Abstract
Large-scale genome-wide association studies (GWAS) have identified many loci associated with body mass index (BMI), but few studies focused on obesity as a binary trait. Here we report the results of a GWAS and candidate SNP genotyping study of obesity, including extremely obese cases and never overweight controls as well as families segregating extreme obesity and thinness. We first performed a GWAS on 520 cases (BMI>35 kg/m2) and 540 control subjects (BMI<25 kg/m2), on measures of obesity and obesity-related traits. We subsequently followed up obesity-associated signals by genotyping the top ∼500 SNPs from GWAS in the combined sample of cases, controls and family members totaling 2,256 individuals. For the binary trait of obesity, we found 16 genome-wide significant signals within the FTO gene (strongest signal at rs17817449, P = 2.5×10−12). We next examined obesity-related quantitative traits (such as total body weight, waist circumference and waist to hip ratio), and detected genome-wide significant signals between waist to hip ratio and NRXN3 (rs11624704, P = 2.67×10−9), previously associated with body weight and fat distribution. Our study demonstrated how a relatively small sample ascertained through extreme phenotypes can detect genuine associations in a GWAS.
Introduction
Obesity is the sixth most important risk factor contributing to the overall burden of disease worldwide [1]. Affected subjects have reduced life expectancy, and they suffer from several adverse consequences such as cardiovascular disease, type 2 diabetes and several cancers. Many studies have shown that body weight and obesity are strongly influenced by genetic factors, with heritability estimates in the range of 65–80% [2], [3]. Genetic variants in several genes are known to influence BMI, but these mutations are rare and often cause severe monogenic syndromes with obesity [4]. With the development of high-throughput genotyping techniques and the implementation of genome-wide association studies (GWAS), common variations, such as those in FTO [5] and MC4R [6], have been associated with obesity and body mass index (BMI). Recent large-scale meta-analysis of multiple GWAS identified additional genes harboring common SNPs that associate with BMI [7]–[10]. GWASs have also found associations with measures of body fat distribution [9], [11], [12]. By far the largest GWAS to date included almost 250 thousand individuals and 2.8 million SNPs [13]. Associations of BMI with 28 loci reached genome wide significance, including 10 that were reported previously and 18 that were newly identified. Four additional loci were associated with body fat distribution, all of which had been identified previously. However, even this major expansion of sample size has not explained much variation, 1.39% for BMI and 0.16% for body fat distribution. On the other hand, confirmation of existing BMI loci, and detailed analysis on their association with obesity as a binary trait and with other obesity-related quantitative traits, are important at the current stage to move GWAS signals forward and understand their functional consequences.
A few studies utilized samples with early-onset or morbid obesity for discovery, and replicated previously reported association signals on BMI [7], [14]–[16], or implicated specific genetic variants such as a recurrent 16p11.2 deletion [17]. Utilizing extreme phenotypes increases the odds ratio of association, with improved power to identify novel association signals under fixed genotyping budgets and fixed sample sizes. We have collected a large cohort of obese cases and families ascertained from tails of BMI distribution together with detailed phenotype measures on multiple obesity-related traits. In addition, we have adult controls who have never been overweight. However, given fixed genotyping budget, instead of genotyping all these samples by whole-genome SNP arrays, we elected to perform a case-control GWAS, and then follow up the top signals by candidate SNP genotyping on the entire set of samples including family members. Therefore, the unique dataset provides an opportunity to examine GWAS associations in a combined sample of cases, family members, and controls.
