Abstract
Background
FFAR1 receptor is a long chain fatty acid G-protein coupled receptor which is expressed widely, but found in high density in the pancreas and central nervous system. It has been suggested that FFAR1 may play a role in insulin sensitivity, lipotoxicity and is associated with type 2 diabetes. Here we investigate the effect of three common SNPs of FFAR1 (rs2301151; rs16970264; rs1573611) on pancreatic function, BMI, body composition and plasma lipids.
Methodology/Principal Findings
For this enquiry we used the baseline RISCK data, which provides a cohort of overweight subjects at increased cardiometabolic risk with detailed phenotyping. The key findings were SNPs of the FFAR1 gene region were associated with differences in body composition and lipids, and the effects of the 3 SNPs combined were cumulative on BMI, body composition and total cholesterol. The effects on BMI and body fat were predominantly mediated by rs1573611 (1.06 kg/m2 higher (P = 0.009) BMI and 1.53% higher (P = 0.002) body fat per C allele). Differences in plasma lipids were also associated with the BMI-increasing allele of rs2301151 including higher total cholesterol (0.2 mmol/L per G allele, P = 0.01) and with the variant A allele of rs16970264 associated with lower total (0.3 mmol/L, P = 0.02) and LDL (0.2 mmol/L, P<0.05) cholesterol, but also with lower HDL-cholesterol (0.09 mmol/L, P<0.05) although the difference was not apparent when controlling for multiple testing. There were no statistically significant effects of the three SNPs on insulin sensitivity or beta cell function. However accumulated risk allele showed a lower beta cell function on increasing plasma fatty acids with a carbon chain greater than six.
Conclusions/Significance
Differences in body composition and lipids associated with common SNPs in the FFAR1 gene were apparently not mediated by changes in insulin sensitivity or beta-cell function.
Introduction
Body mass index is an established independent risk factor for the development of type 2 diabetes (T2D). Non-esterified free fatty acids (NEFA) have been implicated in the development of T2D through effects on insulin release and the development of the metabolic syndrome. The free fatty acid receptor FFAR1 (GPR40 – G-protein-coupled receptor 40) was the first gene product identified to act as an extracellular membrane receptor for FFAs [1]. It is located in the 19q13.1 chromosomal region, which has been linked to T2D and T2D-related phenotypes, in several genome-wide scans [2], [3] and is expressed widely in the pancreas, central nervous system (CNS) and adipocytes, particularly omental adipocytes [1]. Recent in vitro investigations have shown FFAR1 to be activated in pancreatic beta cells by medium- to long-chain FFAs as well as by thiazolidinediones (Rosiglitazone and MCC-555), causing elevated Ca2+ concentrations and subsequent promotion of insulin secretion. Furthermore, mice with over-expression of Ffarr1 show impaired beta cell function and develop diabetes [4], whereas disruption of the gene reduces FFA-stimulated insulin release and, possibly protects from diabetes [5]. Recently two papers have reported that a number of SNPs in the FFAR1 gene mediate effects on insulin secretion, in particular in response to FFA [6], [7]. The role of FFAR1 in the CNS is not known, but it is hypothesised that this may be a mechanism by which FFAs are involved in the hypothalamic regulation of metabolism and its expression in omental adipocytes implicates it in the development of the metabolic syndrome [5]. It has been suggested more recently that FFAR1 plays a role in the taste perception of fatty acids, but this is controversial and needs substantiating [8]. Here we report the relationship between three common FFAR1 SNPs, with BMI, body composition, pancreatic function, insulin sensitivity and plasma lipids, in a cohort of overweight subjects identified to be at increased cardiometabolic risk.
Methods
Cohort
For this enquiry we used the RISCK study cohort. The RISCK study has been described in detail elsewhere [9]. In brief, the RISCK study was a randomized, controlled, parallel trial performed in free-living participants at 5 U.K. centres (University of Reading, Imperial College London, Kings College London, University of Surrey, and the Medical Research Council Human Nutrition Research [MRC-HNR]). A total of 720 participants were recruited, selected on the basis of their increased risk for the metabolic syndrome using a study-specific scoring system. All participants followed a 4-wk run-in period during which they were prescribed a high-saturated fat/high-glycemic index (HS/HGI) “reference” diet before being randomised to the reference diet or one of four isoenergetic dietary interventions to lower saturated fat. The main outcome was a measure of insulin sensitivity with secondary outcomes, including a range of cardiovascular risk markers.
At screening a fasting blood sample was taken and used to measure fasting lipids including total cholesterol; HDL-cholesterol; triglycerides and non-esterified fatty acids (NEFA). LDL-cholesterol was derived from the Friedwald equation [9]. Anthropometry was measured by standard procedures, and body composition by bioelectrical impedance analysis (BIA) see Jebb et al. for details [9]. Insulin sensitivity and beta-cell function were determined by intravenous glucose tolerance test (IVGTT). Insulin sensitivity (Si) and glucose effectiveness (Sg) were estimated using the MINMOD Millennium programme (Version 6.02). The area under the plasma insulin curve up to 19 minutes was computed as an indicator of endogenous insulin secretion (AIRg). The disposition index (DI), a measure of the ability of the beta-cells to compensate for insulin resistance, was calculated from AIRg and Si values [10]. For the purpose of this cross-sectional analysis we investigated the effect of three known common FFAR1 SNPs on BMI, body composition and fasting lipid measures, at entry into the study, and on insulin sensitivity and beta-cell function following 4 weeks of a “reference” high saturated fat, run-in diet.
SNP selection and genotyping
SNPs in the FFAR1 gene region were identified using SNPSelector set for gene SNPs by gene name (http://snpselector.duhs.duke.edu/hqsnp36.html) and cross-checked with information in other databases (Genecards http://www.genecards.org/index.shtml; Entrez-SNP http://www.ncbi.nlm.nih.gov/snp). Due to the small population of the study, we concentrated on common allele variants, and for that reason SNPs with a reported Caucasian minor allele frequency (MAF) of <5% were excluded, and only SNPs in the HapMap were investigated. A resulting 3 SNPs were genotyped for FFAR1 (rs2301151; rs16970264; rs1573611). A further SNP in FFAR1 (rs1978013) which was previously associated with beta-cell function [7] was also genotyped. Selected SNPs were tested for linkage disequilibrium with Haploview Version 4.2 software (http://www.broadinstitute.org/haploview/haploview using the Hapmap download format, version 3, release R2) and using information from previous studies [7] none of the SNPs were in significant LD (r2<5% rs2301151 with rs1573611; r2<5% rs2301151 with rs16970264; r2 1% rs1573611 with rs16970264 see Figure S1).
