Abstract
Often, analysis for pharmacogenomic studies involving multiple drugs from the same class is completed by analyzing each drug individually for association with genomic variation. However, by completing the analysis of each drug individually, we may be losing valuable information. When studying multiple drugs from the same drug class, one may wish to determine genomic variation that explains the difference in response between individuals for the drug class, as opposed to each individual drug. Therefore, we have developed a multivariate model to assess whether genomic variation impacts a class of drugs. In addition to determine genomic effects that are similar for the drugs, we will also be able to determine genomic effects that differ between the drugs (that is, interaction). We will illustrate the utility of this multivariate model for cytotoxicity and genomic data collected on the Coriell Human Variation Panel for the class of anti-purine metabolites (6-mercaptopurine and 6-thioguanine).
Keywords: class of drugs, cytotoxicity, mRNA expression data, statistical analysis, thiopurines
Introduction
Thiopurines, such as 6-mercaptopurine (6-MP), 6-thioguanine (6-TG) and azathioprine, a prodrug that is converted to 6-MP in vivo, are widely used to treat acute lymphoblastic leukemia of childhood and autoimmune disorders.1 Various factors, including polymorphisms and structural variation in DNA, differences in gene expression levels, gender, ethnicity and drug–drug interactions, affect variation in thiopurine drug response. Among these factors, gene expression profiles have been used to identify candidate genes that contribute to variation in drug response on the basis of mRNA expression relative to drug response.2 Traditionally, studies aimed to maximize efficacy and minimize toxicity of chemotherapeutic agents have focused on genes known to have important roles in the pharmacokinetic and pharmacodynamic pathways of a particular drug. 6-MP and 6-TG are prodrugs that must undergo metabolic conversion to form the active drug metabolites, 6-thioguanine nucleotides (6-TGNs), followed by incorporation into DNA to exert their anti-neoplastic and anti-inflammatory effects.1 As these metabolites are critical for the therapeutic effect of thiopurines, 6-TGN concentrations have been used as an index of the therapeutic and toxic effects of these drugs.3, 4, 5
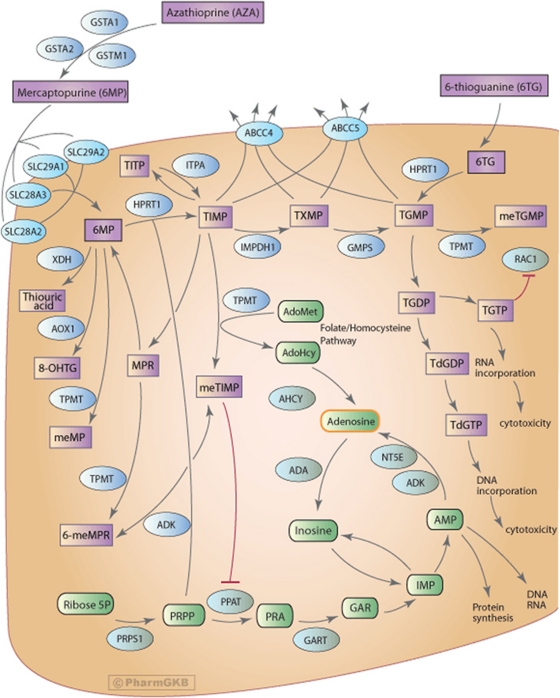
Several genes within the thiopurine-metabolizing pathway have effects on individual variation in the accumulation of 6-TGNs. Figure 1 displays a diagram of the thiopurine pathway as presented in PharmGKB (http://www.pharmgkb.org/index.jsp).6 Thiopurine S-methyltransferase) is a primary factor responsible for variation in the quantity of thiopurine available for enzymatic reactions that lead to the formation of 6-TGNs.7 However, variation in thiopurine S-methyltransferase activity does not account for all of the adverse reactions or poor responses to thiopurine treatment.8 The analyses of the well-studied thiopurine drug-metabolizing pathway genes may provide insights into possible mechanisms responsible for the accumulation of intracellular 6-TGNs involving thiopurine circulation.
Figure 1.
The thiopurine pathway.