Results
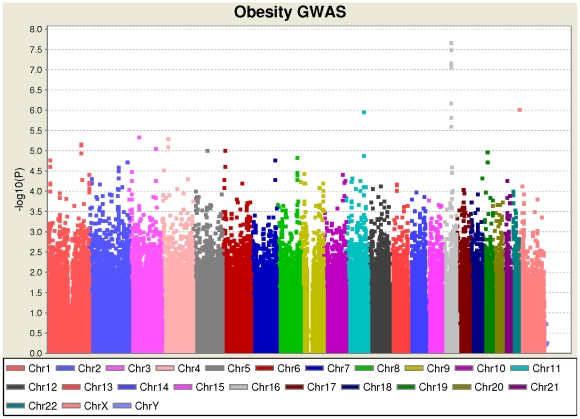
We analyzed genotype data for 520 cases and 540 control subjects, and performed a GWAS on obesity as a binary trait. We observed a strong association of obesity to the FTO gene, with the most significantly associated marker being rs3751812 (P = 2.01×10−8, odds ratio = 1.64). All association signals with P<10−5 are shown in Table 3 , and the Manhattan plot is shown in Figure 1 . No additional loci with genome-wide significance were identified in the GWAS; nevertheless, the fact that FTO readily reached genome-wide significance in a small data set confirmed the high quality of the phenotypes within the sample collection. It also illustrated how a small sample ascertained from extreme phenotypes have high power to detect genuinely associated genes, compared to quantitative trait association analysis conducted on population-based samples.
Table 3. Top association results (P<5×10−8) for GWAS on obesity.
| SNP | Chr | Position | Closest Gene | SNP-gene distance | MAF (case) | MAF (control) | P-value | Odds ratio | P-value (case+control+family) |
| rs3751812 | 16 | 52375961 | FTO | 0 | 0.4836 | 0.3632 | 2.01×10−8 | 1.642 | 4.2×10−12 |
| rs8050136 | 16 | 52373776 | FTO | 0 | 0.4846 | 0.3657 | 3.01×10−8 | 1.631 | 4.7×10−12 |
| rs9941349 | 16 | 52382989 | FTO | 0 | 0.499 | 0.3826 | 6.53×10−8 | 1.607 | 6.1×10−11 |
| rs9930333 | 16 | 52357478 | FTO | 0 | 0.5144 | 0.3983 | 7.90×10−8 | 1.6 | 2.0×10−11 |
| rs10852521 | 16 | 52362466 | FTO | 0 | 0.4088 | 0.5166 | 6.34×10−7 | 0.647 | 1.4×10−11 |
| rs10891096 | 11 | 109767953 | FDX1 | 38 kb | 0.0904 | 0.0383 | 1.03×10−6 | 2.494 | 0.0045 |
| rs7190492 | 16 | 52386253 | FTO | 0 | 0.3064 | 0.4067 | 1.46×10−6 | 0.644 | 7.8×10−9 |
| rs8044769 | 16 | 52396636 | FTO | 0 | 0.4117 | 0.5139 | 2.36×10−6 | 0.662 | 4.9×10−10 |
| rs9813516 | 3 | 60268044 | FHIT | 0 | 0.3004 | 0.2127 | 4.34×10−6 | 1.59 | N/A |
| rs7697609 | 4 | 40651331 | APBB2 | 0 | 0.2587 | 0.1766 | 4.71×10−6 | 1.627 | 0.11 |
| rs6857327 | 4 | 40649029 | APBB2 | 0 | 0.2658 | 0.1848 | 7.76×10−6 | 1.597 | 0.14 |
| rs3912607 | 3 | 163981452 | OTOL1 | 1.3 Mb | 0.3721 | 0.4677 | 8.34×10−6 | 0.675 | N/A |
| rs6921953 | 6 | 16127094 | MYLIP | 110 kb | 0.0212 | 0.0594 | 9.13×10−6 | 0.344 | 0.00054 |
| rs247916 | 5 | 87566009 | TMEM161B | 0 | 0.1965 | 0.2787 | 9.15×10−6 | 0.633 | 0.0022 |
Figure 1. Manhattan plot (logarithm of P-values versus chromosome coordinates) for SNP association on obesity.
The FTO locus on 16q12.2 reached genome-wide significance.
Considering the possibility that some obesity-associated genes may be enriched among the top ranked genes in GWAS, we next followed up selected association signals (∼500 top SNPs) by iSelect genotyping on 2,256 cases, family members and controls. Additionally, we also performed dense genotyping of 49 SNPs in the FTO gene itself, including 7 from the original top 500 and 42 spanning the entire gene. We used the MQLS software for the association analysis, to account for the familial relationships. Interestingly, the significance for FTO SNPs increased by several orders of magnitude (P-values range from 10−9 to 10−12), suggesting that genotyping additional family members increased power to detect genuine associations ( Table 4 ).