DNA was available for 530 participants of the RISCK study. Genotyping was performed by KBioscience using the KASPar genotyping system (Hoddesden, Herts, UK). All SNPs were successfully genotyped with a call rate >97%. Results could not be obtained for 15 participants due to genotyping failure rate in >1 SNP. Deviations from Hardy-Weinburg equilibrium were tested and one SNP (rs1978013) deviated significantly (P<0.001) and was excluded from further analysis. A further three attempts to redesign primers for genotyping this SNP, residing in a region of high homology with surrounding regions in this gene cluster, were unsuccessful.
The RISCK subjects, for which there was DNA available, consisted of an ethnic mix (81% White; 9% S-SE Asian, 7% Black African, 3% other). The allele frequency of all SNPs studied varied significantly by ethnicity, therefore this analysis was restricted to the Caucasian European subjects only, which represented the predominant group (n = 405).
Effect of number of FFAR1 risk alleles carried with change in plasma fatty acid level on metabolic outcome
The RISCK study has been analysed for plasma fatty acid profiles as an independent assessment of dietary change [1]. The fatty acids greater than C-6 are agonists for FFAR1 receptor, recent evidence in-vitro suggest that n-3 fatty acids stimulate the greatest response activation causing a greatest rise in intracellular calcium [1]. To examine the impact of change in plasma fatty acids greater than C-6 and DHA and EPA on metabolic outcome we analysed those subjects that has a positive change in plasma levels of the fatty acids between the baseline visit and end visit at week 24 giving a cohort of 280 of the volunteers.
Statistical analysis
Data were tested for normal distribution and log-transformed for analysis where appropriate, as indicated in tables. Non-normally distributed data were presented as the geometric mean ± 95% confidence intervals. The cohort was separated by genotype and the mean value for each trait presented according to an additive model for each SNP. Each SNP was scored as 0,1,2 according to the number of BMI-increasing alleles carried for rs2301151 and rs1573611. The SNP rs16970264 had no association with BMI and was therefore scored 0,1,2 based on the number of total cholesterol increasing alleles carried. The BMI- or total cholesterol- increasing allele is referred to as the “risk allele”. The association between FFAR1 SNPs and measures of body composition, insulin sensitivity and lipids were carried out using linear regression analysis. Age and gender were included in all models as covariates. For insulin sensitivity and lipid measures the associations were further adjusted for BMI. The effect of genotype on trait was then examined as dominant and recessive models based on the BMI or total cholesterol-increasing risk alleles. The mean of each trait divided by genotype is presented as the recessive and dominant models. The association of risk alleles according to the dominant and recessive models were also tested by linear regression. For the dominant model, a score of 0 was assigned for no risk alleles and 1 for presence (heterozygous or homozygous) of risk alleles, and for the recessive model, a score of 1 was assigned for homozygous for the risk allele and 0 for the others. We used the sum of risk alleles from the dominant models, as the numbers of the rare homozygous genotype were too low to analyse as single genotypes. In the analysis to assess the impact of dietary change metabolic change we used the change in plasma fatty acid profiles, an independent assessment of dietary intake. Uncorrected P-values are presented, however to account for multiple comparisons, we used the False Discovery Rate controlling procedure (q* = 0.05) of Benjamini and Hochburg with BMI, insulin-sensitivity and lipid-related traits treated as different families of hypotheses [11]. It is indicated where P-values satisfied the calculated FDR constraints.
Results
Subject characteristics
The allele frequency, gender and age distribution of the subjects of white ethnic origin included in this study, are shown stratified by genotype for the three SNPs in Table 1 . There were no significant differences in age or gender distributions between the genotypes of any of the three FFAR1 SNPs analysed ( Table 1 ). There was no significant difference in metabolic syndrome or CVD risk, according to the study-specific scoring system) between the genotypes of any of the three FFAR1 SNPs.
Table 1. Characteristics of the study cohort by genotype for three SNPs of FFAR1.
| rs2301151 | rs1573611 | rs16970264 | ||||||||||
| SNP Type/location | Non-synonymous | Near gene 3′ | Near gene 5′ | |||||||||
| AA | AG | GG | P | CC | TC | TT | P | AA | GA | GG | P | |
| n | 233 | 156 | 16 | 247 | 138 | 20 | 3 | 47 | 350 | |||
| Genotype frequency | 0.58 | 0.38 | 0.04 | 0.61 | 0.34 | 0.05 | 0.01 | 0.12 | 0.87 | |||
| Minor allele F | 0.23 | 0.22 | 0.07 | |||||||||
| Age | 52.4 (9.6) | 54.4 (10.3) | 52.4 (11.2) | 0.16 | 53.6 (10.2) | 52.6 (9.5) | 54.4 (10.4) | 0.56 | 56.7 (5.13) | 54.4 (8.9) | 53.0 (10.1) | 0.56 |
| Gender (M%/F%) | 43/57 | 42/58 | 19/81 | 0.16 | 40/60 | 44/56 | 50/50 | 0.89 | 67/33 | 40/60 | 41/59 | 0.91 |
| MS score | 6 [5,7] | 6 [5,8] | 6 [5,8] | 0.26 | 6 [5,8] | 6 [5,8] | 6 [4,6.5] | 0.42 | 7 [6,8] | 7 [5,8] | 6 [5,7] | 0.09 |
| CVD score | 2 [1,4] | 2 [1,5] | 1 [1,3] | 0.35 | 2 [1,5] | 2 [1,4] | 4 [1,7] | 0.43 | 2 [1,9] | 2 [1,5] | 2 [1,4] | 0.83 |
The data are presented as mean (SD) for age, proportion (%) for gender and median [IQR] for metabolic syndrome (MS) and cardiovascular disease (CVD) risk score for each genotype of the three SNPs of FFAR1 which were investigated. Differences (P) in characteristics between genotypes are indicated. The MS and CVD risk score were study specific see Jebb et al. [9]. SNP location data from the NCBI-SNP database.
