Abstract
The fur of common rabbits is constituted of 3 types of hair differing in length and diameter while that of rex animals is essentially made up of amazingly soft down-hair. Rex short hair coat phenotypes in rabbits were shown to be controlled by three distinct loci. We focused on the “r1” mutation which segregates at a simple autosomal-recessive locus in our rabbit strains. A positional candidate gene approach was used to identify the rex gene and the corresponding mutation. The gene was primo-localized within a 40 cM region on rabbit chromosome 14 by genome scanning families of 187 rabbits in an experimental mating scheme. Then, fine mapping refined the region to 0.5 cM (Z = 78) by genotyping an additional 359 offspring for 94 microsatellites present or newly generated within the first defined interval. Comparative mapping pointed out a candidate gene in this 700 kb region, namely LIPH (Lipase Member H). In humans, several mutations in this major gene cause alopecia, hair loss phenotypes. The rabbit gene structure was established and a deletion of a single nucleotide was found in LIPH exon 9 of rex rabbits (1362delA). This mutation results in a frameshift and introduces a premature stop codon potentially shortening the protein by 19 amino acids. The association between this deletion and the rex phenotype was complete, as determined by its presence in our rabbit families and among a panel of 60 rex and its absence in all 60 non-rex rabbits. This strongly suggests that this deletion, in a homozygous state, is responsible for the rex phenotype in rabbits.
Introduction
Rex fur in animals encompasses different abnormal hair coat phenotypes due to spontaneous or induced mutations. Rex coat mutations were described in cats [1],[2], rats [3],[4], mice [5] (several mutations), and rabbits [6]. This heterogeneity in phenotype description, even within species, suggests that different genes and mutations are responsible for these phenotypes. We were interested in deciphering the molecular basis of this trait in strains of rex rabbits bred in our institute for both scientific and commercial purposes. Rabbit (Oryctolagus cuniculus) fur is composed of three different types of hairs: guard hair (3–4 cm long for a diameter of 50–60 µm), awn hair (3–3.5 cm/25–30 µm) and down hair (2.5–3 cm/15 µm). Guard hair and awn hair constitute the physical protection usually called the outer coat, while down hair ensures the thermal protection of the rabbit named the inner coat. Down hair is the most abundant and represents about 90–95% of all hairs. There is variability in the ratio number of the inner and the outer coat hairs, and professionals are looking for a reduction of the coarse hair quantity in rex rabbit furs.
Hair is produced by hair follicles located in the dermis but made of epidermal cells. There is one hair synthesized per follicle and the type of hair depends on the type of follicle. In rabbits, hair follicles are structured into groups: a group is usually constituted of one central primary hair follicle surrounded by 2–4 lateral primary hair follicles and by 20–50 secondary down hair follicles (Figure 1). These three types of hair follicles appear sequentially during fetal development and early after birth. At day 19 of gestation, the central primary hair follicles increase followed at day 25 by the primary side hair follicles. At day 29 of gestation a secondary hair follicle appears for each of the 2 to 4 lateral hair follicles. Finally, secondary derived hair follicles, emerging from the skin by the same hair channel, appear during the early childhood of the animals [7]. In 1919 in the Pays de Loire French region, a mutant phenotype with soft hair was observed by a breeder in a litter of wild gray rabbits. Abbé Gillet, a local priest, considered the trait very fancy and started planning crosses to produce rabbits with this soft hair touching phenotype with guard hair that was no longer than the undercoat. In Germany in 1926 similar, if not identical, phenotypes appeared and the year after in France in Himalayan rabbits. These three phenotypes show simple Mendelian recessive inheritance likely being controlled by three different loci r1, r2 and r3 [8]. This abnormally short hair trait is thought to originate from the degenerescence of primary hair follicles leading to lacking or shortening of guard hair [9]. Interestingly, the whiskers (vibrissae) of the rex animals are bent or curly.
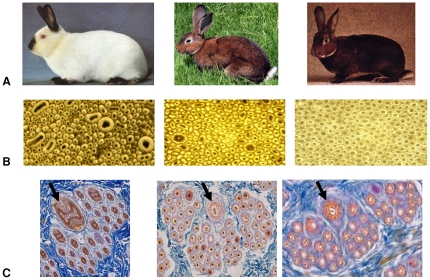
Figure 1. The rex hair trait in rabbit.
(A) A normal rabbit coat (A1), a rex rabbit with a castor coat (A2) and an orylag® with a castor coat (A3). (B) Cross section of a fibre bundle of a normal rabbit coat (B1), a rex rabbit (B2) and an orylag® (B3). (C) Skin cross section of a hair follicle group of a normal rabbit coat (C1), a rex rabbit (C2) and an orylag® (C3). Arrows indicate the differences in shape and diameters of primary central hair follicles and their hair.
In the early nineteen-eighties, the trait was selected at the INRA experimental farm of Le Magneraud to further reduce the number of guard and awn hairs and is now commercialized as orylag® for fur and “Rex du Poitou” for its high quality meat, conferring these animals a high value. In addition, segregation of the rex hair trait was studied in experimental families (G2) using rex and common type rabbit strains (G0). Previous results suggest that these rex experimental rabbits carried only the r1 mutation and that the determinism of this trait is monogenic, autosomal and recessive.
In this study we used a positional cloning approach to identify the gene and the causal mutation of the r1 rex hair trait to help breed rex rabbits and to better understand hair growth and its underlying metabolic pathways.
Results
Phenotype
At first, the phenotype of the rex and normal rabbit coat was determined by visual assessment of the presence of curled whiskers associated with the content and length of coarse fibre. Then fine phenotyping of rex and normal coats was undertaken. Further analysis included objective measurements of the length of both the outer and the inner coat, fibre diameter and coarse fibre content were performed on 44 rex and 54 and normal G2 rabbits.
There was no significant difference in fibre length between the outer and the inner coat in rex individuals (W = 1154; p = 0.1149) while in normal coat rabbits the outer coat is 9.8 mm longer than the inner coat (W = 1666; p = 5.2 10−9). The average length of both the outer coat and the inner coat is shorter than in normal rabbits (H = 72.53, p = 1.64 10−17 and H = 62.95, p = 2.12 10−15).