Table 4. Most significantly associated SNPs in the combined case/control and family cohort.
| SNP | Chr | Loc | Closest Gene | P(MQLS) |
| rs17817449 | 16 | 52370868 | FTO | 2.35×10−12 |
| rs3751812 | 16 | 52375961 | FTO | 4.22×10−12 |
| rs9935401 | 16 | 52374339 | FTO | 4.36×10−12 |
| rs8050136 | 16 | 52373776 | FTO | 4.71×10−12 |
| rs1121980 | 16 | 52366748 | FTO | 9.50×10−12 |
| rs10852521 | 16 | 52362466 | FTO | 1.38×10−11 |
| rs1861866 | 16 | 52361841 | FTO | 1.41×10−11 |
| rs9937053 | 16 | 52357008 | FTO | 1.56×10−11 |
| rs9930333 | 16 | 52357478 | FTO | 2.02×10−11 |
| rs9931494 | 16 | 52384680 | FTO | 4.24×10−11 |
| rs9941349 | 16 | 52382989 | FTO | 6.08×10−11 |
| rs8044769 | 16 | 52396636 | FTO | 4.86×10−10 |
| rs7206790 | 16 | 52355409 | FTO | 6.08×10−10 |
| rs1477196 | 16 | 52365759 | FTO | 9.33×10−9 |
| rs7190492 | 16 | 52386253 | FTO | 9.79×10−9 |
| rs3751813 | 16 | 52376209 | FTO | 1.63×10−8 |
| rs16867321 | 2 | 181070624 | UBE2E3 | 1.63×10−6 |
| rs2887180 | 2 | 181157550 | UBE2E3 | 2.44×10−6 |
| rs4784323 | 16 | 52355066 | FTO | 3.88×10−6 |
| rs925642 | 4 | 187915860 | FAT1/MTNR1A | 7.37×10−6 |
The associated tests were performed by MQLS.
We next completed exploratory analyses of quantitative measures of obesity. In part because of the extreme bimodality of the phenotype distributions based on sample ascertainment, we controlled for case/control status. Moreover, this approach makes it possible to assess the potential effects of genes on the extent of obesity in extremely obese individuals. The complete set of results (P<1×10−6) were given in Table 5 . We note that a few markers reached P<5×10−8, notably waist circumference (chromosome 21, rs11088859, p = 3.75×10−8, nearest gene NCAM2), and waist to hip ratio (chromosome 14, rs11624704, p = 2.67×10−9, nearest gene NRXN3), but the NCAM2 locus cannot be regarded as genome-wide significant considering the need to adjust for multiple phenotypes being tested. Therefore, these exploratory results were provided as potentially interesting findings worthy of additional replication efforts.
Table 5. List of significant associations (P<1×10−6) in quantitative trait analyses.
| phenotype | Chr | SNP | Pos | Gene | SNP-Gene distance | P (adjusting for obesity) |
| BMI | 8 | rs17126232 | 18021930 | ASAH1 | 35143 | 3.62×10−7 |
| BMI | 8 | rs17126237 | 18025369 | ASAH1 | 38582 | 5.57×10−7 |
| HIP | 7 | rs10953454 | 104291049 | LHFPL3 | 0 | 7.20×10−7 |
| HIP | 7 | rs10216243 | 104300860 | LHFPL3 | 0 | 8.07×10−7 |
| HIP | 8 | rs17126232 | 18021930 | ASAH1 | 35143 | 1.50×10−7 |
| HIP | 8 | rs17126237 | 18025369 | ASAH1 | 38582 | 3.45×10−7 |
| HIP | 16 | rs9923451 | 77509940 | WWOX | 0 | 7.95×10−7 |
| HIP | 22 | rs5762430 | 26708472 | PITPNB | 63217 | 7.32×10−7 |
| Waist | 21 | rs11088859 | 21611215 | NCAM2 | 0 | 3.75×10−8 |
| Weight | 8 | rs17126232 | 18021930 | ASAH1 | 35143 | 8.28×10−8 |
| Weight | 8 | rs17126237 | 18025369 | ASAH1 | 38582 | 8.74×10−8 |
| Weight | 13 | rs17081231 | 65865623 | PCDH9 | 0 | 7.18×10−7 |
| WHR | 2 | rs7581710 | 120911651 | INHBB | 85798 | 2.12×10−7 |
| WHR | 6 | rs2807278 | 131851613 | ARG1 | 84445 | 3.18×10−7 |
| WHR | 14 | rs11624704 | 77855830 | NRXN3 | 84016 | 2.67×10−9 |
Table S1 summarizes our results with respect to previously reported associations with obesity related traits. As noted, only FTO and NRXN3 reached genome wide significance. Three genes, including SH2B1, MC4R and KCTD15, showed trends towards significance in the case/control association tests (P-value ranges from 0.015 to 0.065), but they did not pass multiple testing thresholds (based on number of genes tested). For quantitative traits, we cannot estimate the power of our study, since previously published studies utilized population samples for quantitative trait association with BMI [18]. Nevertheless, in our data, it is interesting to see that MC4R and FTO are the two genes with the strongest effect sizes (risk allele odds ratio >1.2), probably explaining why they were the first two genes identified in GWAS for BMI [5], [6].