Effect of FFAR1 polymorphisms on measures of body mass and composition
Carriage of the G allele of rs2301151 was associated with a higher body fat (%) of 1.11% per allele (P = 0.03) when assessed as the additive model ( Table 2 ) and 1.39% higher per risk allele (P = 0.02) when assessed as the dominant model (Table S1), accounting for age and gender, although these associations were not statistically significant when accounting for multiple comparisons by FDR procedure. Carriage of the C allele of rs1573611 was associated with a higher BMI, body fat (%) and waist circumference when examined as the dominant model (Table S1), and with BMI and body fat (%) as an additive model ( Table 2 ). The associations with BMI and body fat (%) but not waist circumference were statistically significant when accounting for multiple comparisons. There was a significant interaction between rs1573611 and gender for waist circumference when examined as the additive (P = 0.03) and the recessive (P = 0.05) models. The effect of SNP on waist circumference was only significant in females (effect = 3.27±1.12 cm higher per C allele, P = 0.02).
Table 2. BMI, waist circumference and body fat by genotype for three SNPs of FFAR1.
| rs2301151 | rs1573611 | rs16970264 | |||||||||||||
| Difference | P | Difference | P | Difference | P | ||||||||||
| AA | AG | GG | TT | TC | CC | AA | AG | GG | |||||||
| BMI (kg/m2) | |||||||||||||||
| mean | 28.71 | 29.41 | 30.01 | 0.67 | 0.11 | 25.61 | 28.86 | 29.31 | 1.06 | 0.009 | 28.53 | 29.53 | 28.85 | 0.55 | 0.41 |
| SE | 0.32 | 0.35 | 1.67 | 0.85 | 0.41 | 0.31 | 2.25 | 0.60 | 0.26 | ||||||
| n | 233 | 156 | 16 | 20 | 138 | 247 | 3 | 47 | 350 | ||||||
| Waist circumference (cm) | |||||||||||||||
| mean | 98.60 | 98.57 | 98.09 | 0.27 | 0.79 | 93.53 | 98.89 | 98.53 | 1.29 | 0.19 | 100.23 | 100.57 | 97.95 | 2.09 | 0.20 |
| SE | 0.85 | 0.95 | 2.72 | 3.02 | 1.09 | 0.76 | 4.41 | 1.68 | 0.66 | ||||||
| n | 233 | 156 | 16 | 20 | 138 | 247 | 3 | 47 | 350 | ||||||
| Body fat (%) | |||||||||||||||
| mean | 33.19 | 34.77 | 37.34 | 1.11 | 0.03 | 28.05 | 33.30 | 34.68 | 1.53 | 0.002 | 28.50 | 35.02 | 33.88 | −0.58 | 0.48 |
| SE | 0.56 | 0.69 | 1.99 | 1.52 | 0.71 | 0.55 | 3.70 | 1.31 | 0.46 | ||||||
| n | 228 | 155 | 16 | 20 | 136 | 244 | 3 | 47 | 345 | ||||||
Data are presented as mean, SEM, n stratified by genotype for each of the three SNPs. For each genotype the risk allele was defined as the BMI-increasing allele and the data are presented as the additive model. The differences in trait by genotype were assessed by linear regression analysis coding the number of risk alleles as 0,1,2. The P-value for the regression is presented and is in bold when reaching significance by the FDR-controlling procedure q* = 0.05. The recessive and dominant models defined according to the risk allele are shown in Supplementary Table S1.
There was no evidence of a SNP-gender interaction for rs2301151 or rs16970264 for any of the variables.
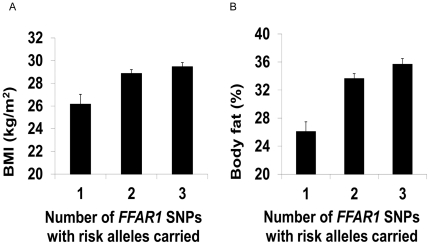
There was a cumulative effect of the number of SNPs of FFAR1 for which risk alleles were carried, on BMI and body fat ( Figure 1 ); for an increasing number of SNPs, where at least one risk allele was carried, there was a higher BMI (effect = 1.04±0.41 kg/m2 per SNP, P = 0.01) and higher body fat-% (effect = 1.75±0.6% per SNP, P = 0.001). These effects were significant when accounting for multiple comparisons. There was no significant SNP×SNP interaction effect examined as either a two-way or three-way interaction using the additive model. There was no evidence of an effect of plasma fatty acid profile integrating with the cumulative number of risk alleles carried to have a significant effect on change in or final BMI, waist measurement and body fat content.
Figure 1. The cumulative effect of carrying risk alleles in three SNPs of FFAR1 on BMI and body fat.
Data presented are mean ± SEM of BMI (Panel a) and body fat (Panel b) for individuals according to the number of SNPs for which risk alleles were present. Each SNP (rs2301151; rs1573611; rs16970264) was scored 0 or 1 based on the presence of risk alleles (dominant model). The presence of risk alleles for each SNP was summed. All individuals had at least 1 SNP with risk alleles present. There was a cumulative effect on BMI of 1.04±0.41 kg/m2 per SNP, P = 0.01 and of body fat (%) of 1.75±0.6% per SNP, P = 0.001 assessed by linear regression analysis with age and gender as covariates. The effects were statistically significant when accounting for multiple comparisons by the Benjamini and Hochberg False Discovery Rate procedure with q* = 0.05.