In the late 1980s, the National Cancer Institute developed a collection of human tumor cell lines (NCI60) from a variety of common solid tumors (lung, colon and breast) for anti-cancer drug screening.9 Although tumor DNA is important for response to chemotherapy, it is hard to come by and increasing evidence has shown that germline DNA is as important as tumor DNA. Therefore, recently, pharmacogenomic research has incorporated non-tumor cell-based model systems that represent common genetic variation among individuals.10, 11 These cell-based model systems have been used to study multiple drugs and thus, a comprehensive set of drug-related endpoints are available for a set of cell lines.
Traditional analysis approaches for pharmacogenomic studies involve analyzing each drug individually for association with genetic variation. However, by completing the analysis of each drug individually, we may be losing valuable information. When studying multiple drugs from the same class of drugs that have similar genetic mechanisms, one may wish to determine genomic variation that explains the difference in cytotoxicity between individuals for the class of drugs as opposed to each individual drug. By analyzing the drugs in the same class together we hope to increase ability to detect genomic associations with both drugs (that is, genetic effects that affect the class of drugs). In addition to a possible increase in power, we will be able to determine whether the genetic variation is associated with the class of drugs or whether the genetic variation affects the drugs differently (that is, interaction).
Therefore, we have developed a multivariate model to assess whether genomic variation impacts a class of drugs and have applied this model to the analysis of thiopurines. These models will allow us to make statements about the class of drugs as opposed to individual drugs. The results from the analysis of a class of drugs may assist researchers in generating hypotheses that will lead to better understanding of the complex nature of the relationship between genomic variation and drug response. This will lead eventually to the development of ‘individualized therapy' for cancer patients, presuming a genomic relationship is found and validated.
Materials and methods
Pharmacogenomic study of anti-purine metabolite drugs
Cell lines, drug and cell proliferation assay
Epstein-Barr virus-transformed lymphoblastoid cell lines derived from 58 Caucasian-American (CA), 53 African-American, 60 Han Chinese-American and 23 Centre d'Etude du Polymorphisme Humain (CA) unrelated subjects were purchased from the Coriell Institute (Camden, NJ, USA). The drugs 6-MP and 6-TG were purchased from Sigma (St Louis, MO, USA). Drugs were prepared in dimethyl sulfoxide immediately before use and further diluted with media. Cells were plated at a density of 5 × 104 cells per well in triplicate in 96-well plates (Corning, Corining, NY, USA). Around 1 h after plating, cells were treated with 6-MP or 6-TG. The CellTiter96 Aqueous Non-Radioactive Cell Proliferation Assays (Promega, Madison, WI, USA) were performed as described by the manufacturer after 72 h incubations. Plates were read in a Safire2 microplate reader (Tecan AG, Mannedorf, Switzerland), with subsequent cytotoxicity measurements recorded at various doses of 6-MP and 6-TG for the cell lines.
Basal affymetrix U133 Plus2.0 GeneChip gene expression data
Whole Genome expression data for cell lines was obtained with Affymetrix U133 plus 2.0 expression array chip. The RNA extraction and the expression array assays were performed following the Affymetrix GeneChip expression technical manual (Affymetrix, Santa Clara, CA, USA). Before the assay, RNA quality was tested using an Agilent 2100 Bioanalyzer. The Affymetrix GeneChip contains over 54 000 probe sets the design of which is based on build 34 of the Human Genome Project. The mRNA expression array data were normalized on the log2 scale using GC Robust Multi-array Average methodologies.12, 13, 14
Model for analysis of a class of drugs
By analyzing the drugs in the same class together in a mixed model framework we hope to increase the ability to detect genomic effects for the class of drugs. We will also be able to determine whether the genomic variation affects the drugs differently (that is, interaction). Below, we outline the model for joint analysis of multiple drugs. The multivariate mixed model proposed for analyzing a class of drugs is
where Yij is the quantitative phenotype value for the ith subject/cell line treated with drug j, Dj is an effect for drug j, Xi is the genomic variable for subject/cell line i, Dj × Xi is the interaction between drug and genomic variable, and a random effect αi to account for the dependency in multiple measurements taken off the same subject/cell line. Lastly, we allow both random variables to follow independent normal distributions with constant variance (αi∼N(0,σα2) and ɛij∼N(0,σ2)). This results in Yij following a normal distribution with mean μij and variance σ2+σα2. The covariance between measurements taken off the subject is σα2, and the covariance between measurements taken off different subjects is 0. This results in the standard mixed model specification for repeated measures, with a covariance matrix that has a compound symmetry structure.15 Likelihood estimates of the fixed effects (genomic main effects and interaction) can be estimated and tested using maximum likelihood methods with estimation of the variance components completed using restricted-maximum likelihood methods.15 If no important interaction between the drug and genomic variable is present, inference for genomic effect will be assessed with a significant effect for the genomic variable (single-nucleotide polymorphism, mRNA expression, copy number and so on) indicating the genomic variable impacts the ‘class of drugs'.