Discussion
In the current study, we performed a GWAS on obesity and obesity-related traits. The FTO gene reached genome-wide significance in this cohort with an odds ratio of 1.6. The MC4R gene is the second gene found by GWAS to be associated with BMI [6], and, while only marginally associated with obesity in our study, its odds ratio was 1.3. Given our modest sample size in GWAS, we estimate that the power to detect association (with perfect SNP tagging) at P<5×10−8 is 78.8% and 0.18% for FTO and MC4R, respectively. These odds ratio estimates in our data are higher than previous reports, for example, odds ratio for FTO is 1.3 in a study for early-onset obesity [5], for FTO and MC4R are 1.46 and 1.02 in a study for extreme obesity [14], or 1.25 and 1.26 in a study on morbidly obese adults with familial obesity [15], or 1.27 and 1.12 for obesity [10]. We note that the Hinney et al report investigated early onset extreme obesity and reported an odds ratio of 1.67 for FTO [16], comparable to our study. Therefore, the increased effect size could be due to the specific sample ascertainment scheme that we have used, that is, we sampled from the extreme tails of a quantitative trait distribution based on BMI. Even with the augmented sample of cases, family members and controls, no other SNPs reached genome wide significance. The results therefore strongly suggest that FTO and MC4R might be the only two major-effect genes for obesity with common variants in populations of European ancestry. Our study also represents an example where enrichment of extreme cases and controls can lead to increased odds ratio, and subsequently leads to improved power to detect associations.
The association between waist to hip ratio and the NRXN3 gene is of interest, as this is the third time the gene has been associated with body fat distribution [12], [13]. Neurexins are expressed in nervous tissue and are thought to be involved in cell adhesion during synapse formation [19]. Besides fat distribution, NRXN3 has been associated with several other traits, including addictions and schizophrenia [20]–[22]. Identifying the specific causal variant may be difficult because NRXN3 is an extremely large gene (∼1.5 Mb) [19]. It is controlled by two promoters and has multiple transcripts. The SNPs associated with weight and fat distribution lie in different parts of the gene and will likely involve different transcripts with potentially different functions. The associated SNP in our study, rs11624704, appears to be about 85 kb upstream of the first exon, while those for the previous two studies, rs10150332 and rs10146997, appear to be about 8 kb apart near exon 11.
In conclusion, we have assayed a sample collection of obese cases, families and never-overweight controls, and performed association analysis on obesity and multiple quantitative phenotype measures. We obtained strong support for FTO as well as suggestive confirmation of several previously identified BMI-associated genes in obesity. Another outcome of our study is the identification of new candidate genes for obesity-related traits. Of particular interest is the association of NRXN3 with body fat distribution among extremely obese individuals.