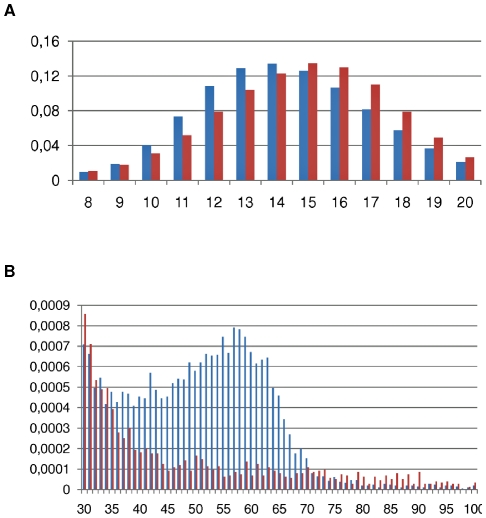
The distribution of hair diameter is bimodal in normal coat rabbits, the first peak corresponding to down hair ([6 µm–20 µm] centered on 14 µm with W = 0.90; p = 0.101), the second to guard hair ([45 µm–65 µm] centered on 55 µm with W = 0.96; p = 0.48). Awn hairs are less abundant and are distributed in between. The situation is quite different in rex animals especially concerning coarse hair. The down hair category (< = 20 µm) also shows a normal distribution as in normal coat but is shifted towards a larger diameter in the rex animals (Figure 2A). The second peak is not detectable due to a decrease in the number of large diameter hairs (Figure 2B).
Figure 2. Distribution of hair diameters in µm (X-axis) in normal (blue) and rex (red) rabbit coats.
Two diameter sizes ranges are represented on two different graphs to cope with the Y-axis scale (frequencies).
Assuming that 30 µm is the value of hair diameters above which humans feel a fur hard at touching, coats of non rex rabbits display a higher coarse fibre content (2.28%) than rex fur (0.96%). In addition, a larger number of secondary hair follicles were observed in rex than in non-rex rabbits by histology on 20 normal and 20 rex skin cross sections (Figure 1C) in accordance with the microscopic observation of fibre bundles (Figure 1B). We also observed that all primary hair follicles from rex rabbits were present in the follicular groups and each of these hair follicles produced a hair with circular shape like downs (Figure 1C).There were very highly significant differences (p<0.001) in the diameter and length of fibers as well as the coarse fiber content between the two coat phenotypes (Table 1).
Table 1. Mean length (± standard deviation) of outer coat, inner coat, fibre diameter and coarse fibre content (defined as the content of fibres having a fibre diameter larger than 30 µm) in rex and normal G2 rabbit coats.
| Presence | Length of | Length of | Diameter of inner | Diameter of outer coat | |||
| of curled | outer coat | inner coat | coat «downs» | «awns» | «guards» | ||
| n | whiskers | (mm) | (mm) | [0 µm; 20 µm] | [20 µm; 45 µm] | >45 µm | |
| Normal coat | 54 | no | 33.4±3.3 | 23.6±2.1 | 93.1% | 5.4% | 1.5% |
| Rex coat | 44 | yes | 18.9±1.9 | 95.9% | 3.7% | 0.4% | |
Finally, this study shows that the percentage of down hairs (< = 20 µm), awns ([20 µm; 45 µm]) and guard hairs (>45 µm) fits with the expected distribution of a follicular group in the common rabbit, one primary central, 2 to 4 primary lateral and 30 to 50 secondary follicles.
Mapping of the rex gene
Genome scan
The segregation of the rex trait in the G2 offspring confirmed an autosomal recessive determinism of this character. A whole genome scan was performed with 109 microsatellite markers from the first genetic map on rabbit families in which the rex trait was segregating [10]. Linkage analysis mapped the rex phenotype on chromosome 14 within an interval of 40 cM.
Fine mapping
In order to reduce the mapping interval of the trait, we produced and characterized microsatellites within the primo-localization region by two methods. First, we produced 19 of these polymorphic markers by screening the rabbit Bacterial Artificial Chromosome (BAC) library [11] for 14 Expressed Sequenced Tags (EST) potentially localized in the 40 cM interval and selected by comparative mapping with the homologous human chromosome region (HSA3). We also retrieved 152 microsatellite markers with a dinucleotide motif of at least 18 repetitions and well distributed in this HSA3 region by in silico screening. Forty-eight of these were selected and polymorphism was tested on a multi-origin rabbit panel and 23 were found segregating in our rabbit families. In total, we produced 171 markers in the first 40 cM interval and 42 were finally genotyped on our families for linkage analysis (19 from BAC library screening and 23 retrieved in silico).
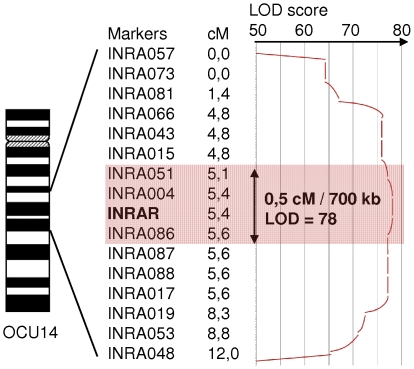
Microsatellite INRA004 shows the most significant linkage with the rex phenotype among the 359 rabbits (LOD = 78; ⊖ = 0). The final interval of localization for rex was estimated at 0.5 cM (700 kb) flanked by INRA051 (LOD = 31; ⊖ = 0.03) and INRA086 (LOD = 40; ⊖ = 0.01) markers. INRA086 belongs to a 4 marker linkage group (INRA086-087-088 and INRA017) with no recombinant and was arbitrarily considered as the lower limit of the interval (Figure 3).
Figure 3. Mapping of the rex coat trait in the rabbit families.
A whole genome scan performed on 187 rabbits for 47 microsatellites localized the rex phenotype (INRAR) on rabbit chromosome 14 within an interval of 40 cM. Fine mapping refined the localization within a 0.5 cM (⊖ = 0 ; LOD = 78) region flanked by microsatellites INRA051and INRA086.
