Abstract
ERα is a ligand-dependent nuclear receptor that is important in breast cancer genesis, behavior and response to hormone-based therapies. A T7 phage display screen against full-length human ERα, coupled with genome-wide exon arrays, was used to identify RAC3 as a putative ERα co-regulator. RAC3 is a rho family small GTPase that is associated with cytoskeletal rearrangement. We demonstrate a novel role for nuclear RAC3 as an ERα transcriptional activator, with prognostic implications for metastatic disease. Through in vitro and cell-based studies, RAC3 was shown to exist in a GTP-bound state and act as a ligand specific ERα co-activator of E2-induced transcription. Over expression of RAC3 induced pro-growth and pro-migratory genes that resulted in increased migration of ERα-positive breast cancer cells. Chemical inhibition and genetic knockdown of RAC3 antagonized E2-induced cell proliferation, cell migration, and ERα mediated gene expression, indicating that RAC3 is necessary for full ERα transcriptional activity. In agreement with the molecular and cellular data, RAC3 over expression in ERα-positive breast cancers correlated with a significant decrease in recurrence free survival and a significant increase in the odds ratio of metastasis. In conclusion, RAC3 is novel ERα co-activator that promotes cell migration and has prognostic value for ERα-positive breast cancer metastasis. RAC3 may also be a useful therapeutic target for ERα-positive breast cancers.
Introduction
Breast cancer is the second leading cause of death among women in North America. Estrogen receptor alpha (ERα) plays an important role in breast development, tumorigenesis and treatment (Johnston) and is a major marker for prognosis and therapeutic response in breast tumors that express ERα (Johnston). ERα-positive (ERα+) tumors make up the majority of breast cancers and are typically treated with antiestrogen or aromatase inhibitor therapies to block ERα activity (Johnston).
Transcriptional regulation is a complex process that involves multiple transiently associated co-regulatory proteins (Muramatsu and Inoue). ERα is a classical nuclear receptor with both genomic and non-genomic functions (Heldring et al.). Its role in transcription has been studied extensively (Heldring et al.). A major mechanism of ERα activation is through the binding of estrogen agonists, which cause the receptor to shed chaperone proteins and become transcriptionally active (Heldring et al.). Since the first ERα interacting proteins were identified (Halachmi et al.), considerable effort has been devoted to understanding the molecular mechanisms by which ERα activities are mediated by diverse interacting proteins (Hall and McDonnell). A number of co-activators and co-repressors have been discovered that have varying degrees of influence on receptor activity (Hall and McDonnell). These co-regulators bind differentially to ERα in the presence of structurally diverse ligands, indicating that the control of ERα action is a complex process that involves multiple cofactors (Shang and Brown). In addition, multiple regions of ERα interact with these proteins, including the N-terminal domain, which contains the Activating Function 1 (AF1) region, and the C-terminal ligand-binding domain, which contains the ligand-sensitive Activating Function 2 (AF2) region (Pike). Co-regulator interactions are common in the AF2 domain and are typically mediated by ligand-induced changes in protein conformation. For estrogen agonists, these conformational changes expose a co-activator-binding pocket that binds the canonical NR binding motif (LXXLL). The AF1, AF2, and other regions of the protein bind a diverse set of co-regulators that directly or indirectly alter the activity of ERα through multiple mechanisms (Kumar et al.). Co-activators such as SRC-3 and TRAP220 are over expressed in some breast cancers and are thought to be an integral part of tumorigenesis (Hall and McDonnell). The identification of novel interacting proteins is key to understanding the normal and abnormal regulation of ERα in tissues and cancer.
Like most cancers, breast cancer mortality is not normally caused by the primary tumor, but rather by metastasis of the initial tumor to areas such as the lymph nodes, brain, lung, and bone (Minn et al.; Weil et al.). While ERα+ tumors have a better short-term prognosis than ERα-tumors, a subset of ERα+ tumors metastasize aggressively and the majority of these metastases continue to express ERα (Harrell et al.). ERα activity likely plays an important role in the establishment and control of tumor metastasis, including regulation of cell proliferation, cell motility, and cell invasion (Gururaj et al.; Neuman et al.; Prest et al.). Co-regulators also play a critical role in E2-induced cell proliferation and cell motility (Lahusen et al.; Saji et al.). In the current study, we used a phage display screen against full-length human ERα, coupled with genome-wide exon array analyses, to identify and characterize a novel co-activator, RAC3, as an integral component of the E2-induced cell proliferative and migratory pathways.
Rho-family GTPases are over expressed in a variety of human tumors and have a wide range of roles throughout the cell, including the regulation of cell migration and adhesion, mitosis, regulation of kinase activity, and regulation of transcription factors through cell signaling pathways (Baugher et al.; Onesto et al.). The RAC family of GTPases consists of three members, RAC1, RAC2, and RAC3. RAC2 expression is myeloid specific, while RAC1 and RAC3 are highly homologous and are expressed in a variety of tissues including the breast (Mira et al.). Previous reports implicate RAC1 and RAC3 in breast pathogenesis (Baugher et al.; Leung et al.; Xie and Haslam). Most existing data describe RAC3 activity in the cytoplasm and membrane. However, recent reports have shown that rho family GTPases and their activating proteins are also present in the nucleus (Sandrock et al.). The role of rho family GTPases in the nucleus is not well understood. In the current study, we characterize RAC3 as an ERα co-activator, demonstrating a novel role for the Rho family of GTPases, including direct activation of a transcription factor and a mechanism for morphological changes associated with RAC3 over expression. RAC3-modulated transcriptional activity may help explain the metastasis of some ERα+ positive breast cancers and the importance of the receptor as a mediator of metastasis.
Material and Methods
Chemicals and Antibodies
17-β Estradiol, ICI 182,780, 4-hydroxytamoxifen, and EHT 1864 were purchased from Sigma-Aldrich (St. Louis, MO). H222.2 ERα and Myc-9E10 antibodies were made by Monoclonal Antibody facility at the University of Chicago. The RAC antibody 23A8 and Polybrene were from Millipore (Billerica, MA). Coelentrazine and Luciferin were from Biosynth (Itasca, IL). Full-length ERα was purchased from Invitrogen (Carlsbad, CA). All western blots were performed using LiCOR secondary antibodies according to manufacturer’s instructions (Lincoln, NE).