Materials and Methods
Study participants
The current GWAS study includes 520 cases and 540 control subjects, who were non-Hispanic Caucasians. Cases were obese (BMI≥35 kg/m2) with a lifetime BMI>40 kg/m2. Among them, 32 were male while the rest were female subjects. Independent controls were selected who had a current and lifetime BMI≤25 kg/m2. The individuals in the samples were of approximately the same age but differed in average BMI by 29 kg/m2 ( Table 1 ). After performing the GWAS, a combined sample of cases, controls and family members (N = 2,256), including all the study participants in the GWAS, were included for genotyping the top ∼500 most significant SNPs based on genotyping budget. Subject characteristics of family members were shown in Table 2 . Note that this is a study originally designed for investigating obesity genes in female subjects, but over time we have included a small fraction of males during the recruitment. All subjects gave written informed consent, and the protocol was approved by the Committee on Studies Involving Human Beings at the University of Pennsylvania.
Table 1. Sample characteristics of 1,060 cases and controls in GWAS.
| cases | N | Minimum | Maximum | Mean | SD |
| 488 females | 32 males | ||||
| BMI1 | 520 | 35.57 | 96.95 | 49.39 | 8.78 |
| %FAT | 467 | 30.70 | 70.70 | 49.88 | 6.03 |
| WEIGHT | 512 | 77.05 | 273.64 | 138.13 | 26.91 |
| HEIGHT | 509 | 135.60 | 198.30 | 167.07 | 7.87 |
| WAIST | 509 | 74.40 | 224.79 | 122.11 | 15.32 |
| HIP | 509 | 67.00 | 276.00 | 148.49 | 18.16 |
| WHR | 508 | 0.61 | 1.46 | 0.83 | 0.09 |
| AGE | 520 | 18 | 64 | 41.06 | 9.35 |
| AGEONSET2 | 411 | .5 | 55.0 | 13.63 | 9.02 |
| controls | |||||
| 532 females | 8 males | ||||
| BMI* | 540 | 15.97 | 24.93 | 20.75 | 1.76 |
| %FAT | 530 | 6.90 | 40.00 | 23.70 | 5.45 |
| WEIGHT | 539 | 37.50 | 90.00 | 55.52 | 6.39 |
| HEIGHT | 539 | 133.90 | 194.00 | 163.49 | 6.53 |
| WAIST | 539 | 40.20 | 100.00 | 72.89 | 5.76 |
| HIP | 539 | 56.90 | 110.10 | 86.51 | 6.51 |
| WHR | 538 | 0.50 | 1.33 | 0.84 | 0.07 |
| AGE | 540 | 16 | 65 | 43.10 | 8.81 |
: Self reported maximum BMI were used in 11 cases and 1 control.
: AGEONSET refers to the age of onset for the obesity diagnosis.
Table 2. Sample characteristics of 2,256 samples in the candidate SNP genotyping.
| N | Minimum | Maximum | Mean | SD | |
| Cases | |||||
| 546 females, 37 males | |||||
| BMI | 583 | 35.08 | 96.95 | 49.41 | 8.81 |
| % FAT | 529 | 30.70 | 70.70 | 50.01 | 5.92 |
| WEIGHT | 574 | 77.05 | 273.64 | 138.19 | 26.62 |
| HEIGHT | 571 | 135.60 | 198.30 | 167.10 | 7.83 |
| WAIST | 571 | 74.40 | 224.79 | 122.46 | 15.17 |
| HIP | 570 | 67.00 | 276.00 | 148.59 | 17.80 |
| WHR | 569 | .61 | 1.46 | .83 | .09 |
| AGE | 583 | 18 | 64 | 41.06 | 9.42 |
| AGEONSET2 | 460 | 1 | 55 | 13.96 | 8.98 |
| Controls | |||||
| 537 females, 8 males | |||||
| BMI | 545 | 15.99 | 24.93 | 20.77 | 1.79 |
| %FAT | 535 | 7.50 | 40.00 | 23.66 | 5.61 |
| WEIGHT | 544 | 37.50 | 90.00 | 55.55 | 6.66 |
| HEIGHT | 544 | 133.90 | 194.00 | 163.43 | 6.82 |
| WAIST | 544 | 40.20 | 100.00 | 72.78 | 5.74 |
| HIP | 544 | 56.90 | 110.30 | 86.84 | 6.94 |