Candidate Gene
In the last 0.5 cM interval, we identified three genes by comparative mapping of this region with mouse and human sequences since no appropriate rabbit sequence was available. Screening the rabbit BAC library identified a BAC sequence containing INRA017 and a part of the SENP2 gene (first gene upstream of the last interval) and a BAC containing the INRA004 and three genes: MAP3K13, TMEM41A and LIPH. MAP3K13 is a member of the serine/threonine protein kinase family which could play a role in the JNK (c-Jun NH(2)-terminal kinase) signaling pathway. TMEM41A is a transmembrane protein of unknown function and not yet characterized. LIPH is a membrane-bound member of the mammalian triglyceride lipase family, the phosphatidic acid-selective phospholipase A1 (PLA1). It specifically hydrolyzes phosphatidic acid (PA) to produce 2-acyl lysophosphatidic acid (LPA), which is a lipid mediator with diverse biological properties that include platelet aggregation, smooth muscle contraction, and stimulation of cell proliferation and motility. Disruption of the LIPH gene in the mouse results in various phenotypes including retarded hair growth and postnatal lethality [12]. Moreover, numerous mutations have been found in the LIPH gene. In 2006, the deletion of the region encompassing exon 4 was identified as being responsible for Hypotrichosis Simplex (HS) in 50 families of Russian people [13]. The Hair of these patients is abnormally short, dystrophic and fragile due to retarded or arrested hair growth. Mutations have also been reported in exon 1 [14], exon 2 [14]–[18],[21], exon 4 [21], exon 5 [17],[19]–[22], exon 6 [23],[24], exon 7–8 [21] and in exon 9 [22]. LIPH was thus considered as a strong candidate gene for the rex phenotype.
Tissue expression
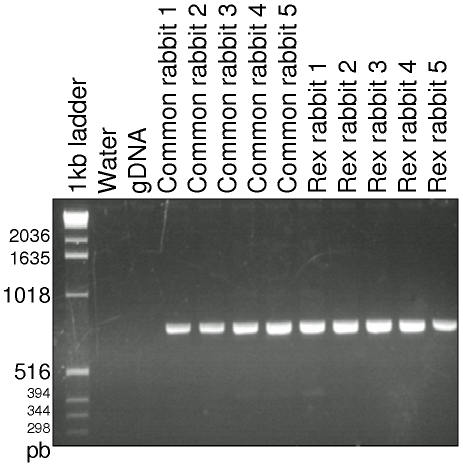
The expression of LIPH was detected in the intestines, muscles, kidney, lung, spleen and liver by RT-PCR (data not shown) and in the skin from the adult wild type and rex rabbits as shown in Figure 4. No alternative transcript was detected in the skin using the four primers covering the coding area (Table 2, Figure 4).
Figure 4. LIPH expression in rabbit skin.
LIPH was detected by RT-PCR (ex 7–10) in rabbit skin of 5 common and 5 rex rabbits. The expected product size is 753 pb.
Table 2. Primers used for RT–PCR.
| cDNA primers | Amplicon size (in pb) | |
| ex1-4 | F1_AACCAGGGGAAACCTAAAGC | 728 |
| R1_TCAGTGTCGGAATGGATGAC | ||
| ex2-8 | F2_CCAGTTTGGTTGCAGGACTT | 889 |
| R2_TGGATTCTGTGGTGTTTCCA | ||
| ex7-10 | F3_TGAGAAAGACCCTCCAATGA | 753 |
| R3_TGCTGATGGACAGCAGAATC | ||
| ex9-10 | F4_TCATCAAGTGAGTCTGCTTGC | 781 |
| R4_CACAGATGTGACACCCATGA |
Gene structure
Genomic DNA and messenger RNA sequencing allowed us to determine the LIPH gene structure in the rabbit. The gene is composed of 10 exons as in other species. The length of the open reading frame is 1932 pb with a predicted protein of 452 amino acids (Figure 5B).
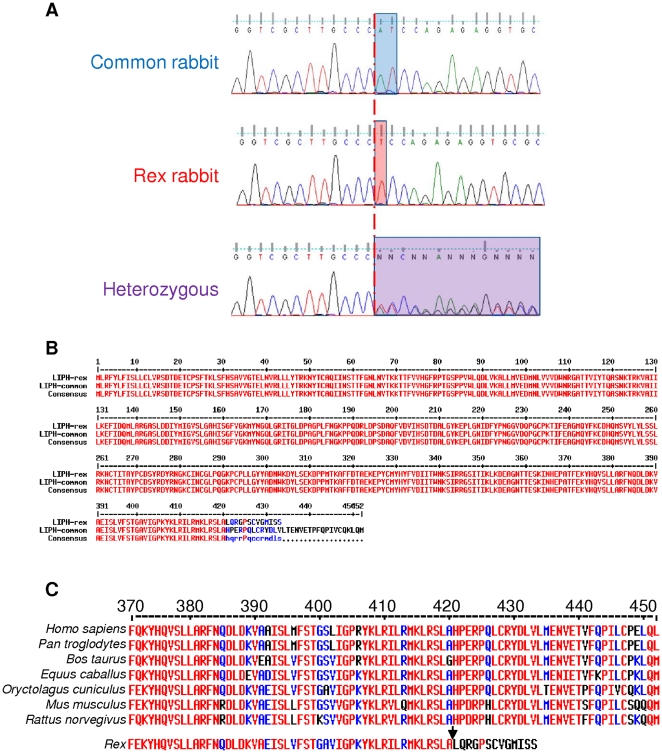
Figure 5. The 1362delA mutation in exon 9 of LIPH in rex rabbits.
(A) Electropherograms of the LIPH exon 9 sequence from a normal common type rabbit (WT), a rex and a heterozygous are shown. The red line indicates the location of the mutation. In heterozygous rabbits there is an overlap of both allele sequences (purple box). (B) Deduced alignment of LIPH proteins between rex and normal common type rabbits. (C) C-terminal conservation in mammalians.