Effect of FFAR1 polymorphisms on measures of insulin sensitivity
There was a nominally significant (unadjusted) association of the G allele of rs2301151 with a ∼73% higher disposition index (DI) and a trend (P = 0.06) towards a ∼52% higher endogenous (1st phase/acute) insulin release (AIRg) when assessed as the recessive model only, accounting for age, gender and BMI (Table S2). There was a nominally significant (unadjusted) dominant effect of the C allele of rs1573611 to be associated with lower fasting plasma glucose (−0.31 mmol/L, P = 0.02) (Table S2) There were no other effects on measures of insulin-sensitivity or beta-cell function, and none of these nominal associations reached significance when accounting for multiple comparisons by FDR procedure. There was no significant effect of rs16970264 on any measures of insulin-sensitivity or beta-cell function ( Table 3 ). There was no evidence of gender-SNP interactions for any of the three SNPs on any measures of insulin-sensitivity or beta-cell function. There was no evidence of a cumulative effect of carrying risk alleles in multiple SNPs, nor was there any evidence of a two- or three-way interaction effect of the SNPs using the additive model (data not shown). There was a cumulative effect of carrying risk alleles in multiple SNPs with change in plasma fatty acid with AIRg measurement taken at 24 weeks. With the AIRg decreasing with the accumulated risk alleles (1 risk allele: 705.3±135 mL.µU−1.min−1, 2 risk alleles: 514.2±39 mL.µU−1.min−1, 3 risk alleles: 456.4±37 mL.µU−1.min−1 p<0.03, uncorrected). There was no other cumulative effect of risk alleles on insulin sensitivity.
Table 3. Measures of insulin sensitivity and beta-cell function by genotype for three SNPs of FFAR1.
| rs2301151 | rs1573611 | rs16970264 | |||||||||||||
| Difference | P | Difference | P | Difference | P | ||||||||||
| AA | AG | GG | TT | TC | CC | AA | AG | GG | |||||||
| Fasting glucose (mmol/L) | |||||||||||||||
| mean | 5.46 | 5.47 | 5.58 | 0.01 | 0.91 | 5.78 | 5.45 | 5.47 | −0.07 | 0.18 | 3.57 | 5.52 | 5.47 | −0.02 | 0.78 |
| SE | 0.04 | 0.05 | 0.52 | 0.13 | 0.06 | 0.04 | 0.20 | 0.08 | 0.03 | ||||||
| n | 233 | 155 | 16 | 20 | 137 | 247 | 3 | 47 | 349 | ||||||
| Fasting insulin (pmol/L) | |||||||||||||||
| Geometric mean | 56.8 | 60.1 | 60.5 | 1.03 | 0.44 | 55.6 | 56.6 | 59.3 | 1.00 | 0.91 | 53.0 | 59.9 | 57.4 | 0.90 | 0.11 |
| 95% CI | [53.3, 64.4] | [56.1, 64.4] | [48.6, 75.4] | [46.9, 65.9] | [52.2, 61.4] | [55.9, 62.9] | [3.22, 873] | [57.8, 74.1] | [54.7, 60.2] | ||||||
| n | 227 | 153 | 13 | 20 | 135 | 238 | 3 | 45 | 340 | ||||||
| Si (×10−4 mL.µU−1.min−1) | |||||||||||||||
| Geometric mean | 2.81 | 2.69 | 3.20 | 1.03 | 0.49 | 3.02 | 2.74 | 2.80 | 1.04 | 0.36 | 1.68 | 2.53 | 2.84 | 1.15 | 0.09 |
| 95% CI | [2.60, 3.05] | [2.43, 2.97] | [2.74, 3.74] | [2.29, 3.98] | [2.44, 3.07] | [2.61, 3.00] | [0.00, 904] | [2.11, 3.03] | [2.66, 3.02] | ||||||
| n | 211 | 146 | 14 | 17 | 125 | 230 | 2 | 43 | 322 | ||||||
| AIRg (mL.µU−1.min−1) | |||||||||||||||
| Geometric mean | 443 | 319 | 521 | 1.01 | 0.73 | 255 | 354 | 335 | 1.01 | 0.83 | 997 | 320 | 337 | 0.96 | 0.42 |
| 95% CI | [403, 483] | [277, 368] | [398, 681] | [155, 418] | [311, 403] | [299, 374] | [0.51, 9185] | [241, 424] | [309, 369] | ||||||
| n | 211 | 146 | 14 | 17 | 125 | 230 | 2 | 43 | 322 | ||||||
| Disposition Index (Arbitrary Units) | |||||||||||||||
| Geometric mean | 972 | 864 | 1667 | 1.06 | 0.27 | 768 | 984 | 933 | 1.04 | 0.7 | 1673 | 808 | 960 | 1.08 | 0.51 |
| 95% CI | [864, 1093] | [737, 1012] | [1236, 2249] | [440, 1342] | [843, 1149] | [829, 1050] | [462, 6057] | [598, 1092] | [598, 1093] | ||||||
| n | 211 | 146 | 14 | 17 | 125 | 230 | 2 | 43 | 322 | ||||||
| Sg (×10−3/min) | |||||||||||||||
| mean | 17.21 | 17.94 | 20.67 | 1.23 | 0.18 | 20.32 | 18.04 | 17.06 | −1.17 | 0.20 | 9.75 | 17.84 | 17.60 | 0.37 | 0.81 |
| SE | 0.42 | 1.15 | 1.76 | 2.56 | 1.24 | 0.45 | 3.96 | 1.63 | 0.55 | ||||||
| n | 211 | 146 | 14 | 17 | 125 | 230 | 2 | 43 | 322 | ||||||
Data are presented as mean, SEM, n (glucose, Sg) or geometric mean, 95% confidence intervals, n (insulin, Si, AIRg, DI) stratified by genotype for each of the three SNPs. For each genotype the risk allele was defined as the BMI-increasing allele and the data are presented as the additive model. The differences in trait by genotype were assessed by linear regression analysis coding the number of risk alleles as 0,1,2. Data for insulin, Si, AIRg and disposition index were logged for regression analysis. The beta-coefficient from the regression was exponentiated which approximates to the percentage difference. The P-value for the regression is presented and no associations reached statistical significance. The recessive and dominant models, defined according to the risk allele, are presented in Supplementary Table S2.