Plasmids
pRK5 RAC3 WT, T17, and V12 were gifts of Ulla Knauss (University College Dublin). PAK1 kinase dead mutant was a gift of Dr. Piers Nash (University of Chicago). ERα S305A and ERα S305E mutants were gifts of Dr. Rakesh Kumar (George Washington University). The ERα S106A, S108A, and S118A mutant was a gift of Dr. Didier Picard (University of Geneva). RAC1, RAC3, and all of the other co-regulators identified by the phage display screen were obtained in pDONR223 from the Human ORFeome collection at the Functional Genomics Facility at the University of Chicago. PGL2-ERE-luciferase and pM-GWB were gifts of Dr. Donald McDonnell (Duke University). pMGWB-RAC1, pMGWB-RAC3, and pDEST40-RAC3 were cloned by Gateway cloning. pRK5-RAC1 and pDEST527 were obtained from Addgene. All other RAC3 and RAC1 mutations were made by site-directed mutagenesis (Stratagene). Full length ERα was cloned into the pACT vector (Promega) by restriction enzyme digestion (Madison, WI). pRL-TK renilla control vector was purchased from Promega (Madison, WI).
Cell Culture
All cells were grown at 37°C in 5% CO2. MCF7 human breast cancer cells, obtained from the ATCC, and ERα negative MCF7 C4-12 cells, obtained from Wade Welshons (University of Missouri), were maintained in high glucose DMEM supplemented with 10% FBS (Oesterreich et al.). T47D cells were obtained from the ATCC and maintained in RPMI supplemented with 10% FBS. MCF7 K1 cells were grown in high glucose DMEM supplemented with 10% stripped media. MCF7 J2 cells were obtained from Essen Biosciences and grown in MEM with 10% FBS and 5μg/mL blasticidin. C4-12 + Flag-ERα cells were made by retroviral infection and grown in high glucose DMEM supplemented with 10% FBS and 0.6μg/mL puromycin.
Mammalian Two-Hybrid
30,000 MCF7 C4-12 cells were seeded in 48 well dishes and transfected with equal amounts of pSG-5 luciferase, GAL-4 vector, VP-16 ER, and RTK using Lipofectamine 2000 from Invitrogen (Carlsbad, CA) according to manufacturer’s instructions. The plates were read using a homemade dual luciferase protocol as described previously (Hampf and Gossen) on a Wallac Microbeta Jet luminometer.
In-vitro Pulldown
Recombinantly expressed HIS-tagged RAC3 was expressed in pDEST-527 obtained from Dr. Dominic Esposito (National Cancer Institute). Full length ERα (Invitrogen) was preincubated with ligands at a final concentration of 100nM. RAC3 protein was preincubated with either GTP or GDP at a final concentration of 100μM. Proteins were combined to achieve a final concentration of 10μM for RAC3 and 100nM for ERα. The mixture was gently rocked at 4C for 2 hours. 50μl of Prewashed Talon beads (Clontech) were then added to each tube and incubated for 1 hour at 4C while rocking gently. The beads were pelleted by centrifugation, washed three times with GBB buffer and bound proteins were eluted in 2x sample buffer. Proteins were resolved by SDS PAGE and analyzed on western blots using the LiCOR system. ERα bands were normalized to the pulled down RAC3 bands.
Luciferase assays
Cells were transfected using Lipofectamine 2000 and Lipofectamine LTX from Invitrogen (Carlsbad, CA) for MCF7 C4-12 and MCF7 cells, respectively, according to the manufacturer’s instructions. Cells were transfected with 0.4ng of pGL2-ERE luciferase, 0.2ng of pRL-TK vector as a transfection control, and 0.4ng of gene constructs per well. Cells were treated with ligands for 24 hours in either 10% stripped serum or 2.5% stripped serum. MCF7 C4-12 cells were read using homemade dual luciferase as described previously (Hampf and Gossen). To increase signal, MCF7 and T47D cells were read using the Promege Dual Luciferase Kit (Madison, WI) according to manufacturer’s instructions.
shRNA Knockdown
Five RAC3 pLKO vector based shRNA clones, six RAC1 GIPZ shRNAmir based clones, and the Non-silencing-GIPZ-shRNAmir control were obtained from Open Biosystems (Huntsville, AL). pLKO-SHC control, VSV-G, and AR plasmids were obtained from Piers Nash (University of Chicago). Five different clones for RAC3 and a SHC control were generated in MCF7 Cells. Cells were infected using the protocol from the RNAi Consortium at the Broad Institute (Moffat et al.).
ChIP
ChIP assays were done as previously described by the Brown lab (Shang et al.). For more detail and primers see Supplemental Methods.
Coimmunoprecipitation
MCF7 C4-12 + FLAG ERα cells were grown in 100mm plates. Cells were transfected with plasmids in 10% stripped serum with Fu-Gene per manufactures instruction. Cells were serum starved overnight, treated in 10% stripped serum with either 10nM E2 or vehicle alone for 1 hour, and lysed in TMN buffer. Cell extracts were analyzed by SDS-Page and Western blots were imaged using the LiCOR system. For a more detailed immunoprecipitation protocol, please see the Supplemental Methods.
Real-time PCR
MCF7 cells were plated in 6-well dishes. Prior to transfections, cells were washed with PBS and grown in 2.5% serum. Cells were transfected overnight using Fu-Gene in 2.5% stripped serum DMEM per manufactures instruction. Cells were then grown in stripped serum for 2 days with daily media changes. For endogenous gene assays, cells were grown in 2.5% stripped serum for 3 days with daily media changes and then serum starved overnight prior to treatment. Cells were preincubated with indicated concentrations of ICI 182,780 or EHT 1864 for 15 minutes prior to the addition of 10nM E2 for 2 hours. cDNA was made using Superscript Vilo cDNA Synthesis kit (Invitrogen) per manufacturers instruction. Real-time PCR was done uisng an Applied Biosystems Step One Plus 96 well Real Time-PCR machine and TAQMAN reagents.
Migration Assays
Migration assays were done using BDFalcon Fluoroblok 96 well inserts. MCF7 cells were transfected in 100mM plates at 60% confluence in 2.5% stripped serum and then serum starved overnight. Cells were grown in Calcein AM media for 30 minute to label the cells. The cells were trypsinized and resuspended in PBS with 5% BSA, washed, and resuspended in serum free media. 100,000 cells were added per well in 70μl of media in serum free media supplemented with ligands. 10% stripped FBS media was placed in the lower wells as a chemoattractant. After a 6-hour incubation in the incubator, cells were detached from the membrane and measured for fluorescence intensity in a black 96 well plate in the Wallac Victor V. For some experiments, the QCM Chemotaxis 96-well Cell Migration Assay kit from Chemicon was used per manufacturer’s instructions (Billerica, MA).