Sequence analysis of this rabbit cDNA shows an identity of 90% compared to other mammalian species. The LIPH protein sequence is also highly conserved along mammalians with more than 80% of identity, including the catalytic domain and the C-terminal region (Figure 5C). We noticed that the rabbit LIPH lid domain is one amino acid longer (Glu at position 239) than that of the other species.
Mutation screening and distribution
By sequencing the complete coding region of the LIPH gene in 4 rex and 4 non rex rabbits, we identified a deletion of one nucleotide (1362delA) located in exon 9 and present in a homozygous state in the rex rabbit gene sequence (Figure 5A). This mutation leads to a frameshift and the appearance of a premature stop codon 32 amino acid downstream potentially resulting in a 19 amino acid truncated protein (Figure 5B). No other sequence differences were detected in the nine other exons of the gene.
The distribution of the 1362delA allele was firstly investigated among rabbits used for the mapping. The 8 G1 and 66 G2 common type rabbits were all heterozygous as expected, and the 8 G1 and 81 G2 rex rabbits were homozygous for the deletion. To confirm this result, the deletion distribution was investigated among unrelated rex animals and strains of standard rabbits. All 60 rex rabbits were tested homozygous for the 1362delA while all 60 rabbits that did not have the rex phenotype were tested homozygous for the wild allele.
Discussion
Macroscopic visual assessment for the presence of curled whiskers, length and content of coarse fibres is efficient to categorize rabbits having a typical defined phenotype in rex or normal coat type as proposed by Castle [8]. However due to variability amongst individuals, it is difficult to classify with certainty few animals having an intermediate coat. Objective measurements of the length of both the inner and the outer coat and coarse fiber content were found to be very helpful. Fine phenotyping using a combination of several criteria including, the length of both guard hair and down, the difference in length between outer and inner coat and measure of fiber diameter distribution greatly improved the classification of the animals. It allowed an accurate description of the rex phenotype in rabbits and was a necessary step to undertake for the positional cloning approach.
Rex rabbit guard hair and down were shorter than in the normal rabbit coat which was in agreement with earlier observations [6],[9]. The decrease in the length of guard hair (−43%) was double than that observed (−20%) in down leading to a closely similar length of both guard hair and down in rex rabbits. This situation can explain the often erroneous description of lacking guard hair in the rex rabbit due to a simple macroscopic view of the fur.
A decrease in hair length could be due to either a shorter duration of hair growth or a slower hair growth rate. By comparing rex and normal rabbit coats, Vrillon [9] did not observe any differences in the duration of the moulting process indicating that hair growth duration was similar in both genotypes and thus strengthening the hypothesis that a decrease in hair length could be due to a decrease in hair growth rate in agreement with observations in humans [13].
Histology allowed us to state that the lack of guard hair in rex rabbits was not due to the degenerescence of primary hair follicles. It also highlighted that primary hair follicles in rex were still active and produce hair. These guard hairs present a modification of their shape and an important decrease of diameter. The hair produced by the primary follicles seem to have disappeared but their reduction in size and shape, especially for guards, made them look like down. This is the reason why guard and awn hairs have been considered as lacking when fur was only visually assessed. It also explains why rex fur is soft touching, plush-like and represents a great value for breeders and the industry.
A positional cloning approach was undertaken in rabbit families segregating for the rex phenotype and for microsatellite markers. Linkage analysis after whole genome scanning and fine mapping allowed us to localize the rex coat trait in a 0.5 cM region on OCU14. In this 700 kb interval, comparative mapping with the human and mouse sequences pointed out LIPH as a strong candidate gene.
A LIPH lpd1 Knock Out mouse exhibited a complex multifactorial phenotype including retarded hair growth [12]. In humans, mutations in a small number of genes have been recently identified for a group of alopecia called Hypotrichosis Simplex (HS). Alopecia is characterized by patchy hair loss on the scalp that can eventually involve the entire scalp (alopecia totalis) or the entire body (alopecia universalis) [25]. Autosomal recessive hypotrichosis is characterized by sparse scalp hair and fragile hair. Three autosomal recessive forms LAH1, LAH2 and LAH3 have been found associated with three different genes, DSG4 [26], LIPH [13] and P2RY5 [27] respectively. Several lanes of evidence were in favour of the LIPH gene. We have shown by RT-PCR that LIPH was expressed in rabbit skin. Then, we established the LIPH gene structure by sequencing all its exons. The normal LIPH mRNA sequence presented in this study differs from that found in rabbit lachrymal glands by 5 nucleotide substitutions, one of which introduces an amino acid change (p.Ala369Val) [28].
A single polymorphism was found as a one base pair deletion in exon 9 (1362delA) of LIPH when comparing sequences from rabbits of normal wild and rex phenotypes. This deletion most probably leads to a premature termination of the rex LIPH protein. Segregation analysis of the 1362delA showed a complete linkage with the rex phenotype across families used for mapping and a total association with the trait on a 120 rabbit panel confirming the recessive determinism of the trait.
LIPH is a membrane-bound member belonging to the mammalian triglyceride lipase family. This gene has a phospholipase A1 activity that catalyses the production of 2-acyl LysoPhosphatidic Acid (LPA) by hydrolyzing Phosphatidic Acid [29]. LPA is a lipid mediator whose biological properties include platelet aggregation, smooth muscle contraction, and stimulation of cell proliferation and motility.
Hair formation is the result of the differentiation of keratinocytes during their migration along the hair shaft. Since LIPH encodes PA-PLA1α with a pla1 activity expressed in the skin, it has been speculated that loss-of-function of LIPH would reduce the production of LPA in hair follicles that could affect the migration, differentiation, or proliferation of keratinocytes, culminating in the arrest of hair growth [13]. PA-PLA 1α has three amino acid residues in the N-terminal domain, Ser154, Asp178 and His248, which form the putative catalytic triad [29]–[32], a β9 loop and a short lid domain which are considered as the structure involved in substrate recognition [29],[33],[34]. All the homozygous mutations in LIPH found in humans directly impact the lipase domain of the PA-PLA1α or lead to a premature termination codon (PTC).