Effect of FFAR1 polymorphisms on plasma lipids
There was a nominally significant association (unadjusted) of carrying the G allele for rs2301151 with higher total cholesterol (0.2 mmol/L per G risk allele, Table 4 ). There was a recessive effect of the G allele which was associated with lower (∼23%, P = 0.004, unadjusted) plasma non-esterified fatty acids (NEFA) (Table S3). There was a recessive effect of the common G allele of rs16970264 on total and LDL cholesterol (Table S3). However, carriage of the G allele was protective for HDL cholesterol (0.09 mmol/L higher per G allele (P<0.05, unadjusted)) and there was no effect on the total∶HDL cholesterol (TC∶HDL) ratio ( Table 4 ). None of these associations were statistically significant when accounting for multiple comparisons by FDR procedure. There was no effect of rs1573611 on lipid measures.
Table 4. Fasting plasma lipids by genotype for three SNPs of FFAR1.
| rs2301151 | rs1573611 | rs16970264 | |||||||||||||
| Difference | P | Difference | P | Difference | P | ||||||||||
| AA | GA | GG | TT | TC | CC | AA | GA | GG | |||||||
| Total Cholesterol (mmol/L) | |||||||||||||||
| mean | 5.57 | 5.77 | 6.14 | 0.2 | 0.01 | 5.62 | 5.59 | 5.71 | 0.08 | 0.33 | 5.67 | 5.39 | 5.72 | 0.3 | 0.02 |
| SE | 0.06 | 0.07 | 0.25 | 0.20 | 0.01 | 0.06 | 0.68 | 0.14 | 0.05 | ||||||
| n | 233 | 155 | 16 | 20 | 137 | 247 | 3 | 47 | 349 | ||||||
| LDL Cholesterol (mmol/L) | |||||||||||||||
| mean | 3.51 | 3.62 | 3.9 | 0.13 | 0.09 | 3.59 | 3.5 | 3.6 | 0.05 | 0.50 | 4.02 | 3.31 | 3.61 | 0.23 | 0.05 |
| SE | 0.06 | 0.07 | 0.22 | 0.19 | 0.08 | 0.05 | 0.63 | 0.13 | 0.05 | ||||||
| n | 233 | 155 | 16 | 20 | 138 | 247 | 3 | 47 | 350 | ||||||
| HDL Cholesterol (mmol/L) | |||||||||||||||
| mean | 1.41 | 1.44 | 1.60 | 0.04 | 0.16 | 1.40 | 1.40 | 1.45 | 0.04 | 0.17 | 1.15 | 1.36 | 1.44 | 0.09 | 0.05 |
| SE | 0.02 | 0.03 | 0.08 | 0.08 | 0.03 | 0.02 | 0.10 | 0.05 | 0.02 | ||||||
| n | 233 | 155 | 16 | 20 | 137 | 247 | 3 | 47 | 349 | ||||||
| Triglycerides (mmol/L) | |||||||||||||||
| Geometric mean | 1.27 | 1.35 | 1.34 | 1.04 | 0.35 | 1.23 | 1.32 | 1.30 | 0.99 | 0.75 | 1.10 | 1.42 | 1.29 | 0.95 | 0.49 |
| 95% CI | [1.20, 1.36] | [1.25, 1.46] | [1.13, 1.59] | [0.98, 1.55] | [1.23, 1.43] | [1.22, 1.39 | [0.71, 1.69] | [1.21, 1.67] | [1.22, 1.35] | ||||||
| n | 233 | 154 | 16 | 20 | 137 | 246 | 3 | 46 | 349 | ||||||
| Non-esterified fatty acids (µmol/L) | |||||||||||||||
| Geometric mean | 651 | 650 | 522 | 0.94 | 0.04 | 630 | 648 | 646 | 0.99 | 0.63 | 603 | 635 | 650 | 1.04 | 0.5 |
| 95% CI | [622, 681] | [610, 693] | [397, 689] | [528, 752] | [611, 687] | [614, 679] | [151, 2403] | [563, 716] | [624, 676] | ||||||
| n | 226 | 149 | 16 | 20 | 132 | 240 | 3 | 46 | 337 | ||||||
| Total: HDL cholesterol ratio | |||||||||||||||
| mean | 4.14 | 4.25 | 3.98 | 0.05 | 0.60 | 4.22 | 4.24 | 4.15 | −0.08 | 0.36 | 4.94 | 4.14 | 4.17 | −0.02 | 0.91 |
| SE | 0.07 | 0.09 | 0.23 | 0.23 | 0.10 | 0.07 | 0.41 | 0.16 | 0.06 | ||||||
| n | 233 | 155 | 16 | 20 | 137 | 247 | 3 | 47 | 349 | ||||||
Data are presented as mean, SEM, n (Total, LDL, HDL and total: HDL cholesterol) or geometric mean, 95% confidence intervals, n (triglycerides, non-esterified fatty acids) stratified by genotype for each of the three SNPs. For each genotype the risk allele was defined as the BMI-increasing allele except for rs16970264 where the risk allele was defined according to the total-cholesterol increasing effect and the data are presented as the additive model. The differences in trait by genotype were assessed by linear regression analysis coding the number of risk alleles as 0,1,2. Data for plasma NEFA and triglycerides were logged for regression analysis. The beta-coefficient from the regression was exponentiated which approximates to the percentage difference. The P-value for the regression is presented. No associations reached statistical significance by the FDR-controlling procedure q* = 0.05. The recessive and dominant models, defined according to the risk allele are presented in Supplementary Table S3.
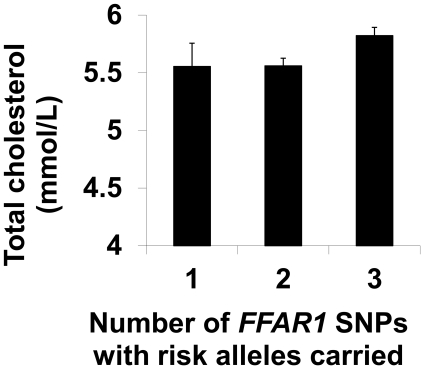
There was a cumulative effect of the number of SNPs for which risk alleles were carried, on total plasma cholesterol ( Figure 2 ). For an increasing number of SNPs where at least one risk allele was carried, the total cholesterol was higher (effect = 0.18±0.08 mmol/L per SNP, P = 0.03), however this was not statistically significant when accounting for multiple comparisons. There was no significant SNP×SNP interaction effect examined as either a two-way or three-way interaction using the additive model. There was no evidence of an effect of change in plasma fatty acid profile integrating with the cumulative number of SNPs for which at least 1 risk allele was carried, to have a significant effect on change in or final level of any lipid parameters.