Results
Identification of RAC3 as an ERα interacting Protein by Phage Display
A phage display approach was used to identify both known and unknown ERα interacting proteins. The screen identified over 300 proteins with p-values under 0.05, including 27% of the previously identified ERα interactors listed in the Human Protein Resource Database (Keshava Prasad et al.). In vitro pull down and mammalian two hybrid approaches confirmed the interaction of ERα with 7 of 10 tested proteins (Supplemental Table 1). The low false positive rate demonstrates that this screening approach is a viable option for identifying potential nuclear receptor co-regulators.
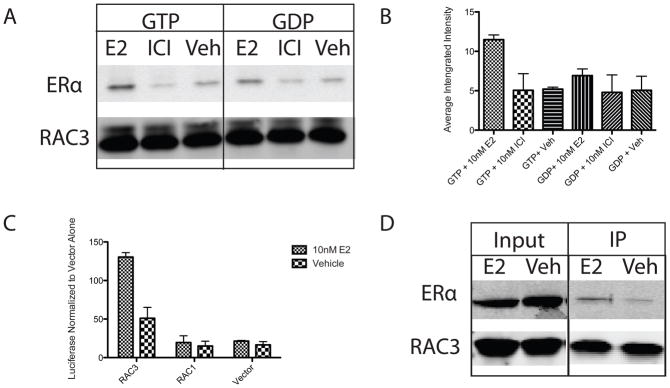
RAC3 was identified as a putative ERα co-regulator by the phage display screen. Direct interaction of the two proteins was confirmed by in vitro pull down experiments using Sf-9 expressed and purified ERα (Invitrogen) as well as recombinantly expressed, His-tagged RAC3 (Figure 1A). Analysis of three independent experiments confirmed that the RAC3/ERα interaction was estradiol dependent, and increased two-fold in the presence of GTP (Figure 1B). Ligand dependent ERα/RAC3 interaction in cells was demonstrated by mammalian two-hybrid and co-immunoprecipitation experiments performed in ERα null, MCF7 C4-12 cells (Oesterreich et al.), with stably over expressed Flag-ERα (Figure 1C and 1D). GFP tagged RAC3 is ubiquitously expressed throughout the cell and localization is not altered by E2 treatment. (Data not shown). Notably, the highly homologous RAC family member RAC1 did not interact with ERα (Figure 1C). In total, these experiments show a direct, ligand dependent, GTP sensitive interaction between ERα and RAC3, but not RAC1.
Figure 1. RAC3 interaction with ERα is GTP and ligand dependent in vitro and in cells.
(A) His-tagged RAC3, pulled down with Talon beads (Clontech), interacts with full length ERα (Invitrogen) in a GTP and estradiol (E2) dependent manner. Western blot was probed with H222.2 and pan RAC antibody. (B) Quantification of western blot by LICOR software. Averaged integrated intensity of ERα bands was normalized to average integrated intensity of RAC3 bands. (C) RAC3 interacts with ERα in ligand dependent manner as measured by mammalian two-hybrid analysis of VP-16-ERα and GAL4 tagged RAC3 and RAC1 in MCF7 C4-12 cells. (D) Coimmunoprecipitation of ERα with RAC3 in MCF7 C4–12 + Flag-ERα cells transfected with WT-RAC3. Western blot was probed with RAC and H222.2 antibodies and all bands were taken from the same blot. Empty wells were spliced out for clarity.
RAC3 is a novel ERα co-activator
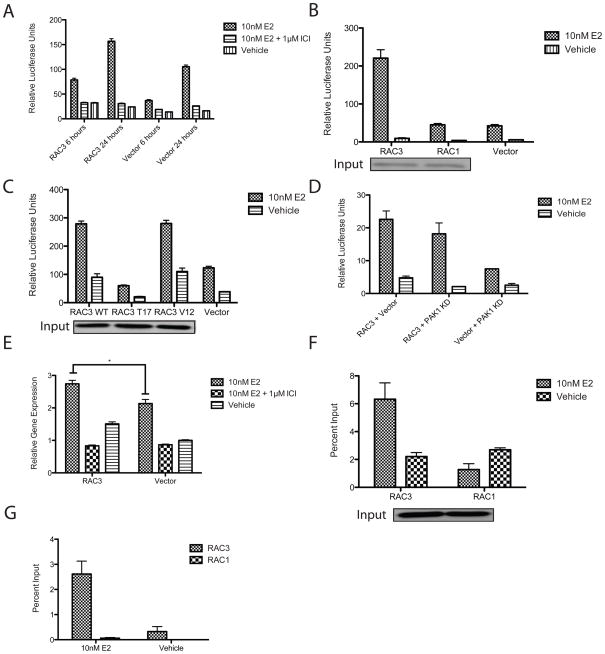
When RAC3 was tested for its effect on ERα transcription using an ERE-Luciferase reporter (Leong et al.), it increased E2 mediated ERE luciferase activity at both 6 and 24 hours in MCF7 cells (Figure 2A). Over expression of RAC3, but not RAC1, increased E2 dependent ERα activation of ERE luciferase (Figure 2B). The dominant negative RAC3 mutant T17N and its constitutively active mutant, RAC3 G12V, were tested for their ability to co-activate ERα (Mira et al.). As shown in Figure 2C, WT RAC3 and RAC3 G12V were comparably able to increase ERE luciferase activity in MCF7 cells. The dominant negative RAC3 T17N was significantly less active than WT RAC3, indicating that GTP is necessary for complete RAC3 induced activation of ERα. Similar results were observed in T47D cells, confirming that this phenotype is not cell-type specific (Supplemental Figure 1A).
Figure 2. RAC3 is an ERα Co-activator.
(A-D) RAC3 over expression increases E2-induced ERE luciferase activity in MCF7 cells transfected with PGL2-ERE-luciferase, RTK, and indicated constructs as described in Material and Methods. MCF7 cells were treated with 10nM E2, 10nM E2 + 1μM ICI, and ethanol for 6–24 hours as indicated. (B,C) Western blots probed with anti-Myc antibody show comparable protein expression of Myc-tagged RAC3 and RAC1 or WT, T17, and V12 RAC3 mutants from an independent transfection. (E) RAC3 over expression increases CCND1 gene expression in MCF7 cells transfected with RAC3 or vector. Gene expression was measure by TAQMAN assay. *, p-value is 0.01. (F,G) RAC3 is present (F) and increases ERα occupancy (G) at the CCND1 promoter in MCF7 C4-12 cells stably transfected with Flag-ERα and transiently transfected with RAC3 or RAC1. Occupancy was determined by ChIP analysis and measured by real-time PCR as percent input. Western blot probed with anti-Myc antibody shows comparable protein expression of Myc-tagged RAC3 and RAC1 from an independent transfection.