In rex animals, the 1362delA mutation in LIPH affects the C-terminal of PA-PLA1α. Since the protein is localized to the plasma membrane [35], we hypothesize that the C-terminal region is involved in the final localization of LIPH where it is active. No specific amino acid sequence pattern was found in this region, but it is noticeable that the C-terminal region is highly conserved in mammalians. Moreover, in humans, a compound mutation including one in the C-terminal has been found leading to HS [22]. The phenotype was that of classic HS suggesting a role of this domain in the PA-PLA1α protein function. Rex rabbits show the first mutation in the C-terminal of PA-PLA1α in a homozygous state involved in a hair growth defect. To that point, it is not possible to exclude other hypotheses such as nonsense mediated decay (NMD) in which most likely aberrant transcripts with a PTC would be largely degraded. The 1362delA mutation could also affect the tertiary structure of PA-PLA1α and then reduce or destroy its activity.
The LIPH mutation found in this study to be associated with the rex coat trait confirms the importance of this gene in the hair follicle metabolism in mammals. Contrarily to the situation encountered in humans, this deletion in rabbits does not lead to sparse hair and is considered as an advantage for professionals. A molecular test is already available for breeders as a tool for improving their breeding programs or for introgression of the rex gene in any other rabbit population.
It is the first time that a mutation in the C-terminal carried in the homozygous state is shown to provoke a short hair phenotype, highlighting a possible important role of this domain in the LIPH function. Further studies are needed to define the role of this mutation in the dysfunction of the LIPH protein and its effect on hair growth retardation.
Materials and Methods
Rabbit families and reference population
Mating plan
Three rabbit INRA strains were used to build three generation families. Two crosses were carried out to produce the G1 generation : (i) eight INRA 2066 wild type (RR) males with 22 rex does (rr) and (ii) three rex males (rr) with 12 rex does (rr). The 53 G1 does (Rr) from the (i) cross were then mated with the 29 G1 males (rr) from the (ii) cross, which produced 853 G2 rabbits with theoretically 50% of homozygous rex (rr) and 50% of heterozygous showing wild phenotype (Rr). All rex rabbits used in this study come from the orylag® strain.
Phenotyping rabbits
Rabbits were phenotyped for coat texture. All G2 rabbits were firstly examined macroscopically by visual assessment for the presence of curled whiskers, the length and the content of coarse fibres. At first, a visual macroscopic observation was considered efficient to categorize rabbits with rex or normal coat type. To improve accuracy, a fleece sample was taken on all animals for objective measurements of the length of the outer and the inner coat with a rule. Finally, a subset of 44 rex and 54 normal coat G2 rabbits were further analyzed for mean fibre diameter, fibre distribution and content of coarse fibres. The latter was defined as the percentage of fibres having a diameter larger than 30 µm by measuring 4000 individual fibre snippets per sample using the Optical Fibre Diameter Analyzer (OFDA) methodology as previously described [36]. Royston's sign for the Shapiro-Wilk test was used to assess the normality of the hair distributions. Kruskall Wallis was then used to determine the difference between normal coat and rex rabbit hairs and inner/outer for both phenotypes.
Blood collection and DNA extraction
Eight milliliters of blood were sampled in vacutainers (K3-EDTA; Becton Dickinson, Rutherford, USA) by cardiac puncture on all rabbits. DNA was extracted from peripheral blood mononuclear cells as described by Jeanpierre [37]. This experiment was licensed under the guidelines of the French Ministry of Agriculture and in agreement with the rules of the National Committee of Animal Experimentation for animal research. Blood samples were drawn by authorized skilled staff in our Inra experimental station of Le Magneraud and all dispositions were taken to minimize suffering (authorization certificate to experiment on living animals N°78–103).
In France, no mandatory ethical committee approval is yet necessary to conduct experiments on the animals of this study.
Mapping of the rex gene
Primo localization
A genome scan was achieved by genotyping 187 G2 rabbits for the 109 microsatellite markers of the genetic rabbit map [10]. A low-cost technique described by Schuelke (2000) was applied for the genotyping of microsatellites. The PCR reaction was carried out with three primers, including a locus specific primer extended with a universal sequence of 17 nucleotides (5′- GACCGGCAGCAAAATTG-3′), a reverse locus specific primer and a universal primer of 17 nucleotides that was 6-Fam, Hex or Tet 5′-labelled (MWG AG Biotech, Ebersberg, Germany). The results were analysed with the Genetic Profilerv1.1 software (ABIPrism377A sequencer; Applied Biosystems, Foster City, CA, USA) or the Genotyper software (MegaBACE1000 sequencer; Amersham Biosciences, New Haven, CT, USA). Genotyping data were checked using an in-house program, which analyzed the consistency of the allele distribution of each marker according to pedigrees.
The CRI-MAP 2.4 software (Green et al., 1990) was used to build the genetic map. The first step identified linked markers by a two-point analysis with the two-point option. In the second step, the linkage groups were examined by multipoint analysis using the build and flipsn options. Linkage data were merged with cytogenetic mapping data to confirm the order of markers or to identify linkage of isolated or weakly informative markers. These various analyses were performed with a LOD score of 3 (lowered to 1.8 in a few cases where the cytogenetic position of the markers made it possible to confirm the proximity). The size of the genetic map was calculated by adding up the genetic distances of all linkage groups plus 15 cM at both ends of each group and including 15 cM on both sides of each unlinked marker.
Fine Mapping
Three-hundred fifty-nine rabbits belonging to a total of 17 families were genotyped with an additional 42 microsatellites. The CRI-MAP software version 2.4 [38] was used to construct the map. We used MultiMap to generate maps and calculate the probabilities for orders obtained from an initial order of reference, by permutation of n adjacent loci.
Characterization of microsatellite markers
Microsatellite markers localized in the primo localization area were produced by molecular genetic approaches in BAC as described by Chantry-Darmon [39].