Figure 2. The cumulative effect of carrying risk alleles in three SNPs of FFAR1 on total cholesterol.
Data presented are mean ± SEM of total cholesterol for individuals according to the number of SNPs for which risk alleles were present. Each SNP in the FFAR1 region which was examined (rs2301151; rs1573611; rs16970264) was scored 0 or 1 based on the presence of risk alleles (dominant model). The number of SNPs with risk alleles was summed. All individuals had at least 1 SNP with risk alleles present. There was a cumulative effect on total cholesterol of 0.18±0.08 mmol/L per SNP, P = 0.03 assessed by linear regression analysis with age, gender and BMI. This was not statistically significant when accounting for multiple comparisons by the Benjamini and Hochberg False Discovery Rate procedure with q* = 0.05.
Discussion
The key findings of the present study were that SNPs of the FFAR1 gene region were associated with cumulative differences in BMI, body composition and total cholesterol in the three SNPs studied, although the effects on total cholesterol were not significant after accounting for multiple comparisons.
The predominant adverse effects on BMI and body fat were mediated by carrying the G allele of rs2301151 and the C allele of rs1573611, with SNP rs1573611 also showing gender specific effects of carrying the C-allele, with an increased waist circumference in females only. The predominant adverse effects on plasma lipids were mediated by the G allele of rs2301151 and the G allele (which was the common allele) of rs16970264. However, the associations with lipids did not remain significant when accounting for multiple comparisons.
SNP rs2301151 is in the coding region of the FFAR1 gene and results in a non-synonymous substitution (Arg211His) located in the intracellular region between transmembrane 5 domain and transmembrane 6 domain of the 7 transmembrane domain protein, [12]. The other SNPs (rs1573611 and rs16970264) are in the non-coding region of gene FFAR1, in this regard we are assuming that these SNPs affect FFAR1 as the closest gene, but cannot exclude the possibility that the observed effects were mediated by another locus in LD with one or other of these SNPs (see Figure S1).
There is no defined metabolic pathway that links FFAR1 with body weight. However, FFAR1 receptor is found throughout the CNS [1], and therefore it may play a role in appetite regulation. Others have hypothesised that FFAR1 may be the receptor that coordinates the appetite suppression in response to FFA [6], [13], [14], [15], and that FFAR1 has a role in the taste perception of fat [8]. If variation in this gene is associated with changes in the latter, it is possible this could exert subtle changes in energy homeostasis. Since FFAR1 has been linked to beta-cell function and type 2 diabetes [2], [3], [7], it is possible that changes in insulin metabolism could impact on energy homeostasis and consequently BMI and body composition. A rare variant, the Gly180Ser mutation, was previously shown to increase in frequency with increasing BMI, providing further support for linkage between variants of the FFAR1 gene and BMI reported in the present study. Vettor et al. [6] suggested FFAR1 may provide a hypothalamic link between the sensing of adequate circulating fatty acid levels and subsequent regulation of energy intake.
The BMI-increasing G allele of rs2301151 was also associated with higher total cholesterol, which was maintained when accounting for the FFAR1-associated changes in BMI. SNP rs16970264 also appeared to modulate blood lipids with the most common GG genotype being associated with higher total and LDL cholesterol, but also being associated with higher, protective levels of HDL cholesterol. However these exploratory findings did not reach significance when accounting for multiple comparisons and would require further investigation in an independent cohort. Interestingly, FFAR1 −/− animals are somewhat protected against the effects of a high fat diet (HFD), with reduced hyperinsulinaemia, glucose-intolerance or insulin-resistance compared to WT mice [5], and without the increases in hepatic steatosis, plasma triacylglycerol or hepatic glucose output seen in the WT mice [5]. Therefore in rodents, FFAR1 mediates metabolic responses to dietary fats. An oral lipid tolerance test was previously found to be associated with suppressed insulin and increased glucose responses in carriers of the rare Gly180Ser mutation, also implicating FFAR1 in lipid handling in humans [6]. Changes in the handling of triacylglycerol can have effects on lipoprotein metabolism. An interesting finding was fasting plasma NEFA levels were also lower (∼77% of AA/AG genotypes, P = 0.004) in subjects who were homozygous for the G allele of rs2301151 (Supplementary Table S3). It is possible that there is an effect on lipid handling at the adipocyte with variation in the FFAR1 gene. Although at a lower level, FFAR1 is expressed in the adipocyte. A recent report suggests that the receptor is present in omental adipose tissue, a key regulator of insulin sensitivity. Genes closely related to FFAR1 are FFAR2 and FFAR3 which both suppress FFA output from the adipocyte when stimulated. Recent reports suggest that there is CNS regulation of adipose tissue metabolism. It could be that the FFAR1 SNPs may change the central signalling to adipocyte reducing FFA output and enhancing beta cell function further. None of the SNPs are in high LD (r2>0.8, supplementary Figure S1) with SNPs of nearby FFAR genes, however SNP rs1573611 is in moderate LD (r2>0.66) with a SNP from the FFAR3 gene. Given this, and the proximity of the genes in this cluster, it is possible these SNPs affect the function of neighbouring FFAR genes.