Talukder et. al. reported that Rho Family GTPases transactivate ERα transcription through p21-activated protein kinase 1 (PAK1) (Talukder et al.). To test if RAC3 coactivation of ERα is independent of the PAK1 pathway, different mutants were used to block the PAK1 pathway. In the presence of a dominant negative PAK1 kinase dead mutant (PAK1 KD), RAC3 increased E2/ERα mediated ERE luciferase activity in MCF7 cells (Figure 2D). PAK1 has been reported to co-activate ERE luciferase by phosphorylating Serine 305 in ERα, which resulted in subsequent phosphorylation of S118 (Rayala et al.). ERα constructs with S305A, S305E, and S104A, S106A, and S118A mutations were co-transfected with RAC3 and ERE-luciferase constructs into MCF7 C4-12 cells. As shown in Supplemental figure 2A-C, phosphorylation mutants that block PAK1 induced phosphorylation and hyperactivation of the PAK1 pathway did not prevent RAC3 enhancement of E2-induced ERα transcription. These results demonstrate that RAC3 can co-activate E2-induced ERα transcription through a mechanism that is independent of the PAK1 pathway.
To confirm the results of the ERE luciferase reporter assays, the effect of RAC3 on E2/ERα induced endogenous gene expression was studied. Cyclin D1 (CCND1) is a known ERα target gene with an important role in cell proliferation and tumor growth (Neuman et al.). As measured by qRT-PCR, RAC3 over expression caused an increase in E2 induced CCND1 transcript in MCF-7 cells (Figure 2E). The presence of RAC3 at the CCND1 promoter was tested using ChIP assays. MCF7 C4-12 cells that stably expressed Flag-ERα were transiently transfected to over express RAC3 WT and RAC1 WT. An E2 dependent recruitment of RAC3 to the CCND1 promoter was observed (Figure 2F), indicating that RAC3 is an important regulator of CCND1 expression. ERα occupancy at the TFF1 gene promoter also increased in the presence of estradiol and over expressed RAC3 (Figure 2G). These results illustrate that RAC3, but not RAC1, over expression is able to enhance E2/ERα mediated transcription.
A single amino acid in RAC3 confers activation of ERα
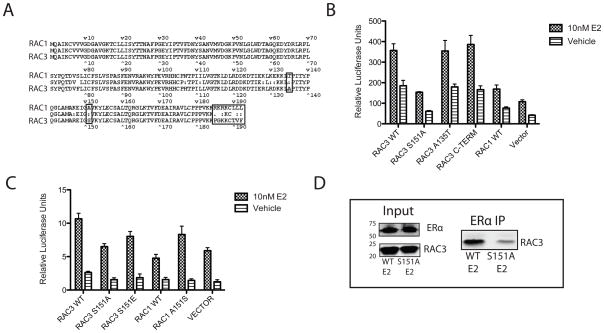
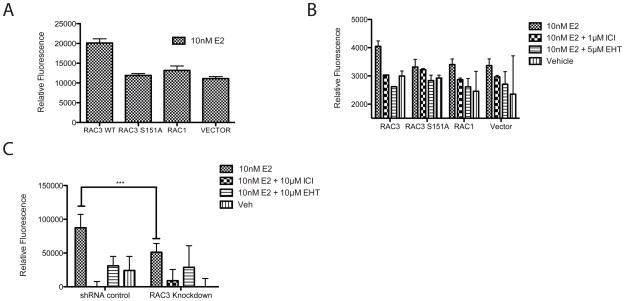
As seen in figure 1C and 2A, RAC1 did not interact with ERα or co-activate E2/ERα mediated ERE luciferase activity, despite its high sequence homology with RAC3. In MCF7 cells, RAC1 and RAC3 mRNA transcripts are expressed at comparable levels, as detected by qRT-PCR (data not shown). RAC2 transcript was not detected in MCF7 cells (data not shown). RAC3 and RAC1 protein alignment shows that the two proteins differ by only 15 amino acids and that most of these differences map to the C-terminus (Figure 3A). Based on the identification of functional motifs by ELM motif scan software, three mutations were introduced to test for differences between the two proteins (Gould et al.). The C-terminus (residues 185–192) of RAC3 was converted to that of RAC1 (RAC3 C-TERM), a putative PKA phosphorylation site in RAC1 (S135) was introduced into RAC3 (RAC3 A135S), and a putative 14-3-3 binding site in RAC3 (S151) was mutated to correspond to RAC1 (RAC3 S151A). As measured by ERE luciferase assays, the only mutant that failed to co-activate ERα above vehicle control was RAC3 S151A (Figure 3B). RAC1 over expression produced a small increase in E2/ERα activity. However, as reported previously (Talukder et al.), RAC1 over expression can activate the PAK1 pathway, thereby increasing ERα transcriptional activity. The effect seen by RAC3 is much more pronounced and is the result of a direct interaction with ERα, as shown in Figure 1. As expected, the reciprocal RAC1 mutation (A151S) increased the ability of RAC1 to activate ERα transcription (Figure 3C). The RAC3 S151A mutation does not altar RAC3 transfection efficiency or protein stability (Figure 3D and data not shown). Co-immunoprecipitation of E2-ERα and RCA3 showed a decreased interaction with the RAC3 S151A mutant compared to RAC3 WT (Figure 3D). These results demonstrate that the ability of RAC3 to act as an ERα co-activator requires Serine 151.
Figure 3. A single amino acid confers RAC3 coactivation of ERα.
(A) Sequence alignment of RAC3 and RAC1. Boxed amino acids were mutated. (B,C) Mutations at S151 of RAC3 and RAC1 alter the ability of the GTPases to activate ERα induced transcription as measured by ERE luciferase assay in MCF7 cells, which were transiently transfected with indicated constructs. (D) RAC3 S1515A mutation decreases the affinity of RAC3 for ERα in MCF7 C4–12 cells stably transfected with Flag-ERα and transiently transfected with RAC3 WT and RAC3 S151A. After a 1hour treatment with 10 nM E2, ERα was pulled down with H222.2 antibody. Western blot was probed with pan-RAC and ERα antibodies; all bands were taken from the same blot with empty wells omitted for clarity.