An in silico approach was also used. The rabbit sequences OryCun1 (2× low-coverage assembly, 777141 sequences in Ensembl comprising traces, EST, contigs, scaffolds and not located on the genome) were compared by BLAST with the human masked sequences of chromosome 3 for the 170 Mb–190 Mb interval of primary location (OCU 14q17). Only sequences with a significant BLAST score set with an “e” value greater than or equal to 10−40 were kept. Microsatellite identification in these sequences has been implemented using Tandem Repeat Finder [40]. Finally, primers to amplify the microsatellites were designed using Primer3 [41]. Three zones of fragment sizes were chosen to allow multiplexing: 100±20 bp, 150±20 and 200±20 bp. We selected uninterrupted microsatellites with at least 18 repetitions in order to improve the probability of observing polymorphism.
Linkage analysis
We used Simwalk2 version 2.91 [42] to perform linkage analysis based on the Z score method taking the family structure, the number of individuals and markers into account. A parametric test assuming a recessive model with complete penetrance and a nonparametric test, with the respective options “parametric linkage analysis” and “non-parametric linkage (NPL) analysis” were used.
Haplotype analysis was also conducted with Simwalk2 v 2.91 and the results were analysed using Haplopainter version 27beta [43] and Ghostscript 8.54.
Gene structure analysis
Total RNA were extracted from different tissues using the RNA Now procedure (Biogentex). Reverse transcriptions (RT) were performed on 5 µg of total RNA using the Superscript First Strand Synthesis System (Invitrogen) following the manufacturer's instructions. RT on rabbit RNA were performed using oligonucleotides(dT)18 followed by PCR using the GoTaq® Flexi DNA Polymerase and reaction buffer (Promega).
The cDNA sequencing of rabbit LIPH was performed after RT-PCR amplification using 4 primer pairs designed to amplify overlapping fragments 500 à 600 bp long covering all the exons of the genes (Table 2). All oligonocleotides were designed using the primer 3 software (http://frodo.wi.mit.edu/primer3/). The PCR amplification reactions were optimized and carried out in a PTC-100 (MJ-Research) using the following cycling conditions: 95°C for 5 min followed by 35 cycles of (95°C for 30 s, annealing temperatures 60°C for 30 s, 72°C for 30 s) and 72°C for 5 min. The resulting PCR products were separated on a 2% agarose gel, purified using Wizard SV Gel and PCR Clean-Up System (Promega, Wisconsin, USA) and sequenced by Qiagen sequencing services. The sequences were assembled in clusters of contiguous sequences using the CAP3 assembler [44] using default parameters.
The resulting sequences were compared to the rabbit and human genome sequences in the NCBI database by means of the BLASTN software (http://www.ncbi.nlm.nih.gov) to deduce the intron/exon structure of the LIPH gene. All LIPH genomic and cDNA sequences generated during the course of this study have been submitted to GenBank databases (HQ845290).
LIPH Polymorphism
All exons of the LIPH gene were amplified using primers in the introns and both sides of the exons for 4 rex and 4 common rabbits. The PCR were performed on genomic DNA extracted from blood of our rabbit families. PCR reactions were optimized using the following cycling conditions: 95°C for 5 min followed by 35 cycles of (95°C for 30 s, annealing temperatures (Table 3) for 30 s, 72°C for 30 s) and 72°C for 5 min. DNA fragments were then analyzed for the gene structure analysis as described above. Primers used are listed in Table 1. The resulting sequences were introduced into the NovoSNP software [45] to find Single Nucleotide Polymorphism in the amplified sequences and compared using “multalign” [46] to find any other polymorphism.
Table 3. Primers used for genomic amplification.
| Exon size | gDNA primers | Amplicon size | Annealing temperature | |
| (in bp) | (in bp) | (in °C) | ||
| Exon 1 | 149 | F1_GCCAGGCACCATTCTAAAAG | 345 | 60°C |
| R1_TGAAAAGAGGAGGAGCCAAA | ||||
| Exon 2 | 368 | F2_TGGGGCTTATTCAGATTTGC | 574 | 60°C |
| R2_TGCAGTGAATAGCAGAGGATTC | ||||
| Exon 3 | 109 | F3_CAGCTAGGACACTTCTCCAAA | 292 | 60°C |
| R3_TGGAAAAGCTGGCTTTGAAC | ||||
| Exon 4 | 102 | F4_ACGAACCATCGAATCAGGAA | 267 | 60°C |
| R4_GCTACCCCCAGGGAGAGACT | ||||
| Exon 5 | 90 | F5_GCTCCCTCTCTCGCTGTAAC | 400 | 60°C |
| R5_CGCAGCTAACTATTGGGGTAA | ||||
| Exon 6 | 171 | F6_AGGGAAACTGCTTGTTGGAC | 208 | 58°C |
| R6_CTCTACAAAGCCAGGGATGC | ||||
| Exon 7 | 96 | F7_AGAAGTGGCTGGGAACCTG | 262 | 60°C |
| R7_CCAATGCGCTGTTCTCATTA | ||||
| Exon 8 | 112 | F8_AGAGCAGAAGTGCAGAACCA | 299 | 60°C |
| R8_GTTATCAGGGGGATGGGTTT | ||||
| Exon 9 | 174 | F9_TCTCCCTGACTTTTCCTACTTCA | 298 | 58°C |
| R9_CCCTCCCCAAATAAATCTTTTAAC | ||||
| Exon 10 | 607 | F10_ACACTGCAGAGAAGGCAGGT | 836 | 60°C |
| R10_AGCGTGGCTCCTGTTCATTA |
Association study
A first panel of rabbits used for mapping was genotyped by direct sequencing for the deletion using primers F9/R9 (Table 3). This panel was constituted by 8 G1 normal and 8 G1 orylag® rex-rabbits and 66 G2 wild type rabbits and 81 G2 orylag® rex-rabbits. A panel of 60 common rabbits and 60 unrelated orylag® rex-rabbits was then genotyped.