Since FFAR1 is predominantly expressed in the pancreas, and to a lesser extent in the brain, it is reasonable to expect polymorphisms in this receptor to exert greater effects on beta-cell function. The GG allele of rs2301151 was associated with a nominally higher disposition index (DI) with a trend (P = 0.06) towards a higher first phase/acute insulin response (AIRg, Supplementary Table S2). It is of interest that there appears to be an interaction between an increase in the number of risk alleles carried, and the change in the receptor agonists (plasma fatty acid greater than C-6) on acute insulin response (AIRg)DI, suggesting a gene-diet interaction, although we did not have sufficient power in this study to formally assess a gene-diet interaction. As there was no difference in insulin sensitivity (Si) or fasting glucose, it appears that this genotype may be associated with a higher insulin secretion for an equivalent degree of insulin sensitivity, ultimately stressing the pancreas. However, these effects were only seen when assessed as a recessive model with a small number of subjects (n = 14) with the GG genotype, so these results should be viewed with caution. SNP rs2301151 investigated in the current study corresponds with the Arg211His polymorphism, which had been found previously to have no effect on glucose or insulin responses to an oral glucose challenge [6]. However, in the study of Vettor et al. [6] there was also no difference in BMI or lipids by genotype, this may have been due to a much higher average BMI, and subsequent differences in metabolic profile in that cohort compared to our cohort (mean BMI ∼37 kg/m2 cf ∼28 in our cohort). Although we were unable to successfully genotype the SNP rs1978013, which had been found previously to be associated with beta-cell function [7], there was moderate linkage disequilibrium with the more common rs1573611 (r2 = 0.31), which was successfully genotyped in the current study. There was no association of this SNP with measures of insulin sensitivity or beta cell function, except for fasting glucose which was lower (−0.31 mmol/L, P = 0.02) in the TT genotype, and was also associated with increased BMI, waist circumference and body fat percent. The protective effects on fasting glucose appear contradictory to the effects on BMI, however the number of subjects with this genotype was 20 (out of a total 404) so these results may be difficult to interpret due to low subject numbers.
There have been very few human studies on the effects of variation in the FFAR1 gene region on metabolic phenotypes. The differences in BMI, body composition and lipids shown in our study of overweight subjects identified to be at increased cardiometabolic risk, were not accompanied by convincing changes in insulin sensitivity or beta cell function. Therefore it appeared that the differences in body composition and lipids were mediated by mechanisms independent of differences in beta-cell function. It is possible that our methods used for measuring insulin sensitivity and beta-cell function were not sensitive enough to detect differences by genotype, however this method was found to be sensitive for detecting changes in insulin sensitivity associated with a small change in weight in these subjects [9]. SNPs in the FFAR1 region have not previously been identified in lipid or BMI associated GWA studies. In a recent meta-analysis of lipid-associated SNPs there were no regions of high association near the FFAR1 gene [16]. This is not unexpected, as previously identified candidate genes have often failed to be identified in GWA studies. In this study we concentrated on variants with a minor allele frequency >0.5 with the intention of studying common variants of this gene. Even with this intention the number of subjects homozygous for the SNPs studied was still small which meant the results were interpreted with some caution. Although not significant, there was a predominant percentage of females, compared to males, who were homozygous for the risk (G) allele of rs2301151. Although gender was included as a covariate for all analyses, it is possible that this gender bias could influence the associations.
The effect of change in dietary fatty acids was assessed by using the data on plasma fatty acids profile which is reflective of dietary intake and reflects the receptor environment. The major agonists of the FFAR1 receptor are fatty acids with a chain length greater than 6. Recent in vitro data suggests the n-3 fatty acids show a greater affinity for the receptor. We were unable to show any relationship between increase in plasma fatty acids and any outcome other than acute insulin response (AIRg) at 24 weeks discussed above.
In summary, we demonstrated that three common SNPs of the FFAR1 gene were associated with body composition and lipid traits. Furthermore, the effects of the 3 SNPs were cumulative on BMI, body fat and total cholesterol. Despite the strong expression of FFAR1 in the pancreas, these differences appeared to be independent of changes in insulin and beta cell function.
Supporting Information
Linkage disequilibrium (LD) plot for the FFAR gene cluster. This region includes FFAR1, FFAR3 and the pseudogene GPR42, with the gene FFAR2 located 78.5 KB downstream of GPR42. LD is expressed as r2 values. This plot was generated in Haploview 4.2. From Hapmap download version 3, release R2.
(TIF)
BMI, waist circumference and body fat by genotype for recessive and dominant models of three SNPs of FFAR1.
(DOC)
Measures of insulin sensitivity and beta-cell function by genotype for recessive and dominant models for three SNPs of FFAR1.
(DOC)
Fasting plasma lipids by genotype for recessive and dominant models for three SNPs of FFAR1.
(DOC)
Acknowledgments
The RISCK Study Group comprised of: Carmel Moore, Mark Chatfield, Christine Williams, Hannah Farrant, Claire Lawrence, Edel Magee, Kit Tsoi, Darren Cole, Steve Austin, Hanneke Mfuni, Kate Guberg, Anna Gent, Celia Greenberg, Caroline Stokes, Mario Siervo, Rosemary Hall, Claire Howard, Namrata Dhopatkar, Bushra Siddiqui, Anne Dornhurst, Fiona Lewis, Samantha Bowen, L Chen, Robert Gray, Nuala Booth, Gary Moore, Roy Sherwood, Anthony Leeds, A Shah, G Saran, J Niehuser-Saran, JA Cockburn, Rachel Gitau, Katie Newens, Sean Lovegrove, Ana Rodriguez-Mateos, John Wright, Margaret Griffin, Nicola Harman. Special thanks to Aseel Al Saleh (Kings College London) for DNA extraction of samples. Thanks to Ashley Olson for statistical advice.
Footnotes
Competing Interests: The authors and their research groups have a number of links with the food industry. In a personal capacity GSF is a consultant to Coca-Cola, Premier Foods, and Unilever; and TABS has acted as a consultant to Seven Seas, is a member of the Scientific Advisory Committee for the Global Dairy Platform, the external scientific review committee of the Malaysian Palm Oil Board, and chairs Cadbury's Global Nutrition Advisory Panel. TABS, BAG, JAL, SAJ, and GSF have received ad hoc honoraria for lectures or writing articles. CGW reported no conflicts of interest. In a non-personal capacity, BAG was formerly a member of an expert group known as the Fat Panel, which was supported by Dairy Crest, Kerry Gold, and Unilever; SAJ is a member of Scientific Advisory Boards for Coca-Cola, Heinz, PepsiCo, Nestlé and Kellogg's; SAJ and JAL sit on government advisory boards that also include food industry members. All research groups received products from a range of food companies gratis for research purposes, including Archer Daniel Mills, Croda, Matthews Foods, Nestle, PepsiCo, Jordan, GSK, and Unilever.