| WHR | 543 | .50 | 1.33 | .84 | .07 |
| AGE | 545 | 16 | 65 | 42.64 | 8.75 |
| Fathers 1 | |||||
| BMI | 400 | 14.68 | 71.89 | 28.66 | 7.27 |
| %FAT | 326 | 3.30 | 59.20 | 25.97 | 8.53 |
| WEIGHT | 378 | 45.46 | 252.27 | 89.77 | 23.93 |
| HEIGHT | 375 | 153.70 | 198.12 | 176.89 | 7.12 |
| WAIST | 359 | 79.00 | 169.00 | 102.14 | 12.86 |
| HIP | 357 | 58.00 | 167.40 | 106.00 | 14.59 |
| WHR | 356 | .72 | 1.66 | .97 | .07 |
| AGE | 400 | 42 | 95 | 66.74 | 9.85 |
| AGEONSET2 | 152 | 1.0 | 74.0 | 40.93 | 18.37 |
| Mothers 1 | |||||
| BMI | 411 | 15.94 | 69.02 | 32.77 | 10.37 |
| %FAT | 338 | 14.70 | 63.30 | 40.69 | 9.39 |
| WEIGHT | 392 | 40.45 | 185.23 | 87.76 | 28.53 |
| HEIGHT | 389 | 142.50 | 181.60 | 163.55 | 6.62 |
| WAIST | 380 | 61.80 | 159.00 | 96.97 | 19.91 |
| HIP | 379 | 60.40 | 174.90 | 116.21 | 21.42 |
| WHR | 379 | .67 | 1.54 | .83 | .08 |
| AGE | 411 | 41 | 90 | 63.47 | 9.77 |
| AGEONSET2 | 247 | .5 | 79.0 | 30.23 | 19.60 |
| Sisters | |||||
| BMI | 278 | 17.01 | 77.58 | 34.34 | 11.23 |
| %FAT | 233 | 15.60 | 59.90 | 39.94 | 11.10 |
| WEIGHT | 268 | 45.68 | 212.95 | 94.74 | 31.57 |
| HEIGHT | 266 | 135.60 | 185.40 | 166.02 | 6.93 |
| WAIST | 260 | 60.90 | 165.80 | 97.48 | 21.45 |
| HIP | 260 | 75.50 | 202.30 | 118.43 | 25.21 |
| WHR | 260 | .63 | 1.02 | .82 | .06 |
| AGE | 278 | 14 | 64 | 37.81 | 9.22 |
| AGEONSET2 | 172 | 1.0 | 48.0 | 18.83 | 9.62 |
| brothers | |||||
| BMI | 90 | 20.99 | 64.82 | 33.67 | 8.08 |
| %FAT | 79 | 4.10 | 49.90 | 29.78 | 9.54 |
| WEIGHT | 89 | 67.05 | 210.11 | 110.27 | 27.48 |
| HEIGHT | 88 | 164.60 | 197.60 | 180.48 | 6.59 |
| WAIST | 88 | 73.10 | 167.00 | 109.44 | 16.65 |
| HIP | 88 | 80.90 | 175.40 | 112.97 | 16.43 |
| WHR | 88 | .80 | 1.35 | .97 | .08 |
| AGE | 90 | 15 | 60 | 38.67 | 9.33 |
| AGEONSET2 | 46 | .5 | 45.0 | 22.76 | 10.82 |
Some obese parents (BMI>40 kg/m2) were selected as cases in Stage 1. All cases were independent and from different families.
AGEONSET refers to the age of onset for the obesity diagnosis.
Phenotype measures
Anthropomorphic phenotypes were directly measured in field settings. Percent fat was estimated using a bioelectric impedance (BIA) measure. The complete list of measures examined in this study is described in Table 1 and Figure S1. Body mass index was calculated from measured height and weight by the standard formula, Weight (kg) divided by Height (m2). Measurements were taken of subjects dressed in light clothing. Height was measured from a standing position using a stadiometer. Weight was measured by a scale with a maximum weight of 600 pounds (270 kg) (Tanita TBF310 Pro Body Composition Analyzer, Tanita, Arlington Heights, IL). Body composition was estimated by bioelectric impedance using the same Tanita scale. Waist circumference was measured while standing at the height of the iliac crest. Hip circumference was taken while standing at the maximum extension of the buttocks. Waist to hip ratio (WHR) was calculated by measured waist circumference divided by measured hip circumference. Age of Obesity Onset was the age at which the subject reported having first become overweight.