Acknowledgments
We thank Aurélie Ducasse for helping in rabbit material collection, Dr Laurent Schibler and Dr Mathieu Gauthier for their advice during the mapping and linkage analysis.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a grant from the Poitou-Charentes region, the Animal Genetics Department and by the Animal Genetics and Integrative Biology unit of INRA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Robinson R. German rex: a rexoid coat mutant in the cat. Genetica. 1968;39:351–352. doi: 10.1007/BF02324474. [DOI] [PubMed] [Google Scholar]
- 2.Gandolfi B, Outerbridge CA, Beresford LG, Myers JA, Pimentel M, et al. The naked truth: Sphynx and Devon Rex cat breed mutations in KRT71. Mamm Genome. 2010;21:509–515. doi: 10.1007/s00335-010-9290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson R. Rex mutant in the Norway rat. J Hered. 1981;72:131–132. doi: 10.1093/oxfordjournals.jhered.a109445. [DOI] [PubMed] [Google Scholar]
- 4.Kuramoto T, Hirano R, Kuwamura M, Serikawa T. Identification of the rat Rex mutation as a 7-bp deletion at splicing acceptor site of the Krt71 gene. J Vet Med Sci. 2010;72:909–912. doi: 10.1292/jvms.09-0554. [DOI] [PubMed] [Google Scholar]
- 5.Fae C. Rex: a dominant autosomal monogenic coat texture character in the mouse. J Genetica. 1939;38:341–344. [Google Scholar]
- 6.Castle WE. Linkage interrelations of three genes for rex (short) coat in the rabbit. Proc Nat Acad Sci. 1933;19:1006–1011. doi: 10.1073/pnas.19.12.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rougeot J, Thebault R-G. Le lapin angora: éditions Point Vétérinaire. 1989. p. 184 p.
- 8.Castle WE. The rex rabbit. Journ Heredity. 1929;20:193–199. [Google Scholar]
- 9.Vrillon JL, Thebault RG, Rochambeau Hd, Dardant P. Photoperiodism effect on fur maturity and fur quality of rabbits, owning or not rex gene. 1988. pp. 244–252. Proceedings “4th Congress of the World Rabbit Science Association”. Budapest, Hungary.
- 10.Chantry-Darmon C, Urien C, de Rochambeau H, Allain D, Pena B, et al. A first-generation microsatellite-based integrated genetic and cytogenetic map for the European rabbit (Oryctolagus cuniculus) and localization of angora and albino. Anim Genet. 2006;37:335–341. doi: 10.1111/j.1365-2052.2006.01462.x. [DOI] [PubMed] [Google Scholar]
- 11.Rogel-Gaillard C, Piumi F, Billault A, Bourgeaux N, Save JC, et al. Construction of a rabbit bacterial artificial chromosome (BAC) library: application to the mapping of the major histocompatibility complex to position 12q.1.1. Mamm Genome. 2001;12:253–255. doi: 10.1007/s003350010260. [DOI] [PubMed] [Google Scholar]
- 12.Wen XY, Bryce DM, Breitman ML. Characterization of lpd (lipid defect): a novel mutation on mouse chromosome 16 associated with a defect in triglyceride metabolism. Hum Mol Genet. 1998;7:743–750. doi: 10.1093/hmg/7.4.743. [DOI] [PubMed] [Google Scholar]
- 13.Kazantseva A, Goltsov A, Zinchenko R, Grigorenko AP, Abrukova AV, et al. Human hair growth deficiency is linked to a genetic defect in the phospholipase gene LIPH. Science. 2006;314:982–985. doi: 10.1126/science.1133276. [DOI] [PubMed] [Google Scholar]
- 14.Naz G, Khan B, Ali G, Azeem Z, Wali A, et al. Novel missense mutations in lipase H (LIPH) gene causing autosomal recessive hypotrichosis (LAH2). J Dermatol Sci. 2009;54:12–16. doi: 10.1016/j.jdermsci.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Ali G, Chishti MS, Raza SI, John P, Ahmad W. A mutation in the lipase H (LIPH) gene underlie autosomal recessive hypotrichosis. Hum Genet. 2007;121:319–325. doi: 10.1007/s00439-007-0344-0. [DOI] [PubMed] [Google Scholar]
- 16.Nahum S, Pasternack SM, Pforr J, Indelman M, Wollnik B, et al. A large duplication in LIPH underlies autosomal recessive hypotrichosis simplex in four Middle Eastern families. Arch Dermatol Res. 2009;301:391–393. doi: 10.1007/s00403-008-0903-9. [DOI] [PubMed] [Google Scholar]
- 17.Petukhova L, Shimomura Y, Wajid M, Gorroochurn P, Hodge SE, et al. The effect of inbreeding on the distribution of compound heterozygotes: a lesson from Lipase H mutations in autosomal recessive woolly hair/hypotrichosis. Hum Hered. 2009;68:117–130. doi: 10.1159/000212504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasternack SM, von Kügelgen I, Müller M, Oji V, Traupe H, et al. In vitro analysis of LIPH mutations causing hypotrichosis simplex: evidence confirming the role of lipase H and lysophosphatidic acid in hair growth. J Invest Dermatol. 2009;129:2772–2776. doi: 10.1038/jid.2009.154. [DOI] [PubMed] [Google Scholar]
- 19.Jelani M, Wasif N, Ali G, Chishti M, Ahmad W. A novel deletion mutation in LIPH gene causes autosomal recessive hypotrichosis (LAH2). Clin Genet. 2008;74:184–188. doi: 10.1111/j.1399-0004.2008.01011.x. [DOI] [PubMed] [Google Scholar]
- 20.Kamran-ul-Hassan Naqvi S, Raza SI, Naveed AK, John P, Ahmad W. A novel deletion mutation in the phospholipase H (LIPH) gene in a consanguineous Pakistani family with autosomal recessive hypotrichosis (LAH2). Br J Dermatol. 2009;160:194–196. doi: 10.1111/j.1365-2133.2008.08822.x. [DOI] [PubMed] [Google Scholar]
- 21.Shimomura Y, Wajid M, Petukhova L, Shapiro L, Christiano AM. Mutations in the lipase H gene underlie autosomal recessive woolly hair/hypotrichosis. J Invest Dermatol. 2009;129:622–628. doi: 10.1038/jid.2008.290. [DOI] [PubMed] [Google Scholar]