Statistical analysis of 6-TG and 6-MP Pharmacogenomic Study
Estimation of the IC50 phenotype (effective dose that kills 50% of the cells) was calculated from a four parameter logistic model for both 6-TG and 6-MP cytotoxicity data for all cell lines.16, 17 The normalized log2 expression data were regressed on gender, race and time since Coriell submission (dichotomized at 10 years). The binary variable of time since Coriell submission was included to adjust for the differences observed in expression values with respect to time since Coriell submission. The residuals from this regression model were then standardized, resulting in a standardized, adjusted, normalized mRNA expression value. The IC50 values were log transformed because of extreme skewness in the distributions, and then in a similar fashion adjusted for gender, race and time since Coriell submission (dichotomized at 10 years) before standardizing. On examination of the distributions for the adjusted standardized IC50 values and the standardized, adjusted, normalized expression values, large outliers were observed in the distributions. As outliers can have a large impact on the results from a mixed model and interaction effects, we removed outliers before analysis. An analysis without removing the outlier points was also conducted and confirmed that the outlier points were highly influential and skewed the results, as seen in Supplementary Figure 1 and Supplementary Table 1. Subjects with standardized IC50 values greater than 4 units in magnitude (outliers with values more than 4 s.d. from the mean) were removed (two Han Chinese-American cell lines and one CA cell line). In addition to removal of cell lines with extreme values for IC50, outlier values for expression were also removed on the basis of a 4 s.d. rule (0.79% removed). Comparison of results from the analysis with no outliers removed was also completed as a sensitivity analysis. Results from the analysis without the removal of outliers are presented in Supplementary Table 1. The multivariate analysis outlined in 'Model for analyses of a class of drugs' was completed using the transformed, adjusted IC50 values and the normalized adjusted mRNA expression probe set values. SAS code used to fit the linear mixed model for the class of drugs analysis is presented in the online Supplementary Material, along with details on the format of the data file and output generated from the mixed model.
Results for analysis of thiopurine drugs 6-TG and 6-MP
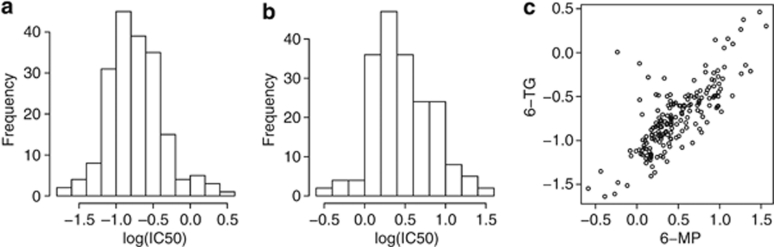
The multivariate model described in 'Model for analyses of a class of drugs' was applied to a pharmacogenomic study of the thiopurine drugs 6-TG and 6-MP as described in ‘Statistical analysis of 6-TG and 6-MP Pharmacogenomic Study.' The correlation between the IC50 for the two drugs was 0.78. Figure 2 displays the distributions for the two drug phenotypes and the relationship between these phenotypes. The analysis was completed first of the probe sets within the thiopurine pathway, followed by an agnostic genome-wide analysis. Before assessing the significance of the expression effects on the class of drugs (that is, main effects), assessment of the interaction effects must be completed. If a significant interaction effect between drug and mRNA expression is observed, the main effect of expression on the IC50 is uninterruptable. Results from the thiopurine pathway analysis, consisting of 30 probe sets, with P-values <0.05 are presented in Table 1. The results showed some evidence that probe sets for genes IMPDH1 (P=0.0002), PRPS1 (P=0.0008), GART (P=0.033) and ABCC5 (P=0.049) have different expression effects on IC50 for 6-TG and 6-MP (that is, interaction effect). However, because of testing 30 probe sets simultaneously, only genes IMPDH1 and PRPS1 were significant after applying a Bonferroni correction (significance level of 0.002). As for genes with a similar effect on 6-TG and 6-MP IC50 (expression main effect), there is evidence of an association with NT5E (P=0.048 and P=0.016) and thiopurine S-methyltransferase (P=0.047). However, none of the probe sets are significant after adjusting for multiple testing.