Knockdown and inhibition of RAC3 block ERα induced gene expression
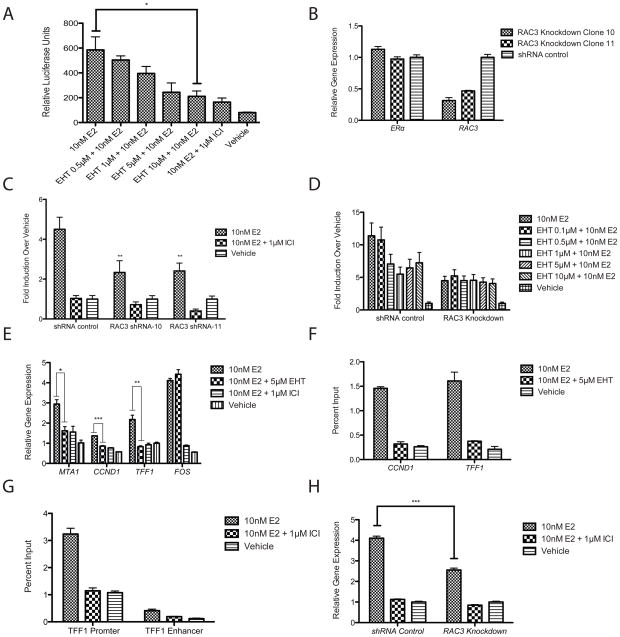
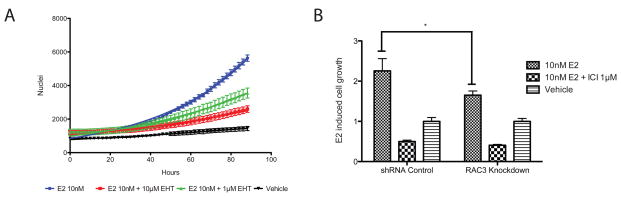
To determine if RAC3 over expression reflects endogenous RAC3 activity, a pan-RAC inhibitor, EHT 1864, was used. This inhibitor has previously been shown to specifically inhibit RAC1, RAC2, and RAC3 activities (Onesto et al.). As shown in figure 4A, EHT 1864 exhibited a dose dependent inhibition of E2/ERα mediated ERE luciferase activity in MCF-7 cells, with maximal inhibition occurring at 5–10 μM EHT 1864. To confirm that this inhibition was mediated by RAC3, two unique RAC3 shRNAs were used to independently knock down RAC3 expression in MCF7 cells. Each shRNA reduced RAC3 mRNA expression by more than 60%, with no effect on ERα or RAC1 expression (Figure 4B and data not shown). E2/ERα mediated ERE transcription was significantly reduced in RAC3 knockdown cells (Figure 4C), compared to control, indicating the importance of endogenous RAC3 as an ERα co-activator in MCF-7 cells. RAC1 knockdown had no effect on E2/ERα mediated ERE transcription (Supplemental Figure 3B). As expected, while EHT 1864 elicited a dose dependent E2/ERα response in control knockdown cells, it was unable to inhibit E2/ERα mediated transcriptional activity in RAC3 knockdown cells (Figure 4D). These data are consistent with the RAC3 over expression data and indicate that EHT 1864 can be used as an inhibitor of endogenous RAC3 co-activation of ERα. As shown in figure 4E, EHT 1864 almost completely inhibited E2/ERα induced expression of endogenous CCND1, TFF1, MTA1 but not FOS, indicating that RAC3 co-activation of ERα is gene selective. The interaction of ERα and RAC3 with the CCND1 and TFF1 promoters was confirmed by ChIP analysis (Figure 4F). Similar to the gene expression data, EHT 1864 completely abrogated E2-induced ERα recruitment to TFF1 and CCND1 promoter sequences (Figure 4F). RAC3 occupancy at the TFF1 promoter and at a previously described TFF1 enhancer sequence was also E2 sensitive (Figure 4G) (Pan et al.). RAC3 recruitment to the TFF1 promoter was blocked both by EHT 1864 and ICI 182,780, demonstrating that occupancy requires active RAC3 and is dependent upon E2/ERα (Figure 4G and data not shown). CCND1 gene expression was also significantly reduced in stable RAC3 knockdown cells, confirming the EHT 1864 inhibition of expression (Figure 4H). EHT 1864 also significantly antagonized E2-induced cell proliferation of MCF7 and T47D cells, confirming that abrogation of E2-induced CCND1 expression has phenotypic consequences (Figure 5A and Supplemental Figure 1B). In agreement with the EHT 1864 induced antagonism, RAC3 knockdown significantly decreased E2 induced cell proliferation (Figure 5B). In total, these results show convincingly that RAC3 is an important co-activator for a subset of ERα target genes and contributes significantly to E2 mediated MCF7 cell proliferation.
Figure 4. Chemical and genetic inhibition of endogenous RAC3 effects on E2/ERα-induced gene expression.
*, p-value <0.05. **, p-value <0.005. ***, p-value <0.0005. (A) EHT 1864 antagonizes E2-induced ERE luciferase transcription in MCF7 cells treated with 10nM E2 and increasing concentrations of EHT 1864. Complete antagonism was observed with 100x treatment with ICI 182,780. B) RAC3 mRNA levels were decreased by over 50% in shRNA knockdown cells as detected by TAQMAN assay. RNA was isolated with Trizol from shRNA mediated stable RAC3 knockdown MCF7 cells. (C) RAC3 knockdown decreases E2-induced activation of ERE luciferase assays in MCF7 cells. (D) RAC3 is required for EHT 1864 mediated antagonism of ERE luciferase. RAC3 knockdown MCF7 Cells were treated with 10nM E2 ± 1μM ICI or increasing concentrations of EHT 1864 (E) EHT 1864 inhibits ERα target genes CCND1, MTA1, and TFF1. Cells were pretreated for 5 minutes with 1μM ICI, 5μM EHT 1864, or vehicle before addition of 10 nM E2. RNA was isolated by Trizol and mRNA expression was determined by qRT-PCR using TAQMAN assays. (F,G) MCF7 C4-12 cells stably transfected with Flag-ERα were grown in 2.5% stripped serum for 1 day and then serum starved overnight. Cells were treated with 10nM E2 and ERα was immunoprecipitated from cell extracts with H222.2 (F), or RAC3 was immunoprecipitated with a pan-RAC antibody (G). Gene occupancy was determined by real time PCR at the CCND1, TFF1 promoter, and TFF1 enhancer (H) RAC3 knockdown decreases CCND1 gene expression as measured by TAQMAN assay.