- 22.Shimomura Y, Wajid M, Zlotogorski A, Lee YJ, Rice RH, et al. Founder mutations in the lipase h gene in families with autosomal recessive woolly hair/hypotrichosis. J Invest Dermatol. 2009;129:1927–1934. doi: 10.1038/jid.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimomura Y, Ito M, Christiano AM. Mutations in the LIPH gene in three Japanese families with autosomal recessive woolly hair/hypotrichosis. J Dermatol Sci. 2009;56:205–207. doi: 10.1016/j.jdermsci.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinkuma S, Akiyama M, Inoue A, Aoki J, Natsuga K, et al. Prevalent LIPH founder mutations lead to loss of P2Y5 activation ability of PA-PLA1alpha in autosomal recessive hypotrichosis. Hum Mutat. 2010;31:602–610. doi: 10.1002/humu.21235. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Mir A, Zlotogorski A, Gordon D, Petukhova L, Mo J, et al. Genomewide scan for linkage reveals evidence of several susceptibility loci for alopecia areata. Am J Hum Genet. 2007;80:316–328. doi: 10.1086/511442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kljuic A, Bazzi H, Sundberg JP, Martinez-Mir A, O'Shaughnessy R, et al. Desmoglein 4 in hair follicle differentiation and epidermal adhesion: evidence from inherited hypotrichosis and acquired pemphigus vulgaris. Cell. 2003;113:249–260. doi: 10.1016/s0092-8674(03)00273-3. [DOI] [PubMed] [Google Scholar]
- 27.Pasternack SM, von Kügelgen I, Aboud KA, Lee YA, Rüschendorf F, et al. G protein-coupled receptor P2Y5 and its ligand LPA are involved in maintenance of human hair growth. Nat Genet. 2008;40:329–334. doi: 10.1038/ng.84. [DOI] [PubMed] [Google Scholar]
- 28.Remington SG, Nelson JD. mRNA encoding a new lipolytic enzyme expressed in rabbit lacrimal glands. Invest Ophthalmol Vis Sci. 2002;43:3617–3624. [PubMed] [Google Scholar]
- 29.Sonoda H, Aoki J, Hiramatsu T, Ishida M, Bandoh K, et al. A novel phosphatidic acid-selective phospholipase A1 that produces lysophosphatidic acid. J Biol Chem. 2002;277:34254–34263. doi: 10.1074/jbc.M201659200. [DOI] [PubMed] [Google Scholar]
- 30.Aoki J, Inoue A, Makide K, Saiki N, Arai H. Structure and function of extracellular phospholipase A1 belonging to the pancreatic lipase gene family. Biochimie. 2007;89:197–204. doi: 10.1016/j.biochi.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 31.Jin W, Broedl UC, Monajemi H, Glick JM, Rader DJ. Lipase H, a new member of the triglyceride lipase family synthesized by the intestine. Genomics. 2002;80:268–273. doi: 10.1006/geno.2002.6837. [DOI] [PubMed] [Google Scholar]
- 32.Kubiak RJ, Yue X, Hondal RJ, Mihai C, Tsai MD, et al. Involvement of the Arg-Asp-His catalytic triad in enzymatic cleavage of the phosphodiester bond. Biochemistry. 2001;40:5422–5432. doi: 10.1021/bi002371y. [DOI] [PubMed] [Google Scholar]
- 33.Aoki J, Inoue A, Okudaira S. Two pathways for lysophosphatidic acid production. Biochim Biophys Acta. 2008;1781:513–518. doi: 10.1016/j.bbalip.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Carrière F, Withers-Martinez C, van Tilbeurgh H, Roussel A, Cambillau C, et al. Structural basis for the substrate selectivity of pancreatic lipases and some related proteins. Biochim Biophys Acta. 1998;1376:417–432. doi: 10.1016/s0304-4157(98)00016-1. [DOI] [PubMed] [Google Scholar]
- 35.Hiramatsu T, Sonoda H, Takanezawa Y, Morikawa R, Ishida M, et al. Biochemical and molecular characterization of two phosphatidic acid-selective phospholipase A1s, mPA-PLA1alpha and mPA-PLA1beta. J Biol Chem. 2003;278:49438–49447. doi: 10.1074/jbc.M213018200. [DOI] [PubMed] [Google Scholar]
- 36.Rafat SA, de Rochambeau H, Brims M, Thebault RG, Deretz S, et al. Characteristics of Angora rabbit fiber using optical fiber diameter analyzer. J Anim Sci. 2007;85:3116–3122. doi: 10.2527/jas.2007-0109. [DOI] [PubMed] [Google Scholar]
- 37.Jeanpierre M. A rapid method for the purification of DNA from blood. Nucleic Acids Res. 1987;15:9611. doi: 10.1093/nar/15.22.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Green P. 1988. Rapid construction of multilocus genetic linkage maps. I. Maximum likelihood estimation.
- 39.Chantry-Darmon C, Bertaud M, Urien C, Chadi-Taourit S, Perrocheau M, et al. Expanded comparative mapping between man and rabbit and detection of a new conserved segment between HSA22 and OCU4. Cytogenet Genome Res. 2005;111:134–139. doi: 10.1159/000086382. [DOI] [PubMed] [Google Scholar]
- 40.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 42.Sobel E, Sengul H, Weeks DE. Multipoint estimation of identity-by-descent probabilities at arbitrary positions among marker loci on general pedigrees. Hum Hered. 2001;52:121–131. doi: 10.1159/000053366. [DOI] [PubMed] [Google Scholar]
- 43.Thiele H, Nürnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics. 2005;21:1730–1732. doi: 10.1093/bioinformatics/bth488. [DOI] [PubMed] [Google Scholar]
- 44.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weckx S, Del-Favero J, Rademakers R, Claes L, Cruts M, et al. novoSNP, a novel computational tool for sequence variation discovery. Genome Res. 2005;15:436–442. doi: 10.1101/gr.2754005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]