Figure 2.
Plots of the standardized IC50 phenotypes; (a) histogram of the 6-TG IC50 values, (b) histogram of the 6-MP IC50 values and (c) scatterplot of the 6-TG and 6-MP IC50 values.
Table 1. Results from multivariate analysis for 30 probe sets within the thiopurine pathway.
| Probe set | Gene | P-value for interaction effect | P-value for expression main effect |
|---|---|---|---|
| 203939_at | NT5E | 0.395 | 0.048 |
| 203672_x_at | TPMT | 0.092 | 0.047 |
| 1553995_a_at | NT5E | 0.331 | 0.016 |
| 204169_at | IMPDH1 | 0.0002a | 0.521 |
| 209440_at | PRPS1 | 0.0008a | 0.735 |
| 212379_at | GART | 0.033 | 0.874 |
| 226363_at | ABCC5 | 0.049 | 0.705 |
Probe sets with effects with P-value <0.05 are displayed.
Statistically significant after adjusting for multiple testing within the pathway (Bonferroni threshold of 0.002).
Next we took an unbiased or agnostic approach and completed a genome-wide multivariate analysis to assess whether variation in mRNA expression for genes outside the drug pathway had an effect on the IC50 for the class of thiopurine drugs. At the 0.0001 significance level, under the null hypothesis of no association we would expect to have 5.5 probe sets with P-values <0.0001. For the analysis of drug by expression interaction effects and expression main effects, we observed 17 and 42 probe sets with P-values <0.0001, respectively. Thus, there appears to be a slight deviation from the null hypothesis for both interaction and main effects, with statistically significant main effects for genes C10orf76 (P=6.19 × 10−9) and HD (1.29 × 10−7) and statistically significant interaction effects for ZNF547 (P=2.79 × 10−7) and SFRS15 (9.0 × 10−7). Upon future investigation of the significant interaction effects, the univariate analysis of 6-TG IC50 with level of mRNA for ZNF547 resulted in a correlation of 0.18 (P=0.01), whereas 6-MP had a correlation of −0.07 (P=0.29); the correlation between level of mRNA for SFRS15 and 6-MP IC50 was −0.07 (P=0.328), whereas 6-TG had a correlation of 0.12 (P=0.096). Thus, for both genes, it appears that there is a positive relationship between mRNA expression level and 6-TG IC50 and no relationship (or slight negative relationship) with 6-MP IC50. The results for probe sets with P-values <10−5 are presented in Table 2. Five probe sets had P-values for expression main effects <10−5 (similar effect of mRNA expression for both drugs), with two probe sets falling in the gene C10orf76. The q-values for these five probe sets ranged from 0.0003 to 0.057. Four probe sets (related to three known genes) were associated with different expression effects for 6-TG and 6-MP IC50, with P-values <10−5 and q-values ranging from 0.013 to 0.066.