Genotyping
DNA was extracted from whole blood or lymphoblastoid cell lines using a high salt method. All cases and control subjects were genotyped on the Illumina HumanHap550 SNP arrays (Illumina, San Diego, CA) with ∼550,000 SNP markers, at the Center for Applied Genomics, Children's Hospital of Philadelphia. Standard data normalization procedures and canonical genotype clustering files were used to process the genotyping signals and generate genotype calls. In addition, the combined sample of cases, controls and family members (N = 2,256, Table 2 ) were genotyped for the top 500 SNPs from the GWAS using the Illumina ISelect platform. All cases, family members, and controls were non-Hispanic Caucasians, and we further utilized multi-dimensional scaling to confirm the ethnicity status of cases and control subjects. A subset of the whole-genome genotype data were previously described in a CNV study on obesity [23].
Association analysis
The PLINK software version 1.07 was used to conduct association tests between SNP genotypes and specific phenotypes of interest. For traits that are approximately normally distributed, we utilized standard linear regression for assessing association but including age, sex and disease status as covariates. We attempted to exclude samples with genotyping rate less than 95% but none of the samples met this criterion. SNPs were excluded in analysis if the minor allele frequency was less than 1% (23298 SNPs were excluded), or if the Hardy-Weinberg Equilibrium P-value was less than 1×10−6 in control subjects (1366 SNPs were excluded), or if the genotype missing rate is higher than 5% (8190 SNPs were excluded). The study participants are of European ancestry as evaluated in previous studies [24]; given whole-genome data, we also performed multi-dimensional scaling analysis on SNPs not in LD (r2<0.2) with each other and confirmed that all cases and control subjects were of genetically inferred European ancestry (Figure S2). The QQ plot for the obesity GWAS is given in Figure S3, and the genomic control inflation factor was 1.05.
The combined dataset of cases, family members and controls was next analyzed using MQLS. MQLS utilizes a quasi-likelihood score test approach developed by Thornton and McPeek [25] that treats the data as a case-control analysis consisting of related and unrelated individuals. This combined approach has substantially more power than separate analyses using either case-control or family based methods. However, we acknowledge that since the candidate SNP genotyping study is not independent of the GWAS, the P-value distributions will be biased and therefore our study cannot be regarded as a standard “2-stage” analysis. The MQLS (b) statistic incorporates parental data in the estimation of case genotypes. We restricted these analyses to obesity status, since the method currently is adapted only for dichotomous phenotypes.
Supporting Information
The distribution of phenotype measures utilized in the current study. The age of onset information is available for cases only. BMI, weight, BIA and waist have bi-modal distribution, so we explored testing on cases only.
(PDF)
Multi-dimensional scaling (MDS) of the SNP genotyping data for samples with whole-genome genotypes, with (left panel) or without (right panel) 30 Asian, 30 African American and 30 Caucasians to seed the graph. A total of 70,593 SNPs not in LD (r2<0.2) and not in sex chromosomes were used in the MDS analysis. All GWAS samples were of genetically inferred European ancestry.
(PDF)
The quantile-quantile (QQ) plot of the association results for the GWAS on obesity. The genomic control inflation factor was 1.05.
(PDF)
Examination of BMI-associated genes in our data set for association with quantitative traits in cases and controls, adjusting for obesity status.