Figure 5. RAC3 affects MCF7 cell proliferation.
(A) EHT 1864 antagonizes E2-induced cell proliferation in MCF7 J2 cells plated in 24 well plate and treated with indicated compounds in 10% stripped serum. (B) RAC3 knockdown significantly decreased E2-induced cell proliferation. *, p-value<0.05. DNA content was measured by Hoescht 22358 fluorescence and values were normalized to vehicle control.
RAC3 is important for E2-induced Cell Migration
RAC3 over expression has been previously shown to increase the migratory potential of breast cancer cells (Baugher et al.). To test for an effect of RAC3 on ERα+ MCF-7 cell migration, cells were transfected with RAC3 WT, RAC3 S151A, RAC1, or vector alone, placed in transwells and treated with 10nM E2. RAC3 WT increased cell migration two-fold over vector alone (Figure 6A). This increase was not observed for the RAC3 S151A mutant and was not seen in cells transfected with RAC1 (Figure 6A). As confirmation that this increase was ERα dependent, E2-induced migration was blocked by co-incubation with the complete ER antagonist ICI 182,780 (Figure 6B). Treatment with the pan-RAC inhibitor EHT 1864 completely inhibited the E2 mediated MCF-7 cell migration, reducing it slightly below vehicle control, suggesting that RAC3 mediates basal migratory activity independent of ERα. In RAC3 knockdown cells, migration was significantly decreased compared to a control cell line (Figure 6C). Also, EHT 1864 was unable to further inhibit migration in MCF7 RAC3 knockdown cells, confirming that endogenous RAC3 is fully responsible for this effect on ERα signaling (Figure 6C).
Figure 6. RAC3 affects MCF7 cell migration.
(A,B) RAC3 over expression increases E2-induced cell migration in MCF7 cells transiently transfected with indicated constructs and serum starved. Migration was measured by labeling cells with Calcein AM and fluorescence was measured. (C)RAC3 knockdown inhibits cell migration in MCF7 cells as measured by QCM Chemotaxis 96-well Cell Migration Assay from Chemicon. Fluorescence was normalized to wells without chemo-attractant. Significance calculated with two-way anova. P-value = 0.002.
RAC3 over expression is an indicator of poor prognosis in human breast tumors
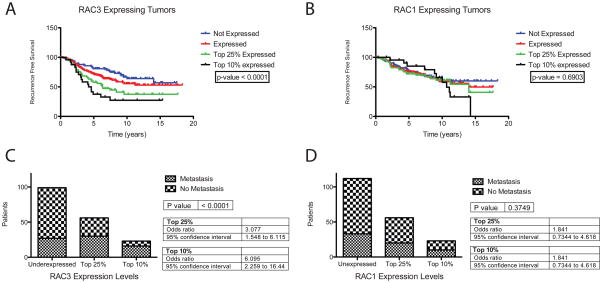
Using Oncomine™ (Compendia Bioscience, Ann Arbor, MI), RAC3 over expression correlates with invasive breast cancer in some datasets. Analysis of the NKI breast cancer patient database revealed the clinical relevance of RAC3 expression in ERα+ tumors (Chang et al.). Recurrence free survival was significantly shorter among women with ERα+ tumors that expressed RAC3 mRNA and this trend increased with increasing RAC3 mRNA expression (Figure 7A). RAC3 over expression also increased the odds ratio for metastatic events by 6.09 in the highest RAC3 expressing ERα+ breast tumors (Figure 7C). However, RAC3 mRNA expression did not correlate significantly with metastasis in ERα-tumor samples, although the sample size was not large enough to completely eliminate this possibility (Data not shown). RAC1 mRNA expression did not correlate with an increase in metastasis or a decrease in recurrence free survival in ERα+ tumors (Figure 7B,7D).
Figure 7. Clinical Consequences of RAC3 expression in ERα positive human breast tumors.
Gene expression data was obtained from the NKI patient database. (Chang et al.) All analysis was done using Prism 5 for Mac OS X from GraphPad (La Jolla, CA). (A,B) Increasing RAC3 expression decreases recurrence free survival shown in Kaplan-Meier curves of recurrence free survival in years. Increasing RAC1 expression had no effect. (C,D) Increasing RAC3 expression increases the proportion of patients with metastatic events. RAC1 expression levels had no statistically significant effect.
Discussion
A phage display screen coupled with exon array analysis identified multiple known and novel ERα interacting proteins, whose interaction could be validated by in vitro pull down and mammalian two hybrid analyses. The low false positive rate demonstrates the efficacy of this technique for the identification of potential ER co-regulators. The technique is also readily applicable to other nuclear receptors or any DNA binding protein. Importantly, this technique allowed us to identify RAC3 as a novel and potent ERα co-regulator.
The data presented here demonstrate a unique role for RAC3 as a critical co-activator of ERα mediated transcription. A combination of over expression, chemical inhibition, and shRNA knockdown data show that RAC3 is necessary for full activation of E2/ERα induced expression of genes known to be involved in cell proliferation, migration, and invasion. In addition, analysis of the NKI database showed that RAC3 expression correlates with poor prognosis for women with ERα+ tumors, and a decrease in recurrence free survival. The clinical data are consistent with the molecular data, which show that RAC3, but not RAC1, is a co-activator of ERα target genes that are involved in the proliferation or migration of hormone-sensitive breast cancers.
RAC3 knockdown in MCF7 cells showed that it was critical for full ERα transcriptional activity. ERα activity in ERE-luciferase reporter assays and for E2-induced CCND1 gene expression was reduced by approximately 50% in RAC3 knockdown cells, compared to control knockdowns. EHT 1864 completely abolished ERα recruitment to the CCND1 promoter as well as E2/ERα mediated CCND1 gene expression. Given the absence of an additional effect of EHT 1864 in RAC3 knockdown cells, RAC3 appears to be both necessary for full E2/ERα activity on CCND1 expression at early time points following E2 exposure.