Table 2. Probe sets with P-value for main effects or interaction effects <10−5 from the multivariate genome-wide expression analysis.
| Probe set | Gene | P-value for interaction effecta | q-Value for interaction effect | P-value for expression main effectb | q-Value for expression main effectb |
|---|---|---|---|---|---|
| 55662_at | C10orf76 | 0.3908 | 0.7611 | 6.19E-09c | 0.0003 |
| 202389_s_at | HD | 0.2812 | 0.7310 | 1.29E-07c | 0.0033 |
| 207268_x_at | ABI2 | 0.9311 | 0.8606 | 4.34E-06 | 0.0571 |
| 214919_s_at | EIF4EBP3 MASK-BP3 | 0.8541 | 0.8498 | 5.28E-06 | 0.0571 |
| 218891_at | C10orf76 | 0.1655 | 0.6825 | 5.47E-06 | 0.0571 |
| 1552794_a_at | ZNF547 | 2.79E-07c | 0.0132 | 0.9270 | 0.9540 |
| 233753_at | SFRS15 | 9.00E-07c | 0.0213 | 0.3153 | 0.8770 |
| 233412_x_at | — | 3.98E-06 | 0.0628 | 0.6796 | 0.9407 |
| 212718_at | PAPOLA | 5.55E-06 | 0.0657 | 0.3304 | 0.8819 |
17 probe sets with P<0.0001 for interaction effect.
42 probe sets with P<0.0001 for main effect.
Statistically significant after adjusting for multiple testing (Bonferroni threshold of 9 × 10−7).
Discussion and conclusions
One of the major challenges facing medicine is to individualize drug therapy. However, the rate at which pharmacogenomics is translated into the clinic is still relatively slow. To identify biologically relevant pharmacogenomic candidate genes and, more importantly, to understand the mechanisms underlying the effects of those genes on drug response phenotypes would be the first step required to successfully translate this information into the clinic. Many therapeutic agents share common mechanisms resulting in similar clinical manifestations in terms of clinical response and adverse drug reactions. The information gained from a multivariate analysis of a class of drugs will enhance our understanding of differences and similarities in drug mechanisms, in turn, making possible the identification of novel pathways, and verification of known pathways, involved in the observed in the pharmacogenomic basis for response to these drugs.
We have outlined and presented the application of a multivariate model for the pharmacogenomic involving mRNA expression data for the analysis of a class of drugs. This model can be easily extended to model the effect of other genomic data types (for example, single-nucleotide polymorphisms, copy number variations and methylation) and their association with a class of drugs. In addition to the analysis of each genomic data type in a ‘one-at-a-time' manner, the multivariate model can be extended to include multiple genomic variants into a single model (that is, multivariable regression).
In the current study, we developed and applied such a multivariate model to analyze the association between gene expression and 6-TG and 6-MP cytotoxicity data, (IC50) generated from 194 lymphoblastoid cell lines. Using the expression data from those probe sets of known ‘thiopurine pathway' genes, our model suggested that expression of the NT5E and thiopurine S-methyltransferase genes might contribute to variation in IC50, that is, cytotoxicity, of both thiopurine drugs studied. In addition, this effect was not due to the interaction between drug and mRNA expression.
NT5E encodes ecto-5′-nucleotidase (EC 3.1.3.5), NT5E/CD73, is anchored to the external side of plasma membrane by glycosyl-phosphatidylinositol.18 NT5E catalyzes the dephosphorylation of extracellular 5′-mononucleotides to nucleosides. In a parallel study designed to address the functional implications of the association between genes and thiopurine drug cytotoxicity (unpublished data), we hypothesized and validated the existence of a cellular ‘thiopurine circulation' which might have an important role in regulating intracellular levels of 6-TGNs, therefore, the cytotoxic effect or efficacy of thiopurine drugs. In this model, NT5E is responsible for the conversion of thiopurine ribonucleotide monophosphates to thiopurine ribonucleosides. The ribonucleotide monophosphates are exported by an ATP-binding cassette transporter and—as a result of the phosphate—are impermeable to cells unless converted to nucleosides by NT5E. The thiopurine ribonucleosides are then able to flow back into the cells through the action of both concentrative and equilibrative transporters on the plasma membrane. Therefore, variation in expression of NT5E could, in theory, influence intracellular levels of 6-TGNs. Studies are on-going to investigate the role of NT5E in response to thiopurine drugs.
