacional de Ciencia y Tecnología Grants 80916 and 106567, and the Consejo de la Investigación Científica 2.26 and 2.14.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006740108/-/DCSupplemental.
References
- 1.Fuqua C, Winans SC, Greenberg EP. Census and consensus in bacterial ecosystems: The LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 2.Telford G, et al. The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-l-homoserine lactone has immunomodulatory activity. Infect Immun. 1998;66:36–42. doi: 10.1128/iai.66.1.36-42.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathesius U, et al. Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc Natl Acad Sci USA. 2003;100:1444–1449. doi: 10.1073/pnas.262672599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: The language of hormones. Proc Natl Acad Sci USA. 2003;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao MM, Teplitski M, Robinson JB, Bauer WD. Production of substances by Medicago truncatula that affect bacterial quorum sensing. Mol Plant Microbe Interact. 2003;16:827–834. doi: 10.1094/MPMI.2003.16.9.827. [DOI] [PubMed] [Google Scholar]
- 6.Pearson JP, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussain MB, et al. The acyl-homoserine lactone-type quorum-sensing system modulates cell motility and virulence of Erwinia chrysanthemi pv. zeae. J Bacteriol. 2008;190:1045–1053. doi: 10.1128/JB.01472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosemeyer V, Michiels J, Verreth C, Vanderleyden J. luxI- and luxR-homologous genes of Rhizobium etli CNPAF512 contribute to synthesis of autoinducer molecules and nodulation of Phaseolus vulgaris. J Bacteriol. 1998;180:815–821. doi: 10.1128/jb.180.4.815-821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Rad U, et al. Response of Arabidopsis thaliana to N-hexanoyl-DL-homoserine-lactone, a bacterial quorum sensing molecule produced in the rhizosphere. Planta. 2008;229:73–85. doi: 10.1007/s00425-008-0811-4. [DOI] [PubMed] [Google Scholar]
- 11.Ortíz-Castro R, Martínez-Trujillo M, López-Bucio J. N-acyl-L-homoserine lactones: A class of bacterial quorum-sensing signals alter post-embryonic root development in Arabidopsis thaliana. Plant Cell Environ. 2008;31:1497–1509. doi: 10.1111/j.1365-3040.2008.01863.x. [DOI] [PubMed] [Google Scholar]
- 12.Spaepen S, Vanderleyden J, Remans R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev. 2007;31:425–448. doi: 10.1111/j.1574-6976.2007.00072.x. [DOI] [PubMed] [Google Scholar]
- 13.Woodward AW, Bartel B. Auxin: Regulation, action, and interaction. Ann Bot (Lond) 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ström K, Sjögren J, Broberg A, Schnürer J. Lactobacillus plantarum MiLAB 393 produces the antifungal cyclic dipeptides cyclo(l-Phe-l-Pro) and cyclo(l-Phe-trans-4-OH-l-Pro) and 3-phenyllactic acid. Appl Environ Microbiol. 2002;68:4322–4327. doi: 10.1128/AEM.68.9.4322-4327.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanoh K, et al. Antitumor activity of phenylahistin in vitro and in vivo. Biosci Biotechnol Biochem. 1999;63:1130–1133. doi: 10.1271/bbb.63.1130. [DOI] [PubMed] [Google Scholar]
- 16.Williams DE, et al. Ambewelamides A and B, antineoplastic epidithiopiperazinediones isolated from the lichen Usnea sp. Tetrahedron Lett. 1998;39:9579–9582. [Google Scholar]
- 17.Gondry M, et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat Chem Biol. 2009;5:414–420. doi: 10.1038/nchembio.175. [DOI] [PubMed] [Google Scholar]
- 18.Pesci EC, Pearson JP, Seed PC, Iglewski BH. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calderón-Villalobos LI, Tan X, Zheng N, Estelle M. Auxin perception—structural insights. Cold Spring Harb Perspect Biol. 2010;2:a005546. doi: 10.1101/cshperspect.a005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oono Y, Chen QG, Overvoorde PJ, Köhler C, Theologis A. age Mutants of Arabidopsis exhibit altered auxin-regulated gene expression. Plant Cell. 1998;10:1649–1662. doi: 10.1105/tpc.10.10.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 23.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 24.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 25.Rahme LG, et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 26.Plotnikova JM, Rahme LG, Ausubel FM. Pathogenesis of the human opportunistic pathogen Pseudomonas aeruginosa PA14 in Arabidopsis. Plant Physiol. 2000;124:1766–1774. doi: 10.1104/pp.124.4.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker TS, et al. Pseudomonas aeruginosa-plant root interactions. Pathogenicity, biofilm formation, and root exudation. Plant Physiol. 2004;134:320–331. doi: 10.1104/pp.103.027888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preston GM. Plant perceptions of plant growth-promoting Pseudomonas. Philos Trans R Soc Lond B Biol Sci. 2004;359:907–918. doi: 10.1098/rstb.2003.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 1999;20:503–508. doi: 10.1046/j.1365-313x.1999.00620.x. [DOI] [PubMed] [Google Scholar]
- 30.Sieberer T, Hauser MT, Seifert GJ, Luschnig C. PROPORZ1, a putative Arabidopsis transcriptional adaptor protein, mediates auxin and cytokinin signals in the control of cell proliferation. Curr Biol. 2003;13:837–842. doi: 10.1016/s0960-9822(03)00327-0. [DOI] [PubMed] [Google Scholar]
- 31.Boisnard-Lorig C, et al. Dynamic analyses of the expression of the HISTONE:YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell. 2001;13:495–509. doi: 10.1105/tpc.13.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holden MTG, et al. Quorum-sensing cross talk: Isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol Microbiol. 1999;33:1254–1266. doi: 10.1046/j.1365-2958.1999.01577.x. [DOI] [PubMed] [Google Scholar]
- 33.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 34.Dharmasiri N, Dharmasiri S, Jones AM, Estelle M. Auxin action in a cell-free system. Curr Biol. 2003;13:1418–1422. doi: 10.1016/s0960-9822(03)00536-0. [DOI] [PubMed] [Google Scholar]
- 35.Degrassi G, et al. Plant growth-promoting Pseudomonas putida WCS358 produces and secretes four cyclic dipeptides: Cross-talk with quorum sensing bacterial sensors. Curr Microbiol. 2002;45:250–254. doi: 10.1007/s00284-002-3704-y. [DOI] [PubMed] [Google Scholar]
- 36.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 37.Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li LL, Malone JE, Iglewski BH. Regulation of the Pseudomonas aeruginosa quorum-sensing regulator VqsR. J Bacteriol. 2007;189:4367–4374. doi: 10.1128/JB.00007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Jennings A, Barlow PW, Forde BG. Dual pathways for regulation of root branching by nitrate. Proc Natl Acad Sci USA. 1999;96:6529–6534. doi: 10.1073/pnas.96.11.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.