The absence of ERα co-regulatory activity for RAC1 versus RAC3 is another example of the previously observed opposing effects of these two highly homologous proteins (Hajdo-Milasinović et al.). In the current study, this difference was mapped to RAC3 S151, which corresponds to A151 in RAC1 (Figure 3). While S151 is also present in RAC2, RAC2 is not expressed in MCF7 or C4-12 cells. As identified by ELM motif analysis, S151 is a putative 14-3-3 binding site. Computational analysis with PROSITE and NetPHOS 2.0 indicate that S151 is a putative protein kinase C (PKC) phosphorylation site that has a high likelihood of being phosphorylated (Blom et al.). S151 has no previously reported effect on the activity of RAC3 and therefore represents a novel functional motif for RAC3.
RAC3 over expression in MCF-7 cells correlated with a pro-growth, pro-migratory phenotype (Figure 5 and Figure 6). E2 induced expression of CCND1, TFF1, and MTA1 has known cell growth, cell motility, and cell invasive consequences in breast cancer cells (Gururaj et al.; Neuman et al.; Prest et al.). In the current study, RAC3-enhanced E2/ERα induced CCND1, TFF1, and MTA1 gene expression correlated with changes in cell proliferation and migration. RAC3 over expression enhanced E2-dependent migration of MCF7 cells and expression of CCND1, whereas inhibition of RAC3 activity either with EHT 1864 or RAC3 shRNA inhibited E2induced migration of MCF7 cells and induction of CCND1, TFF1, and MTA1 mRNA. ERα can induce cell migration through both genomic and non-genomic mechanisms (Sanchez et al.). Although the genomic data presented here clearly demonstrate that RAC3 activity alters the expression of pro-migratory genes, it is also possible that RAC3 can modulate extra-nuclear ERα activities that affect cell proliferation, migration or invasion. This possibility was not explored. Also, because over expression of RAC3 can also increase the migration and invasion of breast cancer cells independent of ERα (Baugher et al.; Chan et al.), at least two pathways must exist for RAC3 mediated tumor cell migration.
While numerous known ERα co-regulators have been described, many of these proteins are not useful drug targets. Previous work on ER/co-regulator based drug discovery focused primarily on the development of selective estrogen receptor modulators (SERMs) that function by binding directly with ERα to recruit or block co-regulator binding to ERα, thereby activating or repressing ERα gene targets that contribute to a particular outcome (Hall and McDonnell). Our data demonstrate that some ERα co-activators, like RAC3, can be inhibited directly with small molecules to block the activity of the receptor. This approach could lead to the establishment of a new, potentially complementary, class of ERα antagonists that act indirectly on the receptor to block E2-induced activation.
Rho family GTPases are best known as integral regulators of the actin cytoskeleton (Heasman and Ridley). As a consequence, Rho family GTPases have important roles in transcriptional regulation through the modulation of actin dynamics and by activating signaling cascades (Jaffe and Hall). The data presented here demonstrate a novel role for Rho family GTPases as direct transcriptional co-activators.
In summary, the small Rho GTPase RAC3 was identified as an ERα co-activator that can promote E2-induced migration and is required for full activation of ERα on a subgroup of genes in MCF7 cells. This activity of RAC3 as a transcriptional co-activator represents a novel function for Rho-family GTPases and a potential new target as a treatment option for ERα positive breast cancers that become therapy resistant.
Supplementary Material
Acknowledgments
This work was supported by NCI 2-RO1-CA089489 (G.L.G), Department of Defense W81XWH-04-1-0791 (G.L.G.), and a gift from the Ludwig Foundation (G.L.G). Core services were supported in part by a 5P30-CA01459935 Cancer Center grant. M.P.W. was partially supported by NIH Grant 5T32GM007183-35.
Footnotes
Conflict of interest: The authors declare no conflicts of interest related to these studies.
Contributor Information
Matthew P Walker, University of Chicago, Committee on Cancer Biology.
Maomao Zhang, University of Chicago, Ben May Department for Cancer Research.
Thien P Le, University of Chicago, Committee on Pathology.
Patricia Wu, University of Chicago, Ben May Department for Cancer Research.
Muriel Lainé, University of Chicago, Ben May Department for Cancer Research.
Geoffrey L Greene, University of Chicago, Ben May Department for Cancer Research.
References
- Baugher PJ, Krishnamoorthy L, Price JE, Dharmawardhane SF. Rac1 and Rac3 isoform activation is involved in the invasive and metastatic phenotype of human breast cancer cells. Breast Cancer Res. 2005;7:R965–74. doi: 10.1186/bcr1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–62. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Chan AY, Coniglio SJ, Chuang Y-y, Michaelson D, Knaus UG, Philips MR, et al. Roles of the Rac1 and Rac3 GTPases in human tumor cell invasion. Oncogene. 2005;24:7821–9. doi: 10.1038/sj.onc.1208909. [DOI] [PubMed] [Google Scholar]
- Chang CY, Norris JD, Jansen M, Huang HJ, McDonnell DP. Application of random peptide phage display to the study of nuclear hormone receptors. Methods Enzymol. 2003;364:118–42. doi: 10.1016/s0076-6879(03)64007-3. [DOI] [PubMed] [Google Scholar]
- Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102:3738–43. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicatiello L, Addeo R, Sasso A, Altucci L, Petrizzi VB, Borgo R, et al. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter. Mol Cell Biol. 2004;24:7260–74. doi: 10.1128/MCB.24.16.7260-7274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goppelt-Struebe M, Golombek M. Fluorometric determination of the DNA content of cells cultured in tissue culture plates. J Immunol Methods. 1992;151:245–8. doi: 10.1016/0022-1759(92)90123-b. [DOI] [PubMed] [Google Scholar]
- Gould CM, Diella F, Via A, Puntervoll P, Gemund C, Chabanis-Davidson S, et al. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 38:D167–80. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururaj AE, Singh RR, Rayala SK, Holm C, den Hollander P, Zhang H, et al. MTA1, a transcriptional activator of breast cancer amplified sequence 3. Proc Natl Acad Sci US A. 2006;103:6670–5. doi: 10.1073/pnas.0601989103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdo-Milasinović A, Ellenbroek SIJ, van Es S, van der Vaart B, Collard JG. Rac1 and Rac3 have opposing functions in cell adhesion and differentiation of neuronal cells. J Cell Sci. 2007;120:555–66. doi: 10.1242/jcs.03364. [DOI] [PubMed] [Google Scholar]
- Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–8. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- Hall JM, McDonnell DP. Co-regulators in nuclear estrogen receptor action: from concept to therapeutic targeting. Mol Interv. 2005;5:343–57. doi: 10.1124/mi.5.6.7. [DOI] [PubMed] [Google Scholar]
- Hampf M, Gossen M. A protocol for combined Photinus and Renilla luciferase quantification compatible with protein assays. Anal Biochem. 2006;356:94–9. doi: 10.1016/j.ab.2006.04.046. [DOI] [PubMed] [Google Scholar]
- Harrell JC, Dye WW, Allred DC, Jedlicka P, Spoelstra NS, Sartorius CA, et al. Estrogen receptor positive breast cancer metastasis: altered hormonal sensitivity and tumor aggressiveness in lymphatic vessels and lymph nodes. Cancer Res. 2006;66:9308–15. doi: 10.1158/0008-5472.CAN-06-1769. [DOI] [PubMed] [Google Scholar]
- Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, et al. Estrogen receptors: how do they signal and what are their targets. Physiol Rev. 2007;87:905–31. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–69. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Johnston SR. New strategies in estrogen receptor-positive breast cancer. Clin Cancer Res. 16:1979–87. doi: 10.1158/1078-0432.CCR-09-1823. [DOI] [PubMed] [Google Scholar]
- Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Gururaj AE, Vadlamudi RK, Rayala SK. The clinical relevance of steroid hormone receptor co-repressors. Clin Cancer Res. 2005;11:2822–31. doi: 10.1158/1078-0432.CCR-04-1276. [DOI] [PubMed] [Google Scholar]
- Lahusen T, Henke RT, Kagan BL, Wellstein A, Riegel AT. The role and regulation of the nuclear receptor co-activator AIB1 in breast cancer. Breast Cancer Res Treat. 2009;116:225–37. doi: 10.1007/s10549-009-0405-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong H, Sloan JR, Nash PD, Greene GL. Recruitment of histone deacetylase 4 to the N-terminal region of estrogen receptor alpha. Mol Endocrinol. 2005;19:2930–42. doi: 10.1210/me.2005-0178. [DOI] [PubMed] [Google Scholar]
- Leung K, Nagy A, Gonzalez-Gomez I, Groffen J, Heisterkamp N, Kaartinen V. Targeted expression of activated Rac3 in mammary epithelium leads to defective postlactational involution and benign mammary gland lesions. Cells Tissues Organs (Print) 2003;175:72–83. doi: 10.1159/000073751. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira JP, Benard V, Groffen J, Sanders LC, Knaus UG. Endogenous, hyperactive Rac3 controls proliferation of breast cancer cells by a p21-activated kinase-dependent pathway. Proc Natl Acad Sci U S A. 2000;97:185–9. doi: 10.1073/pnas.97.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Inoue S. Estrogen receptors: how do they control reproductive and nonreproductive functions? Biochem Biophys Res Commun. 2000;270:1–10. doi: 10.1006/bbrc.2000.2214. [DOI] [PubMed] [Google Scholar]
- Neuman E, Ladha MH, Lin N, Upton TM, Miller SJ, DiRenzo J, et al. Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol Cell Biol. 1997;17:5338–47. doi: 10.1128/mcb.17.9.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterreich S, Zhang P, Guler RL, Sun X, Curran EM, Welshons WV, et al. Re-expression of estrogen receptor alpha in estrogen receptor alpha-negative MCF-7 cells restores both estrogen and insulin-like growth factor-mediated signaling and growth. Cancer Res. 2001;61:5771–7. [PubMed] [Google Scholar]
- Onesto C, Shutes A, Picard V, Schweighoffer F, Der CJ. Characterization of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. Methods Enzymol. 2008;439:111–29. doi: 10.1016/S0076-6879(07)00409-0. [DOI] [PubMed] [Google Scholar]
- Pan YF, Wansa KD, Liu MH, Zhao B, Hong SZ, Tan PY, et al. Regulation of estrogen receptor-mediated long range transcription via evolutionarily conserved distal response elements. J Biol Chem. 2008;283:32977–88. doi: 10.1074/jbc.M802024200. [DOI] [PubMed] [Google Scholar]
- Pike AC. Lessons learnt from structural studies of the oestrogen receptor. Best Pract Res Clin Endocrinol Metab. 2006;20:1–14. doi: 10.1016/j.beem.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Prest SJ, May FE, Westley BR. The estrogen-regulated protein, TFF1, stimulates migration of human breast cancer cells. FASEB J. 2002;16:592–4. doi: 10.1096/fj.01-0498fje. [DOI] [PubMed] [Google Scholar]
- Rayala SK, Talukder AH, Balasenthil S, Tharakan R, Barnes CJ, Wang RA, et al. P21-activated kinase 1 regulation of estrogen receptor-alpha activation involves serine 305 activation linked with serine 118 phosphorylation. Cancer Res. 2006;66:1694–701. doi: 10.1158/0008-5472.CAN-05-2922. [DOI] [PubMed] [Google Scholar]
- Saji S, Kawakami M, Hayashi S, Yoshida N, Hirose M, Horiguchi S, et al. Significance of HDAC6 regulation via estrogen signaling for cell motility and prognosis in estrogen receptor-positive breast cancer. Oncogene. 2005;24:4531–9. doi: 10.1038/sj.onc.1208646. [DOI] [PubMed] [Google Scholar]
- Sanchez AM, Flamini MI, Baldacci C, Goglia L, Genazzani AR, Simoncini T. Estrogen Receptor-{alpha} Promotes Breast Cancer Cell Motility and Invasion via Focal Adhesion Kinase and N-WASP. Mol Endocrinol. doi: 10.1210/me.2010-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrock K, Bielek H, Schradi K, Schmidt G, Klugbauer N. The nuclear import of the small GTPase Rac1 is mediated by the direct interaction with karyopherin alpha2. Traffic. 11:198–209. doi: 10.1111/j.1600-0854.2009.01015.x. [DOI] [PubMed] [Google Scholar]
- Shang Y, Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–8. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–52. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Talukder AH, Meng Q, Kumar R. CRIPak, a novel endogenous Pak1 inhibitor. Oncogene. 2006;25:1311–9. doi: 10.1038/sj.onc.1209172. [DOI] [PubMed] [Google Scholar]
- Weil RJ, Palmieri DC, Bronder JL, Stark AM, Steeg PS. Breast cancer metastasis to the central nervous system. Am J Pathol. 2005;167:913–20. doi: 10.1016/S0002-9440(10)61180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie JW, Haslam SZ. Extracellular matrix, Rac1 signaling, and estrogen-induced proliferation in MCF-7 breast cancer cells. Breast Cancer Res Treat. 2008;110:257–68. doi: 10.1007/s10549-007-9719-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.