The results of this study illustrate the usefulness of analyzing drugs within the same class jointly in a multivariate model, as opposed to individually, which may lead to novel pharmacogenomic hypotheses. The multivariate model enabled us to consider a class of drugs, the thiopurines, and identify genes for which mRNA expression was associated with cytotoxic effect due to a common mechanism of action. Further functional and mechanistic studies are needed to follow-up candidate genes identified through the class of drugs' analysis, in particular, genes C10orf76 and HD, with the ultimate objective that these studies might shed light on the relationship between genomic variation and drug response for classes of drug therapies.
Acknowledgments
The research was supported by the NIH U01 GM61388 (The Pharmacogenetics Research Network), Minnesota Partnership for Biotechnology and Medical Genomics grant H904600431, NIH R21 CA140879 and the Mayo Foundation. Lastly, we would like to thank the PharmGKB and Standard University for use of the diagram depicting the thiopurine pathway (Figure 1).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
Supplementary Material
References
- Lennard L. The clinical pharmacology of 6-mercaptopurine. Eur J Clin Pharmacol. 1992;43:329–339. doi: 10.1007/BF02220605. [DOI] [PubMed] [Google Scholar]
- Potti A, Dressman HK, Bild A, Riedel RF, Chan G, Sayer R, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- Lennard L. Assay of 6-thioinosinic acid and 6-thioguanine nucleotides, active metabolites of 6-mercaptopurine, in human red blood cells. J Chromatogr. 1987;423:169–178. doi: 10.1016/0378-4347(87)80340-7. [DOI] [PubMed] [Google Scholar]
- Lennard L, Rees CA, Lilleyman JS, Maddocks JL. Childhood leukaemia: a relationship between intracellular 6-mercaptopurine metabolites and neutropenia. Br J Clin Pharmacol. 1983;16:359–363. doi: 10.1111/j.1365-2125.1983.tb02178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennard L, Van Loon JA, Lilleyman JS, Weinshilboum RM. Thiopurine pharmacogenetics in leukemia: correlation of erythrocyte thiopurine methyltransferase activity and 6-thioguanine nucleotide concentrations. Clin Pharmacol Ther. 1987;41:18–25. doi: 10.1038/clpt.1987.4. [DOI] [PubMed] [Google Scholar]
- Zaza G, Cheok M, Krynetskaia N, Thorn C, Stocco G, Hebert JM, et al. Thiopurine pathway. Pharmacogenet Genomics. 2010;20:573–574. doi: 10.1097/FPC.0b013e328334338f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Weinshilboum R. Thiopurine S-methyltransferase pharmacogenetics: insights, challenges and future directions. Oncogene. 2006;25:1629–1638. doi: 10.1038/sj.onc.1209372. [DOI] [PubMed] [Google Scholar]
- Gearry RB, Barclay ML, Burt MJ, Collett JA, Chapman BA, Roberts RL, et al. Thiopurine S-methyltransferase (TPMT) genotype does not predict adverse drug reactions to thiopurine drugs in patients with inflammatory bowel disease. Aliment Pharmacol Ther. 2003;18:395–400. doi: 10.1046/j.1365-2036.2003.01690.x. [DOI] [PubMed] [Google Scholar]
- Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Li L, Fridley B, Kalari K, Jenkins G, Batzler A, Safgren S, et al. Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res. 2008;68:7050–7058. doi: 10.1158/0008-5472.CAN-08-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SJ, Dolan ME. Use of CEPH and non-CEPH lymphoblast cell lines in pharmacogenetic studies. Pharmacogenomics. 2005;6:303–310. doi: 10.1517/14622416.6.3.303. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry R, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligobucleotide expression arrays. Journal of the Amrican Statistical Association. 2004;99:909–917. [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- McCulloch CE, Searle SR. Generalized, Linear, and Mixed Models. John Wiley & Sons, Inc.: New York, NY; 2001. [Google Scholar]
- Gallant AR. Nonlinear Statistical Models. Wiley: New York; 1987. [Google Scholar]
- Davidian M, Giltinan DM. Nonlinear Models for Repeated Measurement Data. Chapman & Hall: New York; 1995. [Google Scholar]
- Zimmermann H. 5′-Nucleotidase: molecular structure and functional aspects. Biochem J. 1992;285 (Part 2:345–365. doi: 10.1042/bj2850345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.