(PDF)
Acknowledgments
We thank all the patients and control subjects who donated blood samples for genetic research purposes.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported in part by NIH grants R01DK44073, R01DK56210, and R01DK076023 to R.A.P. and a Scientist Development Grant (0630188N) from the American Heart Association to W.D.L.. Genome-wide genotyping was funded in part by an Institutional Development Award to the Center for Applied Genomics (H.H.) from the Children's Hospital of Philadelphia. No additional external funding was received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Haslam DW, James WP. Obesity. Lancet. 2005;366:1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 2.Malis C, Rasmussen EL, Poulsen P, Petersen I, Christensen K, et al. Total and regional fat distribution is strongly influenced by genetic factors in young and elderly twins. Obes Res. 2005;13:2139–2145. doi: 10.1038/oby.2005.265. [DOI] [PubMed] [Google Scholar]
- 3.Stunkard AJ, Foch TT, Hrubec Z. A twin study of human obesity. Jama. 1986;256:51–54. [PubMed] [Google Scholar]
- 4.Farooqi IS. Genetic aspects of severe childhood obesity. Pediatr Endocrinol Rev. 2006;3(Suppl 4):528–536. [PubMed] [Google Scholar]
- 5.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scherag A, Dina C, Hinney A, Vatin V, Scherag S, et al. Two New Loci for Body-Weight Regulation Identified in a Joint Analysis of Genome-Wide Association Studies for Early-Onset Extreme Obesity in French and German Study Groups. PLoS Genet. 2010;6:e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5:e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 11.Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heard-Costa NL, Zillikens MC, Monda KL, Johansson A, Harris TB, et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS Genet. 2009;5:e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotsapas C, Speliotes EK, Hatoum IJ, Greenawalt DM, Dobrin R, et al. Common body mass index-associated variants confer risk of extreme obesity. Hum Mol Genet. 2009;18:3502–3507. doi: 10.1093/hmg/ddp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyre D, Delplanque J, Chevre JC, Lecoeur C, Lobbens S, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 16.Hinney A, Nguyen TT, Scherag A, Friedel S, Bronner G, et al. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS ONE. 2007;2:e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walters RG, Jacquemont S, Valsesia A, de Smith AJ, Martinet D, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowen L, Young J, Birditt B, Kaur A, Madan A, et al. Analysis of the Human Neurexin Genes: Alternative Splicing and the Generation of Protein Diversity. Genomics. 2002;79:587–597. doi: 10.1006/geno.2002.6734. [DOI] [PubMed] [Google Scholar]
- 20.Hishimoto A, Liu QR, Drgon T, Pletnikova O, Walther D, et al. Neurexin 3 polymorphisms are associated with alcohol dependence and altered expression of specific isoforms. Hum Mol Genet. 2007;16:2880–2891. doi: 10.1093/hmg/ddm247. [DOI] [PubMed] [Google Scholar]
- 21.Lachman HM, Fann CSJ, Bartzis M, Evgrafov OV, Rosenthal RN, et al. Genomewide suggestive linkage of opioid dependence to chromosome 14q. Human Molecular Genetics. 2007;16:1327–1334. doi: 10.1093/hmg/ddm081. [DOI] [PubMed] [Google Scholar]
- 22.Novak G, Boukhadra J, Shaikh SA, Kennedy JL, Le Foll B. Association of a polymorphism in the NRXN3 gene with the degree of smoking in schizophrenia: a preliminary study. World J Biol Psychiatry. 2009;10:929–935. doi: 10.1080/15622970903079499. [DOI] [PubMed] [Google Scholar]
- 23.Wang K, Li WD, Glessner JT, Grant SF, Hakonarson H, et al. Large copy number variations are enriched in cases with moderate to extreme obesity. Diabetes. 2010;59:2690–2694. doi: 10.2337/db10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price RA, Li WD, Zhao H. FTO gene SNPs associated with extreme obesity in cases, controls and extremely discordant sister pairs. BMC Med Genet. 2008;9:4. doi: 10.1186/1471-2350-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thornton T, McPeek MS. Case-control association testing with related individuals: a more powerful quasi-likelihood score test. Am J Hum Genet. 2007;81:321–337. doi: 10.1086/519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The distribution of phenotype measures utilized in the current study. The age of onset information is available for cases only. BMI, weight, BIA and waist have bi-modal distribution, so we explored testing on cases only.
(PDF)
Multi-dimensional scaling (MDS) of the SNP genotyping data for samples with whole-genome genotypes, with (left panel) or without (right panel) 30 Asian, 30 African American and 30 Caucasians to seed the graph. A total of 70,593 SNPs not in LD (r2<0.2) and not in sex chromosomes were used in the MDS analysis. All GWAS samples were of genetically inferred European ancestry.
(PDF)
The quantile-quantile (QQ) plot of the association results for the GWAS on obesity. The genomic control inflation factor was 1.05.
(PDF)
Examination of BMI-associated genes in our data set for association with quantitative traits in cases and controls, adjusting for obesity status.
(PDF)