Funding: The RISCK dietary intervention trial was funded by the Food Standards Agency (http://www.food.gov.uk) project number NO2031. Genetic analyses were funded by the participating centres. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Briscoe CP, Tadayyon M, Andrews JL, Benson WG, Chambers JK, et al. The orphan G protein-coupled receptor GPR40 is activated by medium and long chain fatty acids. J Biol Chem. 2003;278:11303–11311. doi: 10.1074/jbc.M211495200. [DOI] [PubMed] [Google Scholar]
- 2.Panhuysen C, Cupples L, Wilson P, Herbert A, Myers R, et al. A genome scan for loci linked to quantitative insulin traits in persons without diabetes: the Framingham Offspring Study. Diabetologia. 2003;46:579–587. doi: 10.1007/s00125-003-1066-z. [DOI] [PubMed] [Google Scholar]
- 3.van Tilburg JHO, Sandkuijl LA, Strengman E, van Someren H, Rigters-Aris CAE, et al. A Genome-Wide Scan in Type 2 Diabetes Mellitus Provides Independent Replication of a Susceptibility Locus on 18p11 and Suggests the Existence of Novel Loci on 2q12 and 19q13. J Clin Endocrinol Metab. 2003;88:2223–2230. doi: 10.1210/jc.2002-021252. [DOI] [PubMed] [Google Scholar]
- 4.Nagasumi K, Esaki R, Iwachidow K, Yasuhara Y, Ogi K, et al. Overexpression of GPR40 in Pancreatic Beta-Cells Augments Glucose Stimulated Insulin Secretion and Improves Glucose Tolerance in Normal and Diabetic Mice. Diabetes. 2009;58:1067–1076. doi: 10.2337/db08-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan H, Hoos LM, Liu L, Tetzloff G, Hu W, et al. Lack of FFAR1/GPR40 Does Not Protect Mice From High-Fat Diet-Induced Metabolic Disease. Diabetes. 2008;57:2999–3006. doi: 10.2337/db08-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vettor R, Granzotto M, De Stefani D, Trevellin E, Rossato M, et al. Loss-of-Function Mutation of the GPR40 Gene Associates with Abnormal Stimulated Insulin Secretion by Acting on Intracellular Calcium Mobilization. J Clin Endocrinol Metab. 2008;93:3541–3550. doi: 10.1210/jc.2007-2680. [DOI] [PubMed] [Google Scholar]
- 7.Kalis M, Leveen P, Lyssenko V, Almgren P, Groop L, et al. Variants in the FFAR1 Gene Are Associated with Beta Cell Function. PLoS ONE. 2007;2:e1090. doi: 10.1371/journal.pone.0001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cartoni C, Yasumatsu K, Ohkuri T, Shigemura N, Yoshida R, et al. Taste preference for fatty acids is mediated by GPR40 and GPR120. J Neurosci. 2010;30:8376–8382. doi: 10.1523/JNEUROSCI.0496-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jebb SA, Lovegrove JA, Griffin BA, Frost GS, Moore CS, et al. Impact of changing the amount and type of fat and carbohydrate on insulin sensitivity and cardiovascular risk: the RISCK trial. American Journal of Clinical Nutrition. 2010;92:748–758. doi: 10.3945/ajcn.2009.29096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergman RN, Ader M, Huecking K, Van Citters G. Accurate Assessment of Beta-Cell Function. Diabetes. 2002;51:S212–S220. doi: 10.2337/diabetes.51.2007.s212. [DOI] [PubMed] [Google Scholar]
- 11.Benjamini Y, Hochberg Y. CONTROLLING THE FALSE DISCOVERY RATE - A PRACTICAL AND POWERFUL APPROACH TO MULTIPLE TESTING. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 12.Sawzdargo M, George SR, Nguyen T, Xu SJ, Kolakowski LF, et al. A cluster of four novel human G protein-coupled receptor genes occurring in close proximity to CD22 gene on chromosome 19q13.1. Biochemical and Biophysical Research Communications. 1997;239:543–547. doi: 10.1006/bbrc.1997.7513. [DOI] [PubMed] [Google Scholar]
- 13.Lam TK, Pocai A, Gutierrez-Juarez R, Obici S, Bryan J, et al. Hypothalamic sensing of circulating fatty acids is required for glucose homeostasis. Nat Med. 2005;11:320–327. doi: 10.1038/nm1201. [DOI] [PubMed] [Google Scholar]
- 14.Obici S, Feng Z, Morgan K, Stein D, Karkanias G, et al. Central administration of oleic acid inhibits glucose production and food intake. Diabetes. 2002;51:271–275. doi: 10.2337/diabetes.51.2.271. [DOI] [PubMed] [Google Scholar]
- 15.Pocai A, Lam TK, Obici S, Gutierrez-Juarez R, Muse ED, et al. Restoration of hypothalamic lipid sensing normalizes energy and glucose homeostasis in overfed rats. J Clin Invest. 2006;116:1081–1091. doi: 10.1172/JCI26640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Linkage disequilibrium (LD) plot for the FFAR gene cluster. This region includes FFAR1, FFAR3 and the pseudogene GPR42, with the gene FFAR2 located 78.5 KB downstream of GPR42. LD is expressed as r2 values. This plot was generated in Haploview 4.2. From Hapmap download version 3, release R2.
(TIF)
BMI, waist circumference and body fat by genotype for recessive and dominant models of three SNPs of FFAR1.
(DOC)
Measures of insulin sensitivity and beta-cell function by genotype for recessive and dominant models for three SNPs of FFAR1.
(DOC)
Fasting plasma lipids by genotype for recessive and dominant models for three SNPs of FFAR1.
